Submitted:
16 August 2024
Posted:
20 August 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Ribosome Profiling (Ribo-Seq)
2.3. RNA Sequencing (RNA-Seq)
2.4. Ribo-Seq and RNA-Seq Data Processing
2.5. Analysis of Gene Expression at Transcriptional and Translational Levels
2.6. Calculation of Translational Efficiency
2.7. Analysis of Homoeolog Expression Bias (HEB)
3. Results
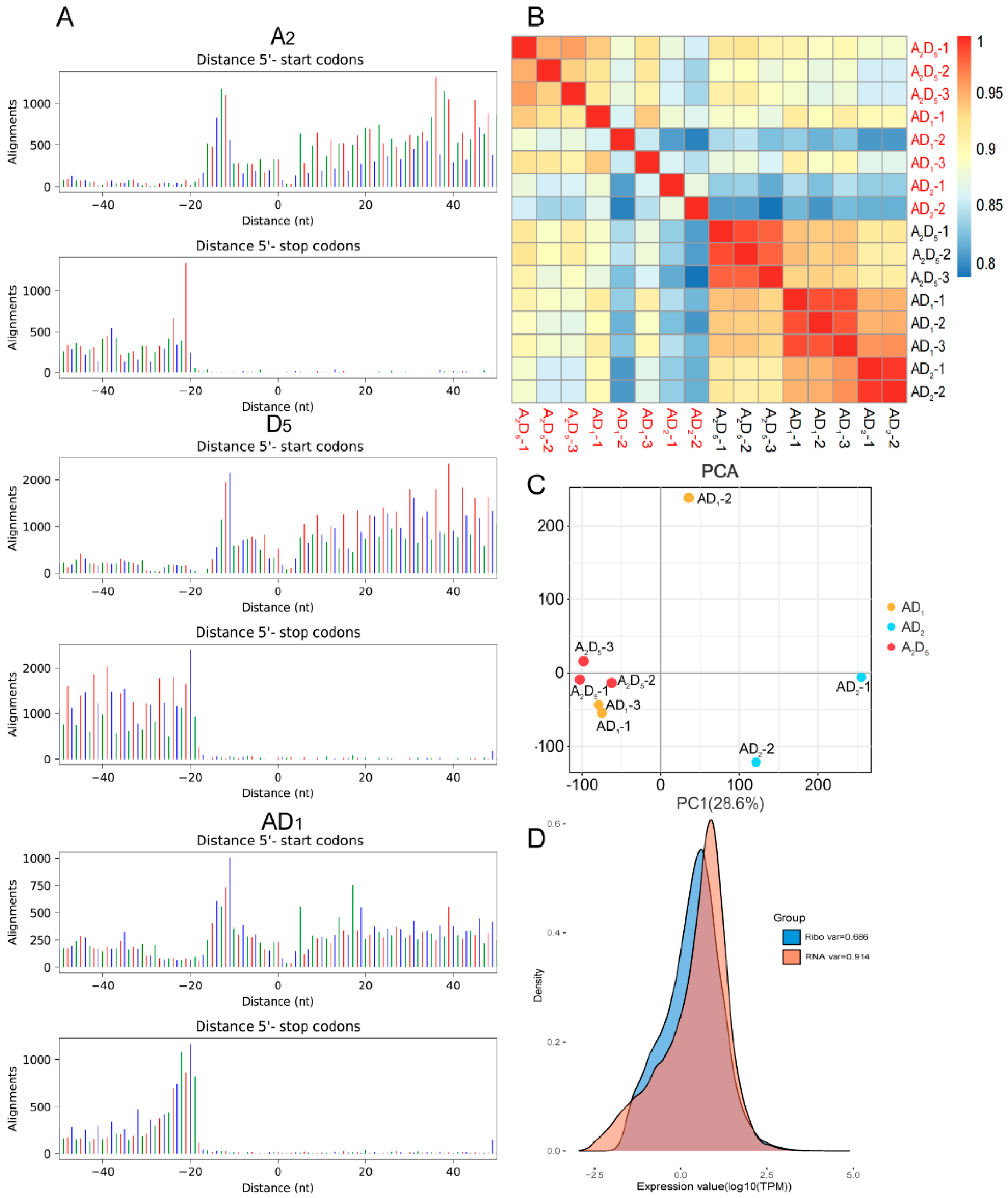
3.1. Transcriptional and Translational Profiling of Diploid and Allopolyploid Cottons
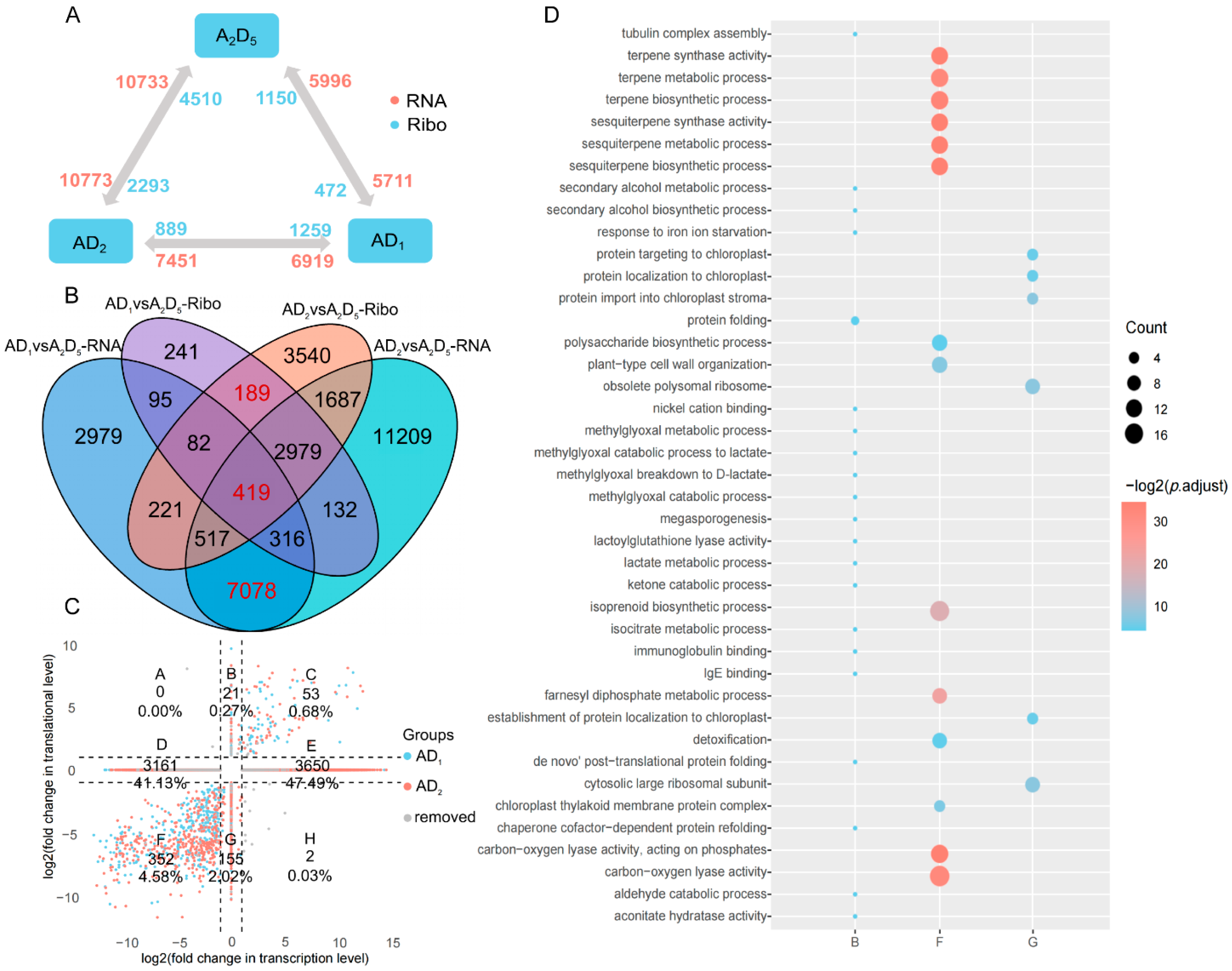
3.2. Concordantly Decreased Transcription and Translation Accompanying Allopolyploidization
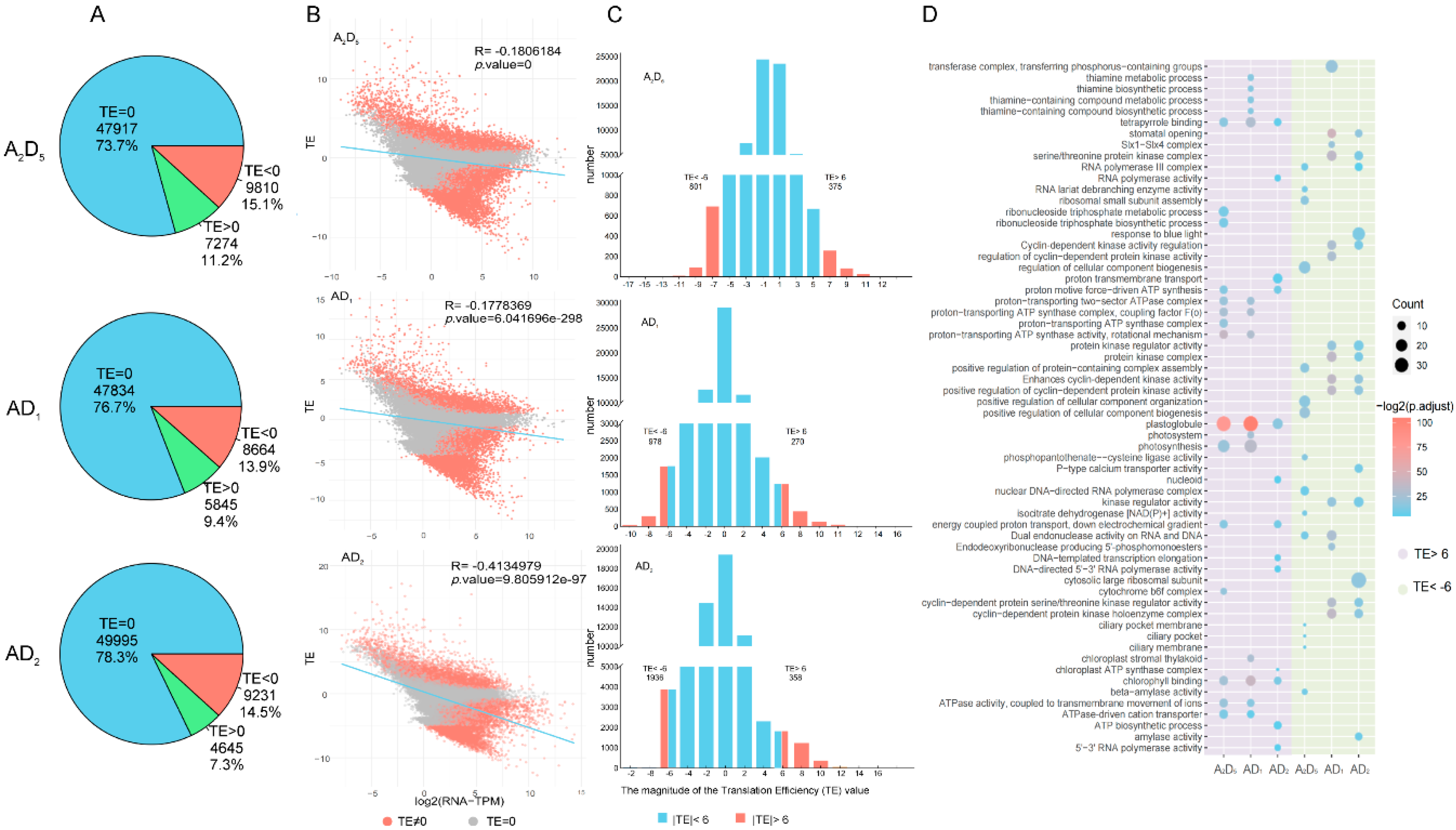
3.3. Impact of Allopolyploidy on Translational Efficiency
3.4. Reduced Amount of Homoeolog Expression Biases at the Translational Level
| A2D5 | AD1 | AD2 | |||||
| Number | Proportion | Number | Proportion | Number | Proportion | ||
| RNA | Total | 5285 | 23.09% | 5237 | 22.88% | 8407 | 36.73% |
| A>D | 2574 | 11.25% | 2576 | 11.25% | 4165 | 18.20% | |
| A<D | 2711 | 11.84% | 2661 | 11.63% | 4242 | 18.53% | |
| p-value* | 0.0595 | 0.2402 | 0.401 | ||||
| Ribo | Total | 2224 | 9.72% | 1013 | 4.43% | 562 | 2.46% |
| A>D | 1115 | 4.87% | 484 | 2.11% | 244 | 1.07% | |
| A<D | 1109 | 4.85% | 529 | 2.31% | 318 | 1.39% | |
| p-value* | 0.8988 | 0.1574 | 0.001799 | ||||
| TE | Total | 1076 | 4.70% | 625 | 2.73% | 547 | 2.39% |
| A>D | 391 | 1.71% | 320 | 1.40% | 237 | 1.04% | |
| A<D | 685 | 2.99% | 305 | 1.33% | 310 | 1.35% | |
| p-value* | 2.20E-16 | 0.5485 | 0.001801 | ||||
4. Discussion
4.1. Ribo-Seq Technical Difficulties
4.2. Cotton Leaf Gene Express Patterns in Transcriptional and Translational Levels
4.3. Expression Bias of Homologous Genes in Cotton
4.4. Translational Regulation that May Be Caused by Polyploidy
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Declaration of interests
References
- Alger, E.I.; Edger, P.P. One subgenome to rule them all: underlying mechanisms of subgenome dominance. Current Opinion in Plant Biology 2020, 54, 108–113. [Google Scholar] [CrossRef]
- Jiao, Y.; Wickett, N.J.; Ayyampalayam, S.; Chanderbali, A.S.; Landherr, L.; Ralph, P.E.; Tomsho, L.P.; Hu, Y.; Liang, H.; Soltis, P.S.; Soltis, D.E.; Clifton, S.W.; Schlarbaum, S.E.; Schuster, S.C.; Ma, H.; Leebens-Mack, J.; Depamphilis, C.W. Ancestral polyploidy in seed plants and angiosperms. Nature 2011, 473, 97–100. [Google Scholar] [CrossRef]
- Liu, Q.; Yuan, H.; Li, M.; Wang, Z.; Cui, D.; Ye, Y.; Sun, Z.; Tan, X.; Schwarzacher, T.; Heslop-Harrison, J.S. Chromosome-scale genome assembly of the diploid oat Avena longiglumis reveals the landscape of repetitive sequences, genes and chromosome evolution in grasses. bioRxiv 2022. [Google Scholar] [CrossRef]
- Van De Peer, Y.; Ashman, T.-L.; Soltis, P.S.; Soltis, D.E. Polyploidy: an evolutionary and ecological force in stressful times. The Plant Cell 2021, 33, 11–26. [Google Scholar] [CrossRef] [PubMed]
- Grover, C.E.; Gallagher, J.P.; Szadkowski, E.P.; Yoo, M.J.; Flagel, L.E.; Wendel, J.F. Homoeolog expression bias and expression level dominance in allopolyploids. New Phytologist 2012, 196, 966–971. [Google Scholar] [CrossRef] [PubMed]
- Hu, G.; Wendel, J.F. Cis–trans controls and regulatory novelty accompanying allopolyploidization. New Phytologist 2018, 221, 1691–1700. [Google Scholar] [CrossRef]
- Yoo, M.-J.; Liu, X.; Pires, J.C.; Soltis, P.S.; Soltis, D.E. Nonadditive gene expression in polyploids. Annual Review of Genetics 2014, 48, 485–517. [Google Scholar] [CrossRef]
- Soltis, D.E.; Misra, B.B.; Shan, S.; Chen, S.; Soltis, P.S. Polyploidy and the proteome. Biochimica et Biophysica Acta (BBA) - Proteins and Proteomics 2016, 1864, 896–907. [Google Scholar] [CrossRef]
- Zhao, J.; Qin, B.; Nikolay, R.; Spahn CM, T.; Zhang, G. Translatomics: the global view of translation. International Journal of Molecular Sciences 2019, 20. [Google Scholar] [CrossRef]
- Ingolia, N.T. Ribosome Footprint Profiling of Translation throughout the genome. Cell 2016, 165, 22–33. [Google Scholar] [CrossRef] [PubMed]
- Burch-Smith, T.; Chotewutmontri, P.; Barkan, A. Multilevel effects of light on ribosome dynamics in chloroplasts program genome-wide and psbA-specific changes in translation. PLOS Genetics 2018, 14. [Google Scholar] [CrossRef]
- Hou, C.Y.; Lee, W.C.; Chou, H.C.; Chen, A.P.; Chou, S.J.; Chen, H.M. Global analysis of truncated RNA ends reveals new insights into ribosome stalling in plants. The Plant Cell 2016, 28, 2398–2416. [Google Scholar] [CrossRef]
- Zhu, X.-T.; Zhou, R.; Che, J.; Zheng, Y.-Y.; Tahir Ul Qamar, M.; Feng, J.-W.; Zhang, J.; Gao, J.; Chen, L.-L. Ribosome profiling reveals the translational landscape and allele-specific translational efficiency in rice. Plant Communications 2023, 4. [Google Scholar] [CrossRef]
- Jian, H.; Wen, S.; Liu, R.; Zhang, W.; Li, Z.; Chen, W.; Zhou, Y.; Khassanov, V.; Mahmoud AM, A.; Wang, J.; Lyu, D. Dynamic translational landscape revealed by genome-wide ribosome profiling under drought and heat stress in potato. Plants 2023, 12. [Google Scholar] [CrossRef]
- Lei, L.; Shi, J.; Chen, J.; Zhang, M.; Sun, S.; Xie, S.; Li, X.; Zeng, B.; Peng, L.; Hauck, A.; Zhao, H.; Song, W.; Fan, Z.; Lai, J. Ribosome profiling reveals dynamic translational landscape in maize seedlings under drought stress. The Plant Journal 2015, 84, 1206–1218. [Google Scholar] [CrossRef] [PubMed]
- Xu, G.; Greene, G.H.; Yoo, H.; Liu, L.; Marqués, J.; Motley, J.; Dong, X. Global translational reprogramming is a fundamental layer of immune regulation in plants. Nature 2017, 545, 487–490. [Google Scholar] [CrossRef] [PubMed]
- Xiong, Q.Z.L.; Du, J.; Zhu, C.; Peng, X.; He, X.; Fu, J.; Ouyang, L.; Bian, J.; Hu, L.; Sun, X.; Xu, J.; Zhou, D.; Cai, Y.; Fu, H.; He, H.; Chen, X. Ribosome profiling reveals the effects of nitrogen application translational regulation of yield recovery after abrupt drought-flood alternation in rice. Plant Physiol Biochem 2020, 155, 42–58. [Google Scholar] [CrossRef]
- Yang, X.; Song, B.; Cui, J.; Wang, L.; Wang, S.; Luo, L.; Gao, L.; Mo, B.; Yu, Y.; Liu, L. Comparative ribosome profiling reveals distinct translational landscapes of salt-sensitive and -tolerant rice. BMC Genomics 2021, 22. [Google Scholar] [CrossRef]
- Balfagón, D.; Zandalinas, S.I.; Dos Reis De Oliveira, T.; Santa-Catarina, C.; Gómez-Cadenas, A. Omics analyses in citrus reveal a possible role of RNA translation pathways and unfolded protein response regulators in the tolerance to combined drought, high irradiance, and heat stress. Horticulture Research 2023, 10. [Google Scholar] [CrossRef]
- Zhu, W.; Chen, S.; Zhang, T.; Qian, J.; Luo, Z.; Zhao, H.; Zhang, Y.; Li, L. Dynamic patterns of the translatome in a hybrid triplet show translational fractionation of the maize subgenomes. The Crop Journal 2022, 10, 36–46. [Google Scholar] [CrossRef]
- Coate, J.E.; Bar, H.; Doyle, J.J. Extensive translational regulation of gene expression in an allopolyploid (Glycine dolichocarpa). The Plant Cell 2014, 26, 136–150. [Google Scholar] [CrossRef]
- Wendel, J.F.; Grover, C.E. Taxonomy and evolution of the cotton genus, Gossypium [M]. Cotton. 2015: 25-44.
- Flagel, L.E.; Wendel, J.F. Evolutionary rate variation, genomic dominance and duplicate gene expression evolution during allotetraploid cotton speciation. New Phytologist 2010, 186, 184–193. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Naoumkina, M.; Thyssen, G.; Fang, D.D.; Hinchliffe, D.J.; Florane, C.; Yeater, K.M.; Page, J.T.; Udall, J.A. The Li2 mutation results in reduced subgenome expression bias in elongating fibers of allotetraploid cotton (Gossypium hirsutum L.). PLoS ONE 2014, 9. [Google Scholar] [CrossRef]
- Hu, G.; Houston, N.L.; Pathak, D.; Schmidt, L.; Thelen, J.J.; Wendel, J.F. Genomically biased accumulation of seed storage proteins in allopolyploid cotton. Genetics 2011, 189, 1103–1115. [Google Scholar] [CrossRef] [PubMed]
- Zheng, D.; Ye, W.; Song, Q.; Han, F.; Zhang, T.; Chen, Z.J. Histone modifications define expression bias of homoeologous genomes in allotetraploid cotton. Plant Physiology 2016, 172, 1760–1771. [Google Scholar] [CrossRef] [PubMed]
- Qanmber, G.; You, Q.; Yang, Z.; Fan, L.; Zhang, Z.; Chai, M.; Gao, B.; Li, F.; Yang, Z. Transcriptional and translational landscape fine-tune genome annotation and explores translation control in cotton. Journal of Advanced Research 2024, 58, 13–30. [Google Scholar] [CrossRef] [PubMed]
- Hsu, P.Y.; Calviello, L.; Wu, H.-Y.L.; Li, F.-W.; Rothfels, C.J.; Ohler, U.; Benfey, P.N. Super-resolution ribosome profiling reveals unannotated translation events in Arabidopsis. Proceedings of the National Academy of Sciences 2016, 113. [Google Scholar] [CrossRef]
- Ingolia, N.T.; Brar, G.A.; Rouskin, S.; Mcgeachy, A.M.; Weissman, J.S. The ribosome profiling strategy for monitoring translation in vivo by deep sequencing of ribosome-protected mRNA fragments. Nature Protocols 2012, 7, 1534–1550. [Google Scholar] [CrossRef]
- Chen, S. Ultrafast one-pass FASTQ data preprocessing, quality control, and deduplication using fastp. iMeta 2023, 2. [Google Scholar] [CrossRef]
- Langmead, B.S.S. Fast gapped-read alignment with Bowtie 2. Nature Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Saski, C.A.; Scheffler, B.E.; Hulse-Kemp, A.M.; Liu, B.; Song, Q.; Ando, A.; Stelly, D.M.; Scheffler, J.A.; Grimwood, J.; Jones, D.C.; Peterson, D.G.; Schmutz, J.; Chen, Z.J. Sub genome anchored physical frameworks of the allotetraploid Upland cotton (Gossypium hirsutum L.) genome, and an approach toward reference-grade assemblies of polyploids. Scientific Reports 2017, 7. [Google Scholar] [CrossRef]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nature Biotechnology 2019, 37, 907–915. [Google Scholar] [CrossRef] [PubMed]
- Shi, W. Rsubread: R package for high-throughput sequencing data processing. Computer software, 2021.
- Dobin, A. STAR aligner software. [Z]. Cold Spring Harbor Laboratory. 2021. [Google Scholar]
- Xiao, Z. RiboCode is a very simple but high-quality computational algorithm to identify genome-wide translated ORFs using ribosome-profiling data. [Z]. 2022. [Google Scholar]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biology 2014, 15. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society Series B: Statistical Methodology 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Yu, G.; Wang, L.-G.; Han, Y.; He, Q.-Y. ClusterProfiler: an R package for comparing biological themes among gene clusters. OMICS: A Journal of Integrative Biology 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Wickham, H. ggplot2: Elegant graphics for data analysis [Z]. Springer-Verlag New York. 2016. [Google Scholar]
- Kolde, R. pheatmap: Pretty heatmaps. 2019.
- Hu, G.; Grover, C.E.; Vera, D.L.; Lung, P.-Y.; Girimurugan, S.B.; Miller, E.R.; Conover, J.L.; Ou, S.; Xiong, X.; Zhu, D.; Li, D.; Gallagher, J.P.; Udall, J.A.; Sui, X.; Zhang, J.; Bass, H.W.; Wendel, J.F.; Purugganan, M. Evolutionary dynamics of chromatin Structure and duplicate gene expression in diploid and allopolyploid cotton. Molecular Biology and Evolution 2024, 41. [Google Scholar] [CrossRef]
- Ingolia, N.T.; Ghaemmaghami, S.; Newman JR, S.; Weissman, J.S. Genome-Wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 2009, 324, 218–223. [Google Scholar] [CrossRef]
- Paulet, D.; David, A.; Rivals, E. Ribo-seq enlightens codon usage bias. DNA Research 2017, 24, 303–210. [Google Scholar] [CrossRef]
- Guttman, M.; Russell, P.; Ingolia Nicholas, T.; Weissman Jonathan, S.; Lander Eric, S. Ribosome profiling provides evidence that large noncoding RNAs do not encodeproteins. Cell 2013, 154, 240–251. [Google Scholar] [CrossRef] [PubMed]
- Juntawong, P.; Girke, T.; Bazin, J.; Bailey-Serres, J. Translational dynamics revealed by genome-wide profiling of ribosome footprints in Arabidopsis. Proceedings of the National Academy of Sciences 2013, 111. [Google Scholar] [CrossRef]
- Chaudhary, B.; Flagel, L.; Stupar, R.M.; Udall, J.A.; Verma, N.; Springer, N.M.; Wendel, J.F. Reciprocal silencing, transcriptional bias and functional divergence of homeologs in polyploid cotton (Gossypium). Genetics 2009, 182, 503–517. [Google Scholar] [CrossRef] [PubMed]
- Hovav, R.; Faigenboim-Doron, A.; Kadmon, N.; Hu, G.; Zhang, X.; Gallagher, J.P.; Wendel, J.F. A transcriptome profile for developing seed of polyploid cotton. The Plant Genome 2015, 8. [Google Scholar] [CrossRef]
- Zhang, T.; Hu, Y.; Jiang, W.; Fang, L.; Guan, X.; Chen, J.; Zhang, J.; Saski, C.A.; Scheffler, B.E.; Stelly, D.M.; Hulse-Kemp, A.M.; Wan, Q.; Liu, B.; Liu, C.; Wang, S.; Pan, M.; Wang, Y.; Wang, D.; Ye, W.; Chang, L.; Zhang, W.; Song, Q.; Kirkbride, R.C.; Chen, X.; Dennis, E.; Llewellyn, D.J.; Peterson, D.G.; Thaxton, P.; Jones, D.C.; Wang, Q.; Xu, X.; Zhang, H.; Wu, H.; Zhou, L.; Mei, G.; Chen, S.; Tian, Y.; Xiang, D.; Li, X.; Ding, J.; Zuo, Q.; Tao, L.; Liu, Y.; Li, J.; Lin, Y.; Hui, Y.; Cao, Z.; Cai, C.; Zhu, X.; Jiang, Z.; Zhou, B.; Guo, W.; Li, R.; Chen, Z.J. Sequencing of allotetraploid cotton (Gossypium hirsutum L. acc. TM-1) provides a resource for fiber improvement. Nature Biotechnology 2015, 33, 531–537. [Google Scholar] [CrossRef]
- Kenan-Eichler, M.; Leshkowitz, D.; Tal, L.; Noor, E.; Melamed-Bessudo, C.; Feldman, M.; Levy, A.A. Wheat hybridization and polyploidization results in deregulation of small RNAs. Genetics 2011, 188, 263–272. [Google Scholar] [CrossRef] [PubMed]
- Hu, G.; Koh, J.; Yoo, M.J.; Grupp, K.; Chen, S.; Wendel, J.F. Proteomic profiling of developing cotton fibers from wild and domesticated Gossypium barbadense. New Phytologist 2013, 200, 570–582. [Google Scholar] [CrossRef] [PubMed]
- Mirouze, M.; Vitte, C. Transposable elements, a treasure trove to decipher epigenetic variation: insights from Arabidopsis and crop epigenomes. Journal of Experimental Botany 2014, 65, 2801–2812. [Google Scholar] [CrossRef]
- Vicient, C.M.; Casacuberta, J.M. Impact of transposable elements on polyploid plant genomes. Annals of Botany 2017, 120, 195–207. [Google Scholar] [CrossRef]
- Ha, M.; Lu, J.; Tian, L.; Ramachandran, V.; Kasschau, K.D.; Chapman, E.J.; Carrington, J.C.; Chen, X.; Wang, X.-J.; Chen, Z.J. Small RNAs serve as a genetic buffer against genomic shock in Arabidopsis interspecific hybrids and allopolyploids. Proceedings of the National Academy of Sciences 2009, 106, 17835–17840. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





