Submitted:
14 August 2024
Posted:
15 August 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Growth Conditions
2.2. Genome De Novo Sequencing, Assembly, and Annotation
2.3. Phylogenetic Analysis
2.4. Comparative Genomic Studies and Whole Genome Relatedness
2.5. Secondary Metabolite and Butirosin BGCs Analysis
3. Results and Discussion
3.1. Sequencing, Assembly, Phylogenetic Analysis, and Genomic Characteristics
3.2. Prediction of Secondary Metabolite Biosynthetic Gene Clusters (BGCs)
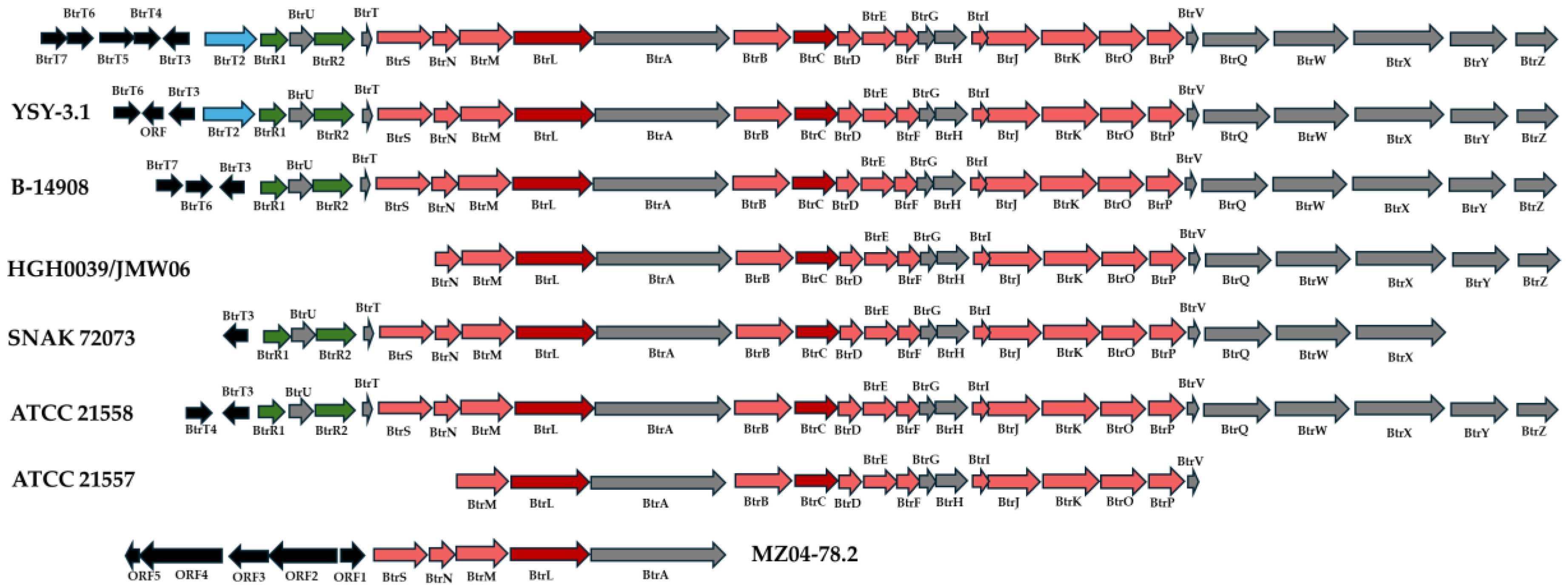
3.3. Comparative Characterization of the Butorosin BGCs
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Krause, K.M.; Serio, A.W.; Kane, T.R.; Connolly, L.E. Aminoglycosides: An Overview. Cold Spring Harb Perspect Med. 2016, 6, a027029. [Google Scholar] [CrossRef] [PubMed]
- Serio, A.W.; Keepers, T.; Andrews, L.; Krause, K.M. Aminoglycoside Revival: Review of a Historically Important Class of Antimicrobials Undergoing Rejuvenation. EcoSal Plus. 2018, 8. [Google Scholar] [CrossRef] [PubMed]
- Park, S.R.; Park, J.W.; Ban, Y.H.; . Sohng, J.K.; Yoon, Y.J. 2-Deoxystreptamine-containing aminoglycoside antibiotics: recent advances in the characterization and manipulation of their biosynthetic pathways. Nat. Prod. Rep. 2013, 30, 11–20. [Google Scholar] [CrossRef] [PubMed]
- Kudo, F.; Numakura, M.; Tamegai, H.; Yamamoto, H.; Eguchi, T.; Kakinuma, K. Extended sequence and functional analysis of the butirosin biosynthetic gene cluster in Bacillus circulans SANK 72073. J. Antibiot (Tokyo), 2005, 58, 373-379. [CrossRef]
- Li, Y.; Llewellyn, N.M.; Giri, R.; Huang, F.; Spencer, J.B. Biosynthesis of the unique amino acid side chain of butirosin: possible protective-group chemistry in an acyl carrier protein-mediated pathway. Chem. Biol. 2005, 12, 665–675. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Zhang, Q.; Deng, Z. Parallel pathways in the biosynthesis of aminoglycoside antibiotics. F1000Res. 2017, 6, F1000 Faculty Rev-723. [CrossRef]
- Arenas, L.A.R.; de Paiva, F.C.F.; Rossini. N. de O.; Li, Y.; Spencer J.; Leadlay, P.; Dias. M.V.B. Crystal structure of BtrK, a decarboxylase involved in the (S)-4-amino-2-hydroxybutyrate (AHBA) formation during butirosin biosynthesis. J. Mol. Struct. 2022, 1267, 133576. [Google Scholar] [CrossRef]
- Dion, H.W.; Woo, P.W.; Willmer, N.E.; Kern, D.L.; Onaga, J.; Fusari, S.A. Butirosin, a new aminoglycosidic antibiotic complex: isolation and characterization. Antimicrob. Agents. Chemother. 1972, 2, 84–88. [Google Scholar] [CrossRef]
- Nakahama, K.; Shirafuji, H.; Nogami, I.; Kida, M.; Yoneda, M. Butirosin 3′-Phosphotransferase from Bacillus vitellinus, a Butirosin-producing Organism, Agric. Biol. Chem. 1977, 41, 2437–2445. [Google Scholar] [CrossRef]
- Espinosa, E.; Bautista, R.; Larrosa, R.; Plata, O. Advancements in long-read genome sequencing technologies and algorithms. Genomics. 2024, 116, 110842. [Google Scholar] [CrossRef]
- Logsdon, G.A.; Vollger, M.R.; Eichler, E.E. Long-read human genome sequencing and its applications. Nat. Rev. Genet. 2020, 21, 597–614. [Google Scholar] [CrossRef]
- Blin, K.; Shaw, S.; Augustijn, H.E.; Reitz, Z.L.; Biermann, F.; Alanjary, M.; Fetter, A.; Terlouw, B.R.; Metcalf, W.W.; Helfrich, E.J.N.; van Wezel, G.P.; Medema, M.H.; Weber, T. antiSMASH 7.0: new and improved predictions for detection, regulation, chemical structures and visualisation. Nucleic. Acids Res. 2023, 51, W46–W50. [Google Scholar] [CrossRef] [PubMed]
- Terlouw, B.R.; Blin, K.; Navarro-Muñoz, JC.; . Avalon, N.E.; Chevrette, M.G.; Egbert, S.; Lee, S.; Meijer, D.; Recchia, M.J.J.; Reitz, Z.L.; et al. MIBiG 3.0: a community-driven. effort to annotate experimentally validated biosynthetic gene clusters. Nucleic. Acids Res. 2023, 51, D603–D610. [Google Scholar] [CrossRef]
- Koren, S.; Walenz, B.P.; Berlin, K.; Miller, J.R.; Bergman, N.H.; Phillippy, A.M. Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 2017, 27, 722–736. [Google Scholar] [CrossRef] [PubMed]
- Hunt, M.; Silva, N.D.; Otto, T.D.; Parkhill, J.; Keane, J.A.; Harris, S.R. Circlator: automated circularization of genome assemblies using long sequencing reads. Genome Biol. 2015, 16, 294. [Google Scholar] [CrossRef] [PubMed]
- Tatusova, T.; DiCuccio, M.; Badretdin, A.; Chetvernin, V.; Nawrocki, E.P.; Zaslavsky, L.; Lomsadze, A.; Pruitt, K.D.; Borodovsky, M.; Ostell, J. NCBI prokaryotic genome annotation pipeline. Nucleic. Acids Res. 2016, 44, 6614–6624. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Meier-Kolthoff, JP.; Carbasse, J.S.; Peinado-Olarte, R.L.; Göker, M. TYGS.and LPSN: a database tandem for fast and reliable genome-based classification and nomenclature of prokaryotes. Nucleic. Acids Res. 2022, 50, D801–D807. [Google Scholar] [CrossRef] [PubMed]
- Chalita, M.; Kim, Y.O.; Park, S.; Oh, H.S.; Cho, J.H.; Moon, J.; Baek, N.; Moon, C.; Lee, K.; Yang, J.; Nam, G.G.; Jung, Y.; Na, S.I.; Bailey, M.J.; Chun, J. EzBioCloud: a genome-driven database and platform for microbiome identification and discovery. Int. J. Syst. Evol. Microbiol. 2024, 74, 006421. [Google Scholar] [CrossRef]
- van Heel, A.J.; de Jong, A.; Song, C.; Viel, J.H.; Kok, J.; Kuipers, O.P. BAGEL4: a user-friendly web server to thoroughly mine RiPPs and bacteriocins. Nucleic. Acids Res. 2018, 46, W278–W281. [Google Scholar] [CrossRef] [PubMed]
- Skinnider, M.A.; Johnston, C.W.; Gunabalasingam, M.; Merwin, N.J.; Kieliszek, A.M.; MacLellan, R.J.; Li, H.; Ranieri, M.R.M.; Webster, A.L.H.; Cao, M.P.T.; Pfeifle, A.; Spencer, N.; To, Q.H.; Wallace, D.P.; Dejong, C.A.; Magarvey, N.A. Comprehensive prediction of secondary metabolite structure and biological activity from microbial genome sequences. Nat. Commun. 2020, 11, 6058. [Google Scholar] [CrossRef]
- Yoon, S.H.; Ha, S.M.; Kwon, S.; Lim, J.; Kim, Y.; Seo, H, Chun, J. Introducing EzBioCloud: a taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int. J. Syst. Evol. Microbiol. 2017, 67, 1613–1617. [Google Scholar] [CrossRef] [PubMed]
- Goris, J.; Konstantinidis, K.T.; Klappenbach, J.A.; Coenye, T.; Vandamme, P.; Tiedje, J.M. DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int. J. Syst. Evol. Microbiol. 2007, 57, 81–91. [Google Scholar] [CrossRef]
- Richter, M.; Rosselló-Móra, R. Shifting the genomic gold standard for the prokaryotic species definition. Proc. Natl. Acad. Sci. U S A. 2009, 106, 9126–9131. [Google Scholar] [CrossRef] [PubMed]
- Kudo, F.; Mori,, A. ; Koide M.; Yajima, R.; Takeishi, R.; Miyanaga, A.; Eguchi, T. One-pot enzymatic synthesis of 2-deoxy-scyllo-inosose from d-glucose and polyphosphate. Biosci. Biotechnol. Biochem. 2021, 85, 108–114. [Google Scholar] [CrossRef] [PubMed]
- Wehmeier, U.F.; Piepersberg,W. Enzymology of aminoglycoside biosynthesis-deduction from gene clusters. Methods Enzymol. 2009, 459, 459–491. [Google Scholar] [CrossRef]
- Gao, Q.; Thorson, J.S. The biosynthetic genes encoding for the production of the dynemicin enediyne core in Micromonospora chersina ATCC53710. FEMS Microbiol. Lett. 2008, 282, 105–114. [Google Scholar] [CrossRef]
- Bililign, T.; Hyun,C. G.; Williams, J.S.; Czisny, A.M.; Thorson,.JS. The hedamycin locus implicates a novel aromatic PKS priming mechanism. Chem. Biol. 2004, 11, 959–969. [Google Scholar] [CrossRef]
- Ahlert, J.; Shepard, E.; Lomovskaya, N.; Zazopoulos, E.; Staffa, A.; Bachmann, B.O.; Huang, K.; Fonstein, L.; Czisny, A.; Whitwam, R.E.; Farnet, C.M.; Thorson, J.S. The calicheamicin gene cluster and its iterative type I enediyne PKS. Science. 2002, 297, 1173–1176. [Google Scholar] [CrossRef]
- Hyun, C.G.; Bililign, T.; Liao, J.; Thorson, S. The biosynthesis of indolocarbazoles in a heterologous E. coli host. Chembiochem. 2003, 4, 114–117. [Google Scholar] [CrossRef]
- Hyun, C.G.; Kim, S.S.; Sohng, J.K.; Hahn, J.; Kim, J.; Suh, J. An efficient approach for cloning the dNDP-glucose synthase gene from actinomycetes and its application in Streptomyces spectabilis, a spectinomycin producer. FEMS Microbiol. Lett. 2000, 183, 183–189. [Google Scholar] [CrossRef]

| Region | Type | From | To | Most similar known cluster | Similarity |
|---|---|---|---|---|---|
| Region 1 | NRPS, LAP | 305619 | 356240 | ||
| Region 2 | RRE-containing | 747520 | 767798 | ||
| Region 3 | RiPP-like | 851869 | 864055 | ||
| Region 4 | Crocagin, HR-T2PKS | 945752 | 1014754 | ||
| Region 5 | TransAT-PKS, NRPS | 2461872 | 2549008 | ||
| Region 6 | TransAT-PKS, NRPS | 2563881 | 2640650 | Pelgipeptin | 25% |
| Region 7 | Cyclic-lactone-autoinducer | 2734390 | 2754992 | ||
| Region 8 | Terpene | 2821778 | 2843727 | ||
| Region 9 | Betalactone | 3032661 | 3062918 | ||
| Region 10 | TransAT-PKS, NRPS | 3175761 | 3243813 | ||
| Region 11 | Phosphonate | 3441175 | 3454343 | ||
| Region 12 | Amglyccycl | 3489373 | 3517357 | Butirosin A/B | 84% |
| Region 13 | NRPS | 3540249 | 3611046 | Octapeptin C4 | 29% |
| Region 14 | NRP-metallophore, NRPS | 3614412 | 3666638 | Bacillibactin | 100% |
| Region 15 | Opine-like-metallophore | 3687196 | 3729044 | Bacillopaline | 100% |
| Region 16 | T3PKS | 3837645 | 3878793 | ||
| Region 17 | Proteusin | 5176800 | 5197111 | ||
| Region 18 | TransAT-PKS-like, NRPS | 5276428 | 5336092 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





