1. Introduction
The COVID-19 pandemic, caused by the SARS-CoV-2 virus, was a global health crisis. The virus spreads primarily through respiratory droplets and causes symptoms ranging from mild respiratory problems to severe pneumonia and acute respiratory distress syndrome [
1]. Vaccination plays a central role in the adaptive immunity and management of the COVID-19 pandemic [
2] by stimulating the immune system to mount a protective response, including both cellular and humoral immunity [
3].
The rs671 variant, a known East Asian-specific genetic diversity, is a missense mutation in aldehyde dehydrogenase 2 (
ALDH2). An amino acid substitution from glutamic acid (GAA) to lysine (AAA) at position 504 or 487 of the immature or mature ALDH2 enzyme (Glu504Lys or Glu487Lys) results in a collapsed three-dimensional structure [
4]. ALDH2 plays a crucial role in alcohol metabolism by oxidizing acetaldehyde, a toxic product of ethanol oxidation, into the less harmful substance, acetate [
5,
6,
7]. Individuals carrying the rs671 variant, found almost exclusively in East Asians, have substantially reduced ALDH2 enzymatic activity, leading to accumulation of acetaldehyde in the body when they consume alcohol [
8]. This accumulation causes vasodilation, resulting in skin flushing, increased heart rate, nausea, and other unpleasant symptoms [
9], a phenomenon known as the Asian flush [
10,
11]. The variant allele of rs671 is associated with various phenotypes [
12,
13,
14,
15,
16,
17], including alcohol-irrelevant disease risks, such as vasospastic angina [
18] and cognitive impairment [
19].
Based on our previous findings on the association between rs671 and immune function (i.e., rs671 influences basal T cell subpopulation [
20] and the efficacy of immune checkpoint inhibitors [
15]), we launched a prospective cohort study to investigate the immunogenicity of the COVID-19 mRNA vaccine and found an inverse association between the rs671 variant and IgG production [
21]. Subsequently, a web-based retrospective cohort study revealed lower susceptibility to COVID-19 among rs671 variant carriers [
22]. These seemingly contradictory findings suggest the involvement of cellular immunity which plays a greater role than humoral immunity in protecting against COVID-19 [
23] as shown by the successful recovery of patients with X-linked agammaglobulinemia and genetic deficiency of mature B lymphocytes from COVID-19 [
24].
To test our hypothesis, we designed an additional study using frozen blood cells from the abovementioned study on vaccine immunogenicity to evaluate SARS-CoV-2-specific T lymphocyte counts before and after COVID-19 vaccination.
2. Materials and Methods
2.1. Study Design and Participants
The current study used a database of frozen blood cells collected from participants of our prospective and observational study [
25]. To maximize statistical power with limited financial resources, rs671 minor allele carriers were prioritized over major allele carriers. The participants were employees and students of Saga University, Japan, aged 20 years or older, who voluntarily received the COVID-19 vaccine administered at Saga University. Pregnant women and those who had been infected or suspected of being infected with COVID-19 were excluded from the study.
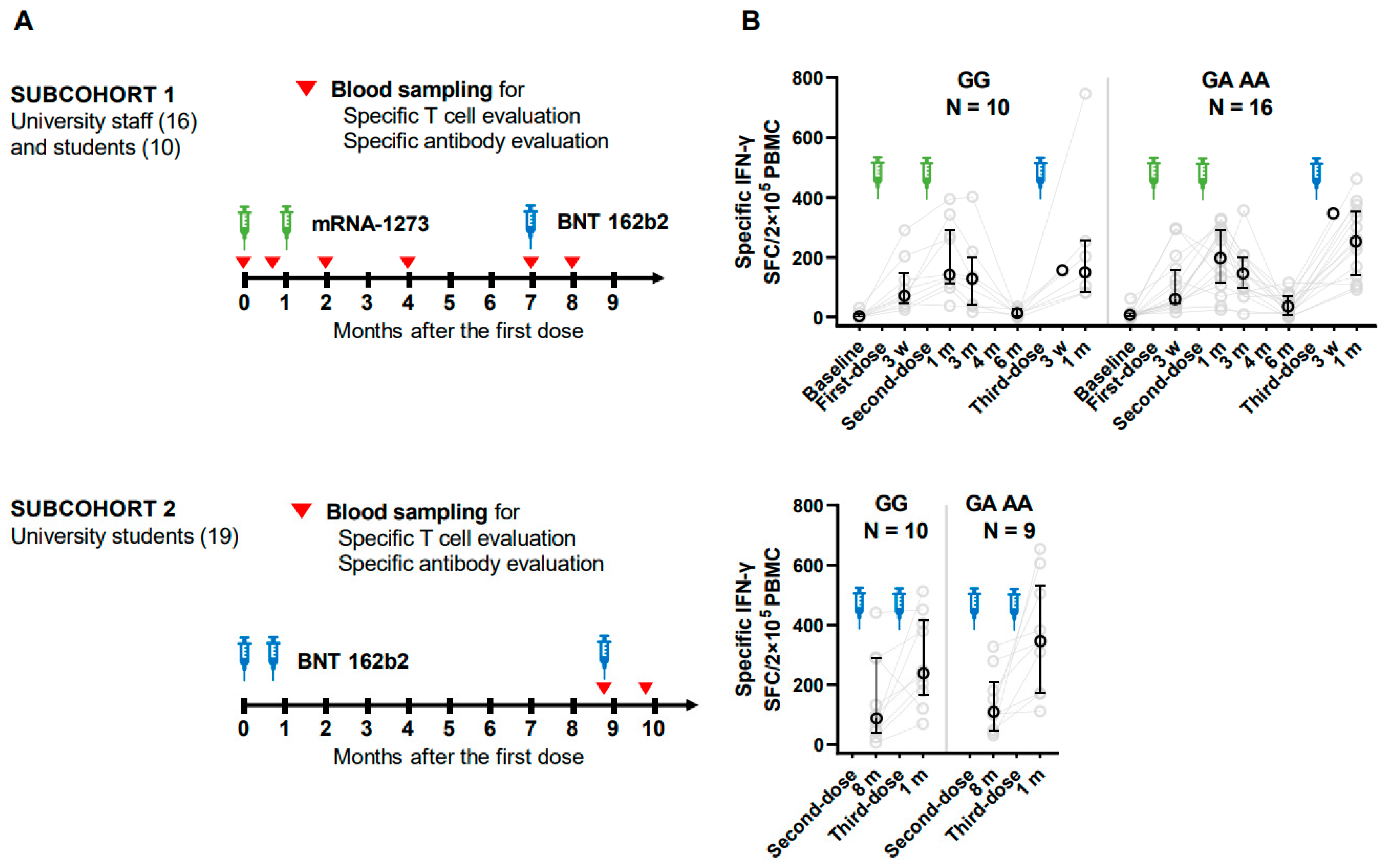
As shown in
Figure 1, university employees (N = 16) and some of the students (N = 10) completed two doses of mRNA-1273 (Moderna Inc., Cambridge, MA, USA/Takeda Pharmaceutical Co., Ltd., Tokyo, Japan) (100 µg) starting August 9–11, 2021, and a booster dose of BNT 162b2 (Pfizer Inc., New York, NY, USA/BioNTech SE, Mainz, Germany) (30 µg) (subcohort-1), and the rest of university students (N = 19) received three doses of BNT 162b2 (Pfizer) starting May 19, 2021 (subcohort-2). Vaccination history was recorded using a vaccination certificate issued by the local government of Saga Prefecture, Japan. mRNA-1273 and BNT 162b2 were completed at an interval of three or four weeks, and booster vaccination with BNT 162b2 was given six or eight months after the second dose for subcohorts 1 and 2, respectively. Blood samples were collected at baseline, three weeks after the first dose, one, three, and six months after the second dose (two, four, and seven months after the first dose), and one month after the third dose (8 months after the first dose) for subcohort 1. For subcohort 2; eight months after the second dose and one month after the second dose. Blood sample collection for subcohort 2 began before the booster dose because of a change in the budget plan.
2.2. Questionnaire
A self-administered questionnaire was used to obtain participants’ general information, as previously reported [
21]. Briefly, sex, date of birth, disease (chronic lung disease, heart disease, stroke, kidney disease, liver disease, blood disease, diabetes, immunodeficiency, malignancy, collagen disease, and allergic disease), treatment history, cigarette smoking, drinking, and exercise habits were recorded. Ethanol intake in the previous six months was estimated, adjusted per 60 kg of body weight, and then categorized into <1, <20, and ≥20 g/day. Exercise habit was chosen from no habit, <1 day/week, 1 to 3 days/week, and ≥3 days/week. The question “Do you feel psychological stress?” was asked to evaluate perceived stress on a five-point scale: no (0), mostly no (1), unsure (2), quite often (3), and yes (4).
2.3. Genotyping
The
ALDH2 rs671 genotype was determined using the DNA extracted from blood clots using a TaqMan® SNP genotyping assay system (Thermo Fisher Scientific, Waltham, MA, USA) as described previously [
25].
2.4. Separation of Peripheral Blood Mononuclear Cells (PBMCs)
Peripheral blood was drawn into EDTA-2K vacuum blood sampling tubes. To collect PMBCs, blood samples were subjected to gradient separation with Lymphoprep™ (Serumwerk Bernburg, Burnburg, Germany) within the day of sampling, according to manufacturer’s instruction. The cell suspension extracted at the border of the top and second layers was collected, washed with > 5 times the volume of saline solution, and dissolved in KM Banker II (Kohjin Bio Co., Ltd., Saitama, Japan), a culture medium suitable for the long-term cryopreservation of lymphocytes. A small portion of the PBMC suspension was subjected to cell counting after 0.4 w/v% trypan blue. Finally, the PBMC samples were gently cooled and stored at −80 °C until the enzyme-linked immunospot (ELISPOT) assay.
2.5. ELISPOT Assay
A human IFN-γ ELISPOT kit (Mabtech AB, Nacka Strand, Sweden) was used to evaluate cellular immunity against SARS-CoV-2 using lyophilized 315-peptide mixtures of SARS-CoV-2 spike glycoprotein, PepMixTM SARS-CoV-2 (JPT Peptide Technologies, Berlin, Germany), the original strain (PM-WCPV-S-2) as antigens. The peptide mixture was dissolved in DMSO at a concentration of 0.5 ug/eptide, and a DMSO addition group was provided as a negative control. Similarly, an NF-kB activator, phorbol myristate acetate, and ionomycin addition group were provided as positive controls. The assay was performed according to the manufacturer's instructions using an ImmunoSpot S6 VERSA spot counter (Cellular Technology Limited, Shaker Heights, Ohio, USA).
2.6. Statistical Analysis
SARS-CoV-2 infection during follow-up was detected by an increase in anti-SARS-CoV-2 S1 protein IgN levels or specific IgG, and T-cell counts without vaccination, and the corresponding observation points were removed after suspected infection. Spearman’s rank correlation ρ was calculated to examine non-linear association between rank or continuous variable, such as rs671 variant number, ethanol intake, ELISPOT counts, and IgG levels. Mixed models were used to compute the association between the rs671 variant allele numbers and the log-transformed specific T cell levels to account for repeated measurements and the random effect of the subpopulation (proc mixed by SAS9.4 TS Level 1M5 for Windows, SAS Institute, Cary, NC, USA). ELISPOT counts were converted into log values to approximate a normal distribution. Statistical significance was set at p < 0.05.
3. Results
3.1. Baseline Characters of the Participants
Table 1 shows the baseline characteristics of each subcohort, including 30 males and 15 females with confirmed rs671 genotypes: wild-type homozygous (GG type, N = 20), heterozygous (GA type, N = 17), and variant homozygous (AA type, N = 8). Daily ethanol intake (g/day) was low in the variant allele carriers (GA and AA) (ρ = −0.42, p < 0.01). The distribution of exercise habits, perceived stress, and allergic diseases did not differ among the three groups (p > 0.4, Fisher’s exact test). Steroid use was reported only by two participants of the GG-type.
3.2. Changes in Cellular Immune Response after Vaccination
As depicted in
Figure 1, we conducted an IFN-γ ELISPOT assay to quantify the number of T cells producing IFN-γ in response to SARS-CoV-2 specific antigens. The variant carriers tended to have higher counts than wild-type carriers at most time points. For example, at baseline in subcohort 1, wild and variant types had median counts of 0.5 and 5.5 spot- forming cells per 2×10
5 PBMC, respectively. One month after the second dose, the medians reached 139.5 and 196, respectively. Six months after the second dose, the number decreased to 36 in the wild-type group, showing an indistinguishable level from baseline (p = 0.10, Wilcoxon rank-sum test), whereas 116 in the variant-type group were higher than baseline (p < 0.01).
As shown in
Table 2, the effects of rs671 were estimated using multivariate mixed models with repeated measures. Model 1, including categorical time-points, vaccine type, age, sex, and the number of variant alleles, estimated a positive effect of the variant (β = 0.27 per allele, p = 0.01). Model 2, additionally including lifestyle, perceived stress, steroid use, and allergic disease history, gave a similar estimation (β = 0.3, p = 0.007).
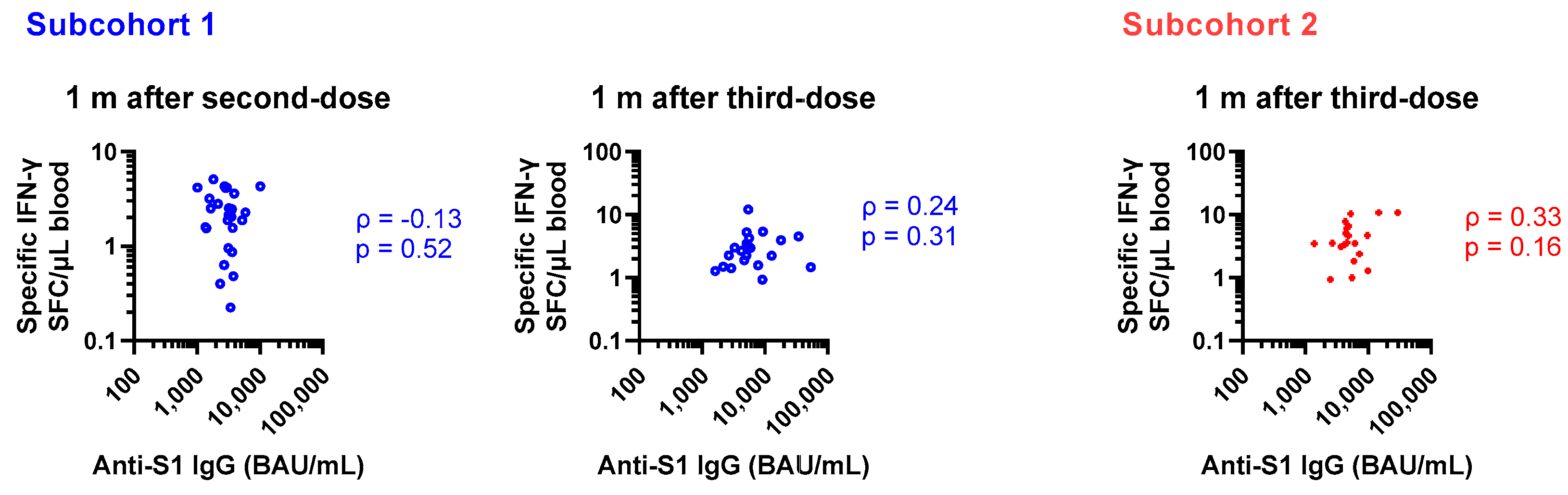
As reported previously, anti-SARS-CoV-2 spike 1 IgG (anti-S1 IgG) and the rs671 variant were inversely associated in the current cohort [
25]. Therefore, humoral and cellular immunity were correlated with rs671 in the opposite direction. As shown in
Figure 2, no rank correlation was observed between specific IgG and T cell responses obtained from the same blood samples. In a mixed model to estimate log-transformed specific T cell count by categorical time-points and log-transformed anti-S1 IgG considering repeated measures and subcohorts, anti-S1 IgG was not a predictor of specific T cell count (partial correlation coefficient = −0.05, p = 0.7, 183 observations).
4. Discussion
Confirming our hypothesis, we found for the first time a positive association between the rs671 variant and cellular immunity characterized by SARS-CoV-2 spike protein specific IFN-γ+ T cell count after COVID-19 vaccination in a prospective observation of the general population in Japan. Cellular immunogenicity remained detectable six months after the second dose only in the rs671 variant group. The humoral immune reaction measured using anti-S1 IgG was not associated with the cellular immunity observed in the current study.
Although ALDH2 is known as an alcohol-metabolizing enzyme, its essential role is the metabolism of endogenous aldehydes, such as formaldehyde and 4-hydroxy-2-nonenal (4-HNE), which are produced in the body as byproducts of normal cellular processes, such as spontaneous generation during one-carbon metabolism [
26], demethylation of sarcosine [
27,
28] for formaldehyde, and peroxidation of arachidonic acid [
29] for 4-HNE. The accumulation of these aldehydes due to low ALDH2 activity is a potential pathogen via adduct formation with DNA [
30,
31] and proteins [
32], resulting in carcinogenesis and functional alterations of the molecules [
33,
34].
Endogenous formaldehyde and 4-HNE inhibit mechanistic target of rapamycin (mTOR) signaling by modulating mTOR complex components [
35]. The mTOR protein kinase serves as a central regulator of cell growth, proliferation, metabolism, and survival. It exists in two distinct multiprotein complexes: mTOR complex 1 (mTORC1) and mTOR complex 2 (mTORC2). mTORC1 primarily regulates the processes related to cell growth, protein synthesis, and metabolism in response to nutrients, growth factors, and energy status. Inhibition of mTORC1 signaling by formaldehyde and 4-HNE may have implications for the maintenance of memory T cells, which are a small population of long-lived immune cells derived from effector T cells that provide rapid and specific responses upon re-exposure to previously encountered pathogens or antigens [
36]. In our observation, the variant type showed higher T cell counts than the wild type, however, multiple testing of rank correlation between the variant number and specific T cell count for each time-point gave only significance at six months after second dose (ρ = 0.5, p = 0.02). This finding is consistent with the hypothesis that accumulation of endogenous aldehydes inhibits mTOR signaling, providing acquired immunity via survival of memory T cells.
Another possible mechanism is the inhibition of T cell glycolysis by endogenous aldehydes, as observed in acetaldehyde-exposed T cells [
37]. By interfering with glycolytic activity, endogenous formaldehyde and 4-HNE may shift cellular metabolism towards alternative pathways such as fatty acid oxidation, the pentose phosphate pathway, and oxidative phosphorylation for energy production. Memory T cells favor these alternative pathways, which are crucial for providing energy, biosynthetic precursors, and maintaining the redox balance, ensuring the metabolic fitness, longevity, and functionality of memory T cells.
Although the COVID-19 vaccines in our study were designed for the original strain, PBMCs in the current study reacted with original, delta, and omicron variants (r squared = 0.71 and 0.82 for original vs delta and original vs omicron, respectively,
Figure S1). In contrast, humoral immunity is expected to escape, particularly for omicron variants [
38,
39]. Given this evidence, our hypothetical benefit of the rs671 variant on the survival of memory T cells may hold throughout the pandemic; the rs671 variant may promote COVID-19 protection via cellular immunity induced by the vaccine or exposure history to similar virus. The lower occurrence of COVID-19 and related hospitalizations in our previous report [
22] is thus explainable by the current findings; however, the reported strong effect in the early phase (hazard ratio 0.2) may not be fully explained, suggesting the presence of additional defense mechanisms, such as antimicrobial effects of metabolic intermediate aldehydes [
40]. For instance, we propose that the accumulation of formaldehyde in individuals with the rs671 variant may protect against COVID-19 through its bacteriostatic effect at the physiological level (100 µM) [
41]. This infection defense phenotype of the rs671 variant is a possible reason for its spread in East Asia following major lifestyle changes associated with the infectious hazards of rice cultivation [
42,
43,
44,
45].
To address the limitation of the validity of the current study owing to the small sample size, we attempted to maximize the detection power using statistical modeling to obtain all observation points. Targeting single cytokine production, IFN-γ, is another limitation, because multi-cytokine evaluation gives us background information, IL-4 for example, an indicator of antibody production systems [
46]. In addition, the use of PBMC without subpopulation sorting limits the specificity of findings because several types of immune cells can produce IFN-γ, including CD4+ T helper cells, CD8+ cytotoxic T cells, and natural killer T cells.
5. Conclusions
This study revealed for the first time a positive association between the rs671 variant and enhanced cellular immunity following COVID-19 vaccination. Reduced ALDH2 activity may favor the development and maintenance of COVID-19 specific memory T cells. The current findings support the previously reported lower susceptibility to COVID-19 infection in rs671 variant carriers. These findings may help in designing effective vaccination strategies; however, the underlying mechanisms require further investigation.
6. Patents
This section is not mandatory but may be added if there are patents resulting from the work reported in this manuscript.
Supplementary Materials
The following supporting information can be downloaded at the website of this paper posted on Preprints.org, Figure S1: Cross-reactivity to SARS-CoV-2 delta and omicron variants
Author Contributions
Conceptualization, A. M.; Study design, A.M. and M.H.; Investigation, M.H., A.M., C.I., T.S., T.F., G.Y., and M.T.; Data Curation, M.H. and A.M.; Statistical Analysis, A.M. and S.B.; Writing-Original Draft Preparation, S.B.; Writing-Review and Editing, All authors; Visualization, A.M. and S.B.; Supervision, A.M., M.H, K.K., Y.M., and Y.H. All authors have read and agreed to the published version of the manuscript.
Funding
This study was funded by a research grant for Research on Emerging and Re-emerging Infectious Diseases, Health and Labour Science Research Grants from the Ministry of Health, Labour and Welfare, Japan (R2-SHINKOGYOSEI-SHITEI-003 and 20HA2001), and the JSPS KAKENHI Grant (No. P21K19652) of the Japanese Ministry of Education, Culture, Sports, Science and Technology. The funding bodies had no role in the design of the study; collection, analysis, and interpretation of data; or in writing the manuscript.
Institutional Review Board Statement
This study was approved by the Research Review Committee and Human Genome Ethics Review Committee of the Faculty of Medicine, Saga University, Japan (approval numbers: R2-24, R2-44, R3-4, R3-9, and R3-39), and conducted in accordance with the Declaration of Helsinki.
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
The datasets generated and/or analyzed in the current study are not publicly available to protect the privacy of the participants; however, they are available from the corresponding author upon reasonable request.
Acknowledgments
We thank Mikako Horita, Kazuhiro Kawamoto, and Ayako Ajishi for their administrative and technical support. We also thank the students, healthcare workers, and employees of Saga University for their participation in the present study and for their donation of blood samples for the evaluation of cellular immune responses after vaccination.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Zhu, N., Zhang, D., Wang, W., Li, X., Yang, B., Song, J., Zhao, X., Huang, B., Shi, W., Lu, R.; et al. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N Engl J Med 2020, 382, 727-733. [CrossRef]
- Vabret, N., Britton, G.J., Gruber, C., Hegde, S., Kim, J., Kuksin, M., Levantovsky, R., Malle, L., Moreira, A., Park, M.D.; et al. Immunology of COVID-19: Current State of the Science. Immunity 2020, 52, 910-941. [CrossRef]
- Polack, F.P., Thomas, S.J., Kitchin, N., Absalon, J., Gurtman, A., Lockhart, S., Perez, J.L., Pérez Marc, G., Moreira, E.D., Zerbini, C.; et al. Safety and Efficacy of the BNT162b2 mRNA Covid-19 Vaccine. N Engl J Med 2020, 383, 2603-2615. [CrossRef]
- Farres, J.; Wang, X.; Takahashi, K.; Cunningham, S.J.; Wang, T.T.; Weiner, H. Effects of changing glutamate 487 to lysine in rat and human liver mitochondrial aldehyde dehydrogenase. A model to study human (Oriental type) class 2 aldehyde dehydrogenase. J Biol Chem 1994, 269, 13854-13860. [CrossRef]
- Wang, M.F.; Han, C.L.; Yin, S.J. Substrate specificity of human and yeast aldehyde dehydrogenases. Chem Biol Interact 2009, 178, 36-39. [CrossRef]
- Peng, G.S.; Chen, Y.C.; Tsao, T.P.; Wang, M.F.; Yin, S.J. Pharmacokinetic and pharmacodynamic basis for partial protection against alcoholism in Asians, heterozygous for the variant ALDH2*2 gene allele. Pharmacogenet Genomics 2007, 17, 845-855. [CrossRef]
- Isse, T.; Matsuno, K.; Oyama, T.; Kitagawa, K.; Kawamoto, T. Aldehyde dehydrogenase 2 gene targeting mouse lacking enzyme activity shows high acetaldehyde level in blood, brain, and liver after ethanol gavages. Alcohol Clin Exp Res 2005, 29, 1959-1964. [CrossRef]
- Chen, C.H.; Ferreira, J.C.; Gross, E.R.; Mochly-Rosen, D. Targeting aldehyde dehydrogenase 2: new therapeutic opportunities. Physiol Rev 2014, 94, 1-34. [CrossRef]
- Moh, I.; Simon, D.; Gross, E.R. The Alcohol Flush Response. Graph Med Rev 2024, 4,. [CrossRef]
- Brooks, P.J.; Enoch, M.A.; Goldman, D.; Li, T.K.; Yokoyama, A. The alcohol flushing response: an unrecognized risk factor for esophageal cancer from alcohol consumption. PLoS Med 2009, 6, e50. [CrossRef]
- Hishimoto, A.; Fukutake, M.; Mouri, K.; Nagasaki, Y.; Asano, M.; Ueno, Y.; Nishiguchi, N.; Shirakawa, O. Alcohol and aldehyde dehydrogenase polymorphisms and risk for suicide: a preliminary observation in the Japanese male population. Genes Brain Behav 2010, 9, 498-502. [CrossRef]
- Matsumoto, A. [Fundamental Properties of Aldehyde Dehydrogenase 2 (ALDH2) and the Importance of the ALDH2 Polymorphism]. Nihon Eiseigaku Zasshi 2016, 71, 55-68. [CrossRef]
- Matsumoto, A. [Importance of an Aldehyde Dehydrogenase 2 Polymorphism in Preventive Medicine]. Nihon Eiseigaku Zasshi 2018, 73, 9-20. [CrossRef]
- Matsumoto, A. The bidirectional effect of defective ALDH2 polymorphism and disease prevention. Adv Exp Med Biol 2019, 1193, 69-87. [CrossRef]
- Matsumoto, A.; Nakashima, C.; Kimura, S.; Sueoka, E.; Aragane, N. ALDH2 polymorphism rs671 is a predictor of PD-1/PD-L1 inhibitor efficacy against thoracic malignancies. BMC Cancer 2021, 21, 584. [CrossRef]
- Matsumoto, A.; Thompson, D.C.; Chen, Y.; Kitagawa, K.; Vasiliou, V. Roles of defective ALDH2 polymorphism on liver protection and cancer development. Environ Health Prev Med 2016, 21, 395-402. [CrossRef]
- Hayashida, H.; Matsumoto, A.; Nanri, H.; Nishida, Y.; Takagi, Y.; Hara, M. ALDH2 rs671 variant allele is associated with higher energy intake in middle-aged and elderly Japanese who routinely consume alcohol. Environ Health Prev Med 2023, 28, 29. [CrossRef]
- Mizuno, Y., Harada, E., Morita, S., Kinoshita, K., Hayashida, M., Shono, M., Morikawa, Y., Murohara, T., Nakayama, M., Yoshimura, M.; et al. East asian variant of aldehyde dehydrogenase 2 is associated with coronary spastic angina: possible roles of reactive aldehydes and implications of alcohol flushing syndrome. Circulation 2015, 131, 1665-1673. [CrossRef]
- Yu, R.L.; Tan, C.H.; Lu, Y.C.; Wu, R.M. Aldehyde dehydrogenase 2 is associated with cognitive functions in patients with Parkinson's disease. Sci Rep 2016, 6, 30424.
- Matsumoto, A., Terashima, Y., Toda, E., Akao, C., Miyake, Y., Matsuo, K., Shimanoe, C., Nishida, Y., Hara, M., Tanaka, K.; et al. Reduced T cell immunity in variant ALDH2 allele carriers. in The 90th Annual Meeting of the Japanese Society for Hygiene. 2020. Iwate, Japan.
- Matsumoto, A.; Hara, M.; Ashenagar, M.S.; Tokiya, M.; Sawada, T.; Iwasaka, C.; Furukawa, T.; Kitagawa, K.; Miyake, Y.; Hirota, Y. Variant Allele of ALDH2, rs671, Associates with Attenuated Post-Vaccination Response in Anti-SARS-CoV-2 Spike Protein IgG: A Prospective Study in the Japanese General Population. Vaccines (Basel) 2022, 10,. [CrossRef]
- Takashima, S.; Tokiya, M.; Izui, K.; Miyamoto, H.; Matsumoto, A. Asian flush is a potential protective factor against COVID-19: a web-based retrospective survey in Japan. Environ Health Prev Med 2024, 29, 14. [CrossRef]
- Rydyznski Moderbacher, C., Ramirez, S.I., Dan, J.M., Grifoni, A., Hastie, K.M., Weiskopf, D., Belanger, S., Abbott, R.K., Kim, C., Choi, J.; et al. Antigen-Specific Adaptive Immunity to SARS-CoV-2 in Acute COVID-19 and Associations with Age and Disease Severity. Cell 2020, 183, 996-1012.e1019. [CrossRef]
- Devassikutty, F.M.; Jain, A.; Edavazhippurath, A.; Joseph, M.C.; Peedikayil, M.M.T.; Scaria, V.; Sandhya, P.; Govindaraj, G.M. X-Linked Agammaglobulinemia and COVID-19: Two Case Reports and Review of Literature. Pediatr Allergy Immunol Pulmonol 2021, 34, 115-118. [CrossRef]
- Matsumoto, A.; Hara, M.; Ashenagar, M.S.; Tokiya, M.; Sawada, T.; Iwasaka, C.; Furukawa, T.; Kitagawa, K.; Miyake, Y.; Hirota, Y. Variant allele of ALDH2, rs671, associates with attenuated post-vaccination response in anti-SARS-CoV-2 spike protein IgG: A prospective study in the Japanese general population. Vaccines (Basel) 2022, 10, 1035. [CrossRef]
- Burgos-Barragan, G., Wit, N., Meiser, J., Dingler, F.A., Pietzke, M., Mulderrig, L., Pontel, L.B., Rosado, I.V., Brewer, T.F., Cordell, R.L.; et al. Mammals divert endogenous genotoxic formaldehyde into one-carbon metabolism. Nature 2017, 548, 549-554.
- Wagner, M.A.; Jorns, M.S. Monomeric sarcosine oxidase: 2. Kinetic studies with sarcosine, alternate substrates, and a substrate analogue. Biochemistry 2000, 39, 8825-8829. [CrossRef]
- Ai, L., Tan, T., Tang, Y., Yang, J., Cui, D., Wang, R., Wang, A., Fei, X., Di, Y., Wang, X.; et al. Endogenous formaldehyde is a memory-related molecule in mice and humans. Commun Biol 2019, 2, 446.
- Shoeb, M.; Ansari, N.H.; Srivastava, S.K.; Ramana, K.V. 4-Hydroxynonenal in the pathogenesis and progression of human diseases. Curr Med Chem 2014, 21, 230-237.
- Yukawa, Y., Ohashi, S., Amanuma, Y., Nakai, Y., Tsurumaki, M., Kikuchi, O., Miyamoto, S., Oyama, T., Kawamoto, T., Chiba, T.; et al. Impairment of aldehyde dehydrogenase 2 increases accumulation of acetaldehyde-derived DNA damage in the esophagus after ethanol ingestion. Am J Cancer Res 2014, 4, 279-284.
- Mulderrig, L., Garaycoechea, J.I., Tuong, Z.K., Millington, C.L., Dingler, F.A., Ferdinand, J.R., Gaul, L., Tadross, J.A., Arends, M.J., O'Rahilly, S.; et al. Aldehyde-driven transcriptional stress triggers an anorexic DNA damage response. Nature 2021, 600, 158-163.
- Traverso, N.; Menini, S.; Maineri, E.P.; Patriarca, S.; Odetti, P.; Cottalasso, D.; Marinari, U.M.; Pronzato, M.A. Malondialdehyde, a lipoperoxidation-derived aldehyde, can bring about secondary oxidative damage to proteins. J Gerontol A Biol Sci Med Sci 2004, 59, B890-895. [CrossRef]
- Yang, M.; Zhang, Y.; Ren, J. ALDH2 Polymorphism and Ethanol Consumption: A Genetic-Environmental Interaction in Carcinogenesis. Adv Exp Med Biol 2019, 1193, 229-236. [CrossRef]
- Mali, V.R.; Ning, R.; Chen, J.; Yang, X.P.; Xu, J.; Palaniyandi, S.S. Impairment of aldehyde dehydrogenase-2 by 4-hydroxy-2-nonenal adduct formation and cardiomyocyte hypertrophy in mice fed a high-fat diet and injected with low-dose streptozotocin. Exp Biol Med (Maywood) 2014, 239, 610-618. [CrossRef]
- Zhang, H.; Forman, H.J. 4-hydroxynonenal-mediated signaling and aging. Free Radic Biol Med 2017, 111, 219-225. [CrossRef]
- Inoue, S.; Niikura, M.; Mineo, S.; Kobayashi, F. Roles of IFN-γ and γδ T Cells in Protective Immunity Against Blood-Stage Malaria. Front Immunol 2013, 4, 258. [CrossRef]
- Cao, J.; Liao, S.; Zeng, F.; Liao, Q.; Luo, G.; Zhou, Y. Effects of altered glycolysis levels on CD8(+) T cell activation and function. Cell Death Dis 2023, 14, 407.
- Cao, Y., Wang, J., Jian, F., Xiao, T., Song, W., Yisimayi, A., Huang, W., Li, Q., Wang, P., An, R.; et al. Omicron escapes the majority of existing SARS-CoV-2 neutralizing antibodies. Nature 2022, 602, 657-663.
- Planas, D., Saunders, N., Maes, P., Guivel-Benhassine, F., Planchais, C., Buchrieser, J., Bolland, W.H., Porrot, F., Staropoli, I., Lemoine, F.; et al. Considerable escape of SARS-CoV-2 Omicron to antibody neutralization. Nature 2022, 602, 671-675.
- Darwin, K.H.; Stanley, S.A. The aldehyde hypothesis: metabolic intermediates as antimicrobial effectors. Open Biol 2022, 12, 220010. [CrossRef]
- Berry, S.B., Espich, S., Thuong, N.T.T., Chang, X., Dorajoo, R., Khor, C.C., Heng, C.K., Yuan, J.M., Fox, D., Anaya-Sanchez, A.; et al. Disruption of aldehyde dehydrogenase 2 protects against bacterial infection. bioRxiv 2023,. [CrossRef]
- Oota, H., Pakstis, A.J., Bonne-Tamir, B., Goldman, D., Grigorenko, E., Kajuna, S.L., Karoma, N.J., Kungulilo, S., Lu, R.B., Odunsi, K.; et al. The evolution and population genetics of the ALDH2 locus: random genetic drift, selection, and low levels of recombination. Ann Hum Genet 2004, 68, 93-109. [CrossRef]
- Deiana, G.; Sun, R.; Huang, J.; Napolioni, V.; Ciccocioppo, R. Infection burden and ALDH2 rs671, East Asian genetic diversity: A reply. Alcohol Clin Exp Res (Hoboken) 2024,. [CrossRef]
- Matsumoto, A. Infection burden and ALDH2 rs671: East Asian genetic diversity. Alcohol, Clinical and Experimental Research 2024,. [CrossRef]
- Deiana, G.; Sun, R.; Huang, J.; Napolioni, V.; Ciccocioppo, R. Contribution of infectious diseases to the selection of ADH1B and ALDH2 gene variants in Asian populations. Alcohol Clin Exp Res (Hoboken) 2024,. [CrossRef]
- Liu, M., Yang, Z., Wu, Q., Yang, Y., Zhao, D., Cheng, Q., Li, Y., Liu, G., Zhao, C., Pan, J.; et al. IL-4-secreting CD40L(+) MAIT cells support antibody production in the peripheral blood of Heonch-Schönlein purpura patients. Inflamm Res 2024, 73, 35-46. [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).