Main Text
Neuropsychiatric disorders, such as schizophrenia (SCZ) and bipolar disorder (BPD), pose a significant global health burden, affecting millions of individuals worldwide [
1]. Precise prevalence estimates of these disorders are impossible to obtain due to clinical and methodological factors, such as the complexity of neuropsychiatric diagnosis, their overlap with other disorders, and varying methods for determining diagnoses. Given these complexities, SCZ and other psychotic disorders are often combined in prevalence estimation studies [
2].
To address the critical need for objective diagnostic markers in neuropsychiatry, this study explores the clinical application of novel digital technologies. By leveraging cerebral organoids (COs) derived from patient-specific induced pluripotent stem cells (iPSCs) [
3] and multi-electrode array technology (MEA), we have developed a digital analysis pipeline (DAP) that builds upon the foundation established by electroencephalography (EEG) analyses [
4,
5,14,15,16,19]. This approach aims to uncover distinct electrophysiological signatures associated with SCZ and BPD, offering a physiologically relevant and comprehensive assessment of neural network dynamics.
COs recapitulate key aspects of human brain development and provide a physiologically relevant in vitro model system to investigate the neurophysiological mechanisms underlying complex psychiatric disorders [
5]. The application of MEA in patient-derived COs enables comprehensive analysis of neural network dynamics and extraction of digital biomarkers that can serve as objective diagnostic indicators, complementing insights gleaned from traditional EEG techniques [
6]. By bridging the gap between advanced in vitro models, cutting-edge electrophysiological analysis, and machine learning-driven feature extraction, this research presents a promising approach to addressing the critical need for validated digital markers in the field of neuropsychiatry.
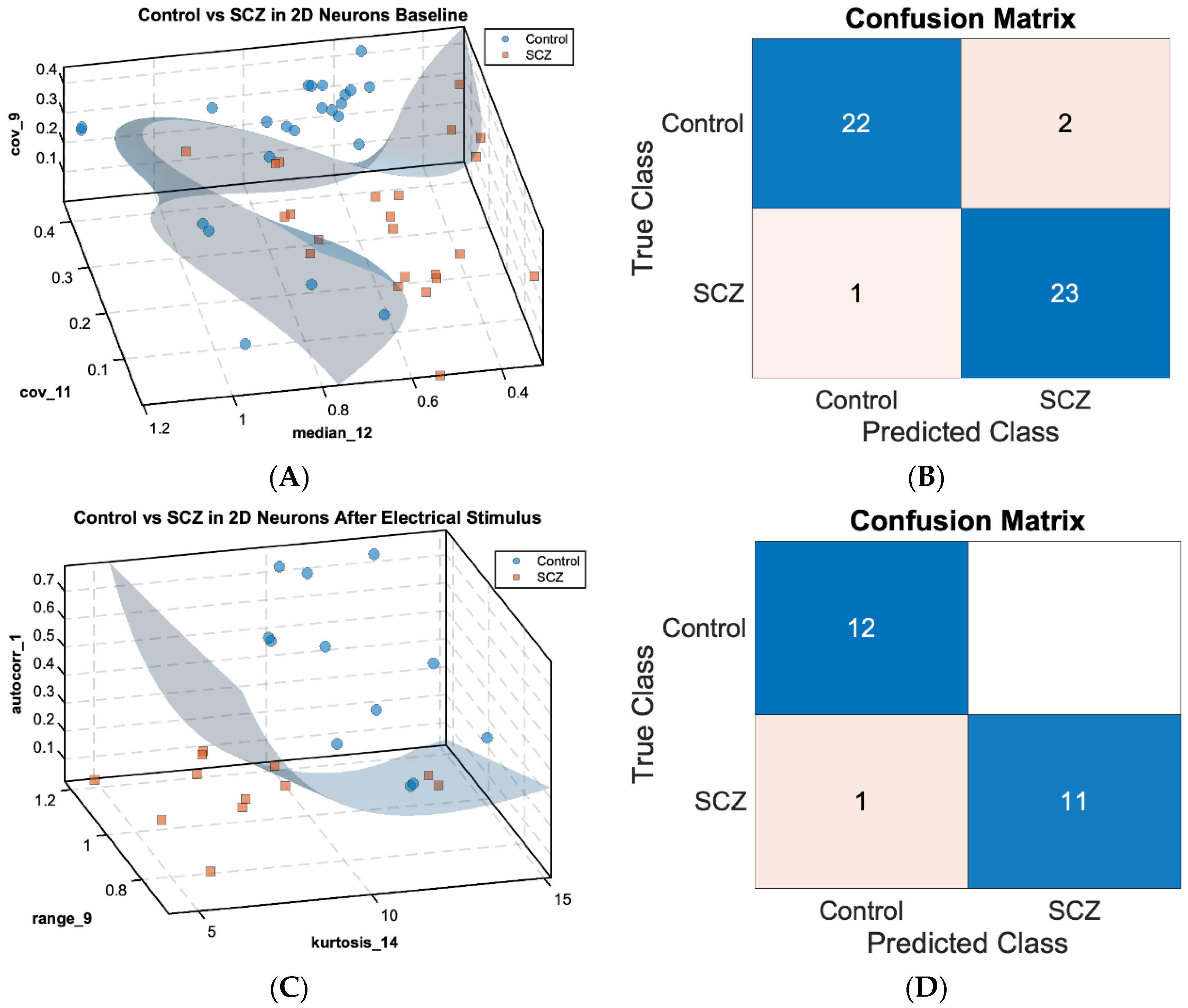
The DAP designed in this study investigates the electrophysiological properties of COs and cortical neurons in monolayer culture (2DNs) derived from patients with SCZ and BPD. Utilizing MEA and Stimulus-Response Dynamic Network Modeling (SRDNM), we analyzed neural recordings to identify influential nodes in the neural network. An extensive feature map of the sink index dynamics was computed for each channel and screened by a feature selection algorithm [20] to identify the features most significant to cohort classification. These features were then used to train a Support Vector Machine (SVM) classifier to distinguish different patient-derived organoids from healthy controls (
Figure 1A,C).
Raw MEA recordings were retrieved from our previously published studies [
7,
8,
9]. These studies compared cortical neurons in monolayer, including co-cultured excitatory and inhibitory neurons [
10], derived from iPSCs from SCZ patients and healthy control individuals. Our studies also compared iPSC-derived COs from SCZ and BPD patients and healthy control individuals that were cultured for nine months. COs have increased cellular diversity and network functionality with complex electrophysiological properties which can enable a more comprehensive understanding of neural dynamics underlying neuropsychiatric disorders.
Figure 1A,C show scatterplots in a 3-dimensional feature space with the SVM decision boundary for 2DNs separating the healthy control cohort (n = 24 for baseline, n = 12 for post-electrical stimulus (PES) condition, as blue dots) versus the SCZ cohort (n = 24 for baseline, n = 12 for PES condition, as orange squares) under baseline (
Figure 1A) and PES conditions (
Figure 1C). The scatterplots illustrate the separation of control and SCZ neurons based on significant features of the sink index, indicating that features related to channel-wise sink index dynamics can distinguish healthy and SCZ populations with high accuracy.
In the control versus SCZ baseline (
Figure 1A), three sink index features were identified as critical to cohort separation: median of channel 12
and covariance of channels 9
and 11
, achieving a classification accuracy of 0.958. In the PES condition (
Figure 1C), sink index features identified include the autocorrelation of channel 1 (
), range of channel 9
, and kurtosis of channel 14
, achieving the same accuracy of 0.958 in the validation test. The respective confusion matrices (
Figure 1B,D) provide a visual representation of classification performance. Only three instances among the 48 samples in the baseline condition and one instance among the 24 samples in the PES conditions were misclassified, indicating a robust separation between control and SCZ classes under both paradigms.
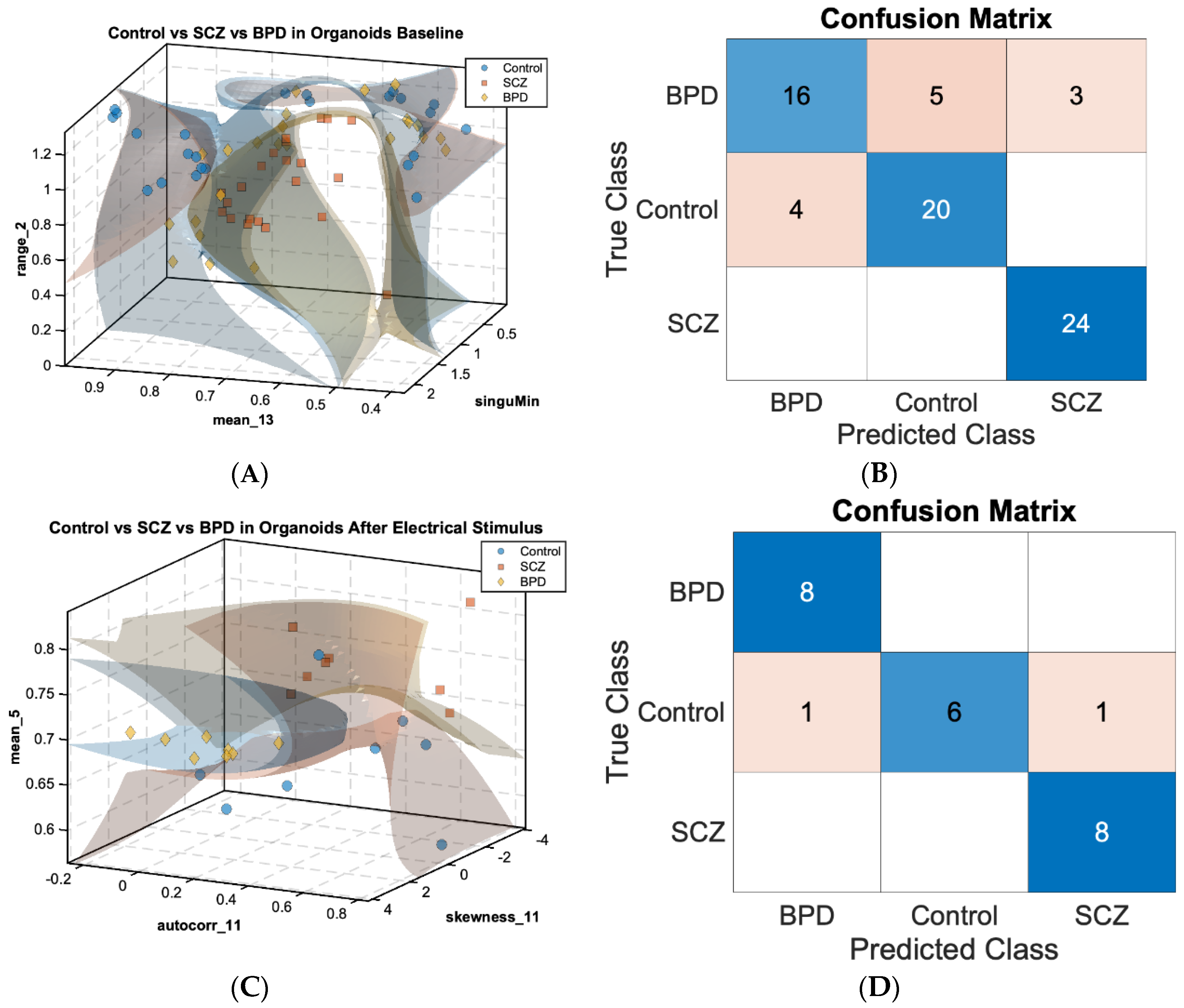
Similarly,
Figure 2A,C show 3D scatterplots displaying SVM decision boundaries for the three cohorts: control (n = 24 for baseline, n = 8 for PES condition, as blue dots), SCZ (n = 24 for baseline, n = 8 for PES condition, as orange squares), and BPD (n = 24 for baseline, n = 8 for PES condition, as yellow diamonds) in COs under the baseline (
Figure 2A) and PES conditions (
Figure 2C), respectively.
In the baseline condition (
Figure 2A), the MRMR feature selection algorithm identified the sink index features significant to classification as the range of channel 2
, mean of channel 13
, and all-channel-minimum singular value
. These features were able to separate control, SCZ, and BPD COs with an accuracy of 0.833, despite an imbalance in cohort sizes. In the PES condition (
Figure 2C), selected sink index features were the mean of channel 5
, autocorrelation of channel 11
, and skewness of channel 11
, with a significant improvement in classification accuracy to 0.916. The respective confusion matrices (
Figure 2B,D) demonstrate the classification performance. These results show that most misclassified instances under baseline conditions were distributed in the BPD class. However, we were able to improve the classification accuracy significantly in the PES condition as the BPD cohort was densely clustered in the identified feature space; only two within control cohorts among 24 instances that were misclassified as diseased cohorts. Hence, while we were not able to clearly distinguish BPD cohorts and control cohorts under baseline conditions, we were able to distinguish between healthy cohorts and disease cohorts (Control vs SCZ, Control vs BPD, SCZ vs BPD) using analyses of MEA recordings from the PES conditions.
These results demonstrate that for the 2DN and CO cohorts an SVM classifier can effectively distinguish healthy control and patient-derived organoids and neurons, utilizing extracted cellular-digital biomarkers regarding sink index features from MEA data. More importantly, symptoms of diseased cohorts were significantly manifested in the selected feature space under the PES condition for both 2DNs and COs thus we were able to distinguish more clearly among all cohorts, especially in COs. The clear separation of classes in the scatter plots and the high classification accuracy depicted in the confusion matrices underscore the potential of this DAP for identification of electrophysiological signatures associated with neuropsychiatric disorders.
The application of MEA technology in patient-derived COs offers a relevant and controlled environment for studying electrophysiological correlates of neuropsychiatric disorders. Clinical research [
11,
12] has identified significant barriers to the effective diagnosis and treatment of neuropsychiatric disorders, including the subjective nature of current diagnostic methods and the lack of reliable biomarkers. Given the challenge of subjective diagnostic methods [
11,
12], our DAP provides ways to improve diagnostic accuracy by utilizing a quantifiable and objective method for assessing neural activity. Moreover, it manages and interprets complex, high-dimensional, time-varying data using advanced machine learning techniques [
13], facilitating the extraction of meaningful features that distinguish SCZ and BPD. Since traditional animal models have limitations in replicating human brain physiology [
6], studies of human COs can bridge the gap between in vitro studies and clinical applications. Our study addresses the critical need for validated digital biomarkers, offering a set of objective, reproducible indicators that improve diagnostic precision and inform personalized treatment strategies.