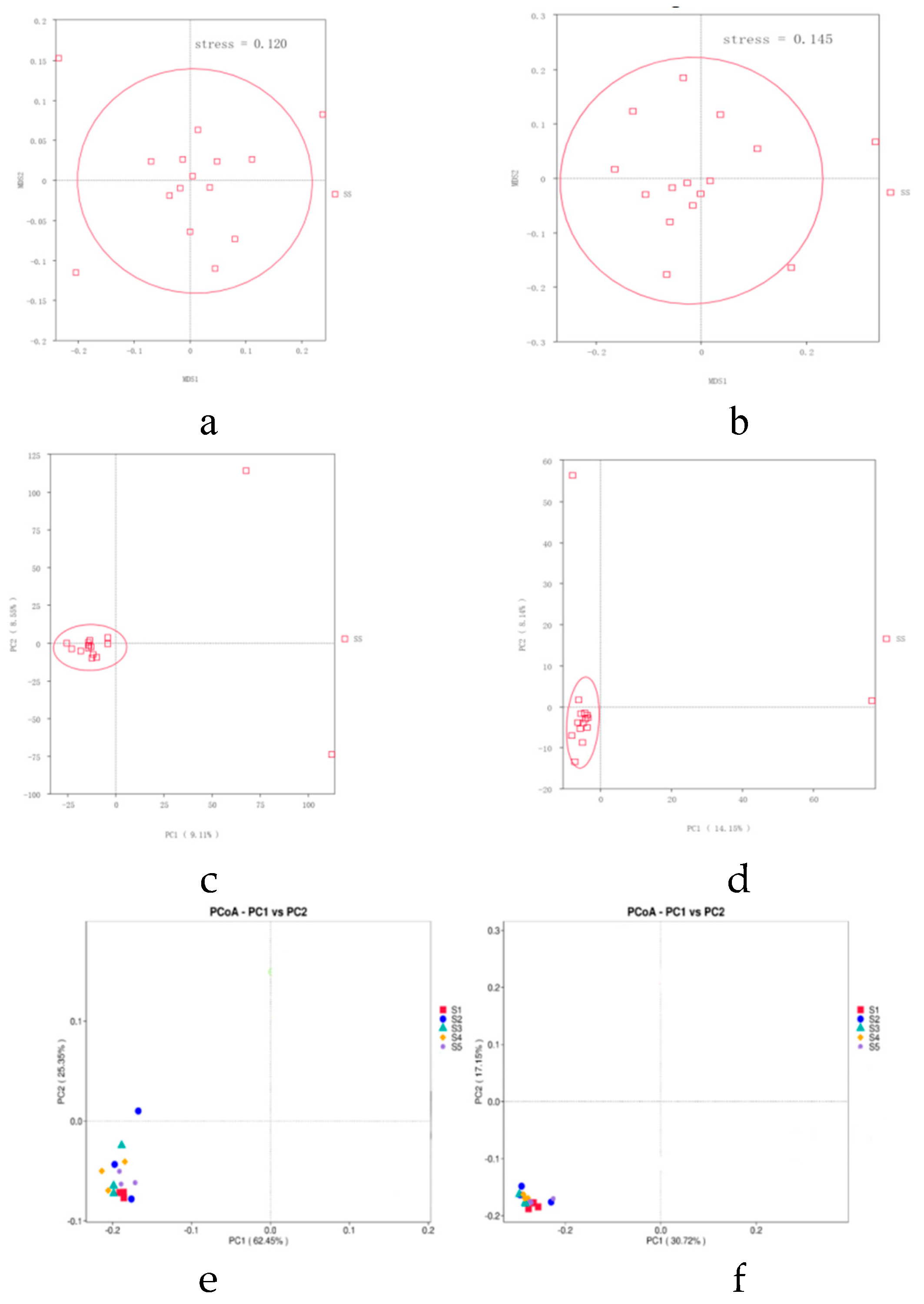
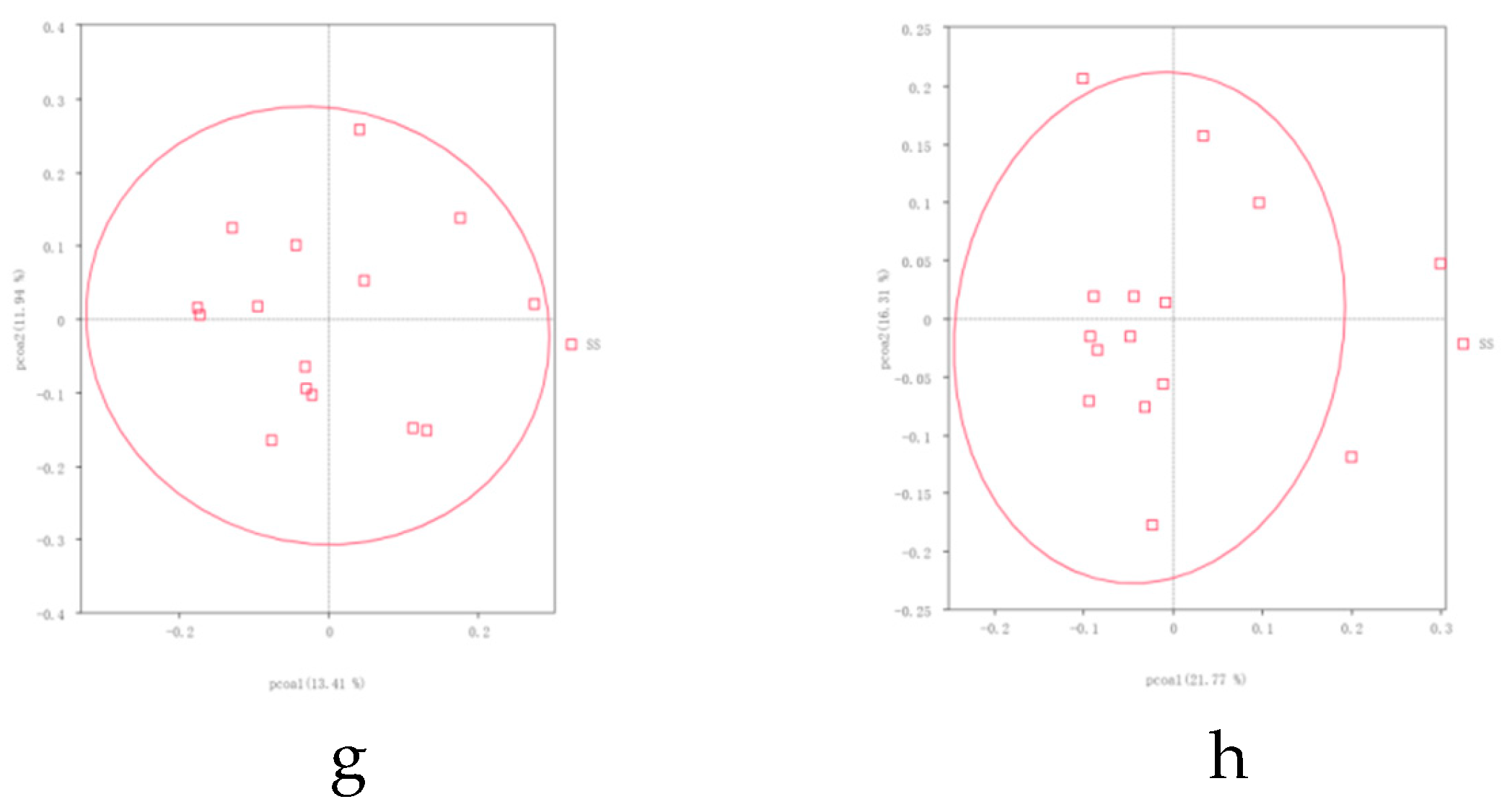
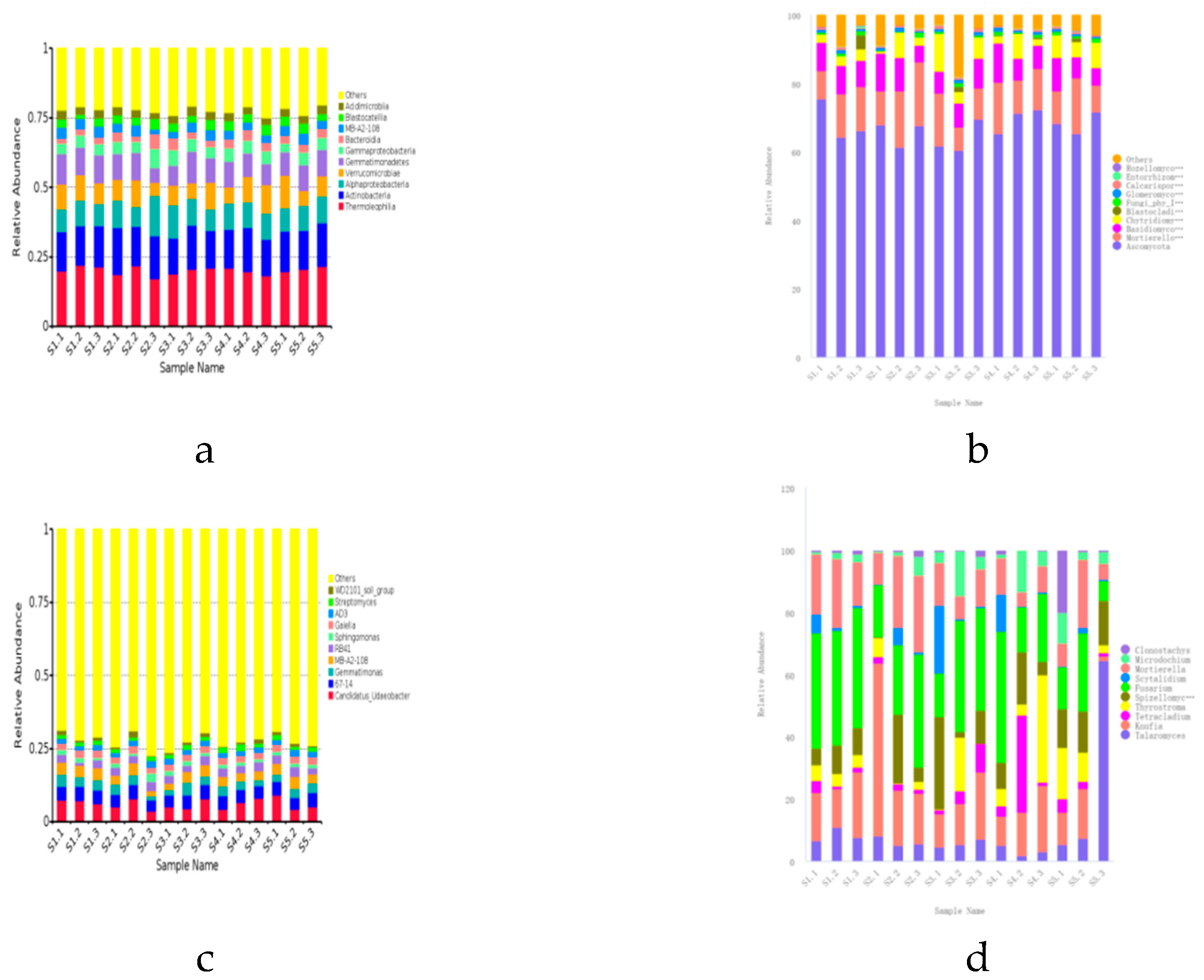
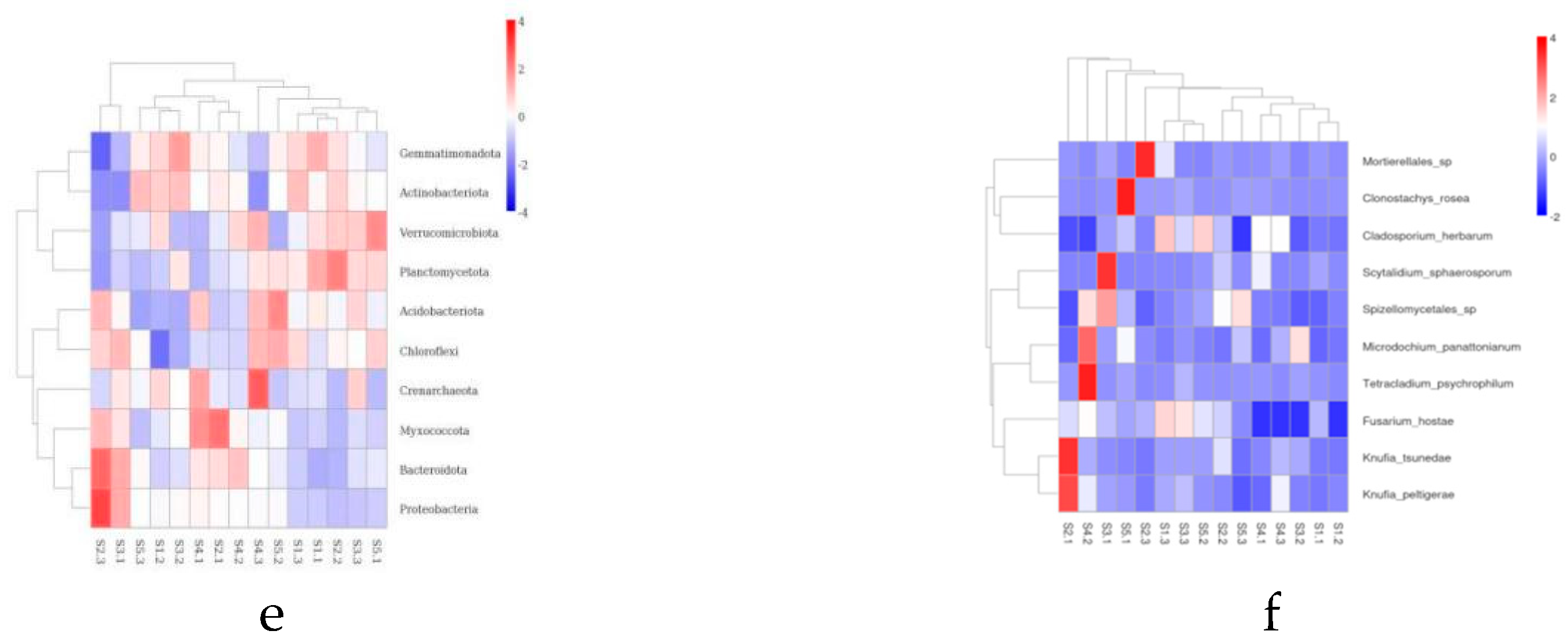
The microbial community structure in the rhizosphere soil is crucial for
H. arenarium (L.) Moench. to obtain nutrients. So, we studied and analyzed the microbial community structure in the rhizosphere soil as well as the screening of growth promotion and antagonistic bacteria. Then we found that in the rhizosphere soil environment, bacteria had a higher total number of species, community diversity and abundance than fungi, indicating that the root environment was still bacteria-dominated, which was consistent with the view that soil microorganisms were mainly bacteria[
19,
20], higher bacterial diversity indicates higher soil resistance[
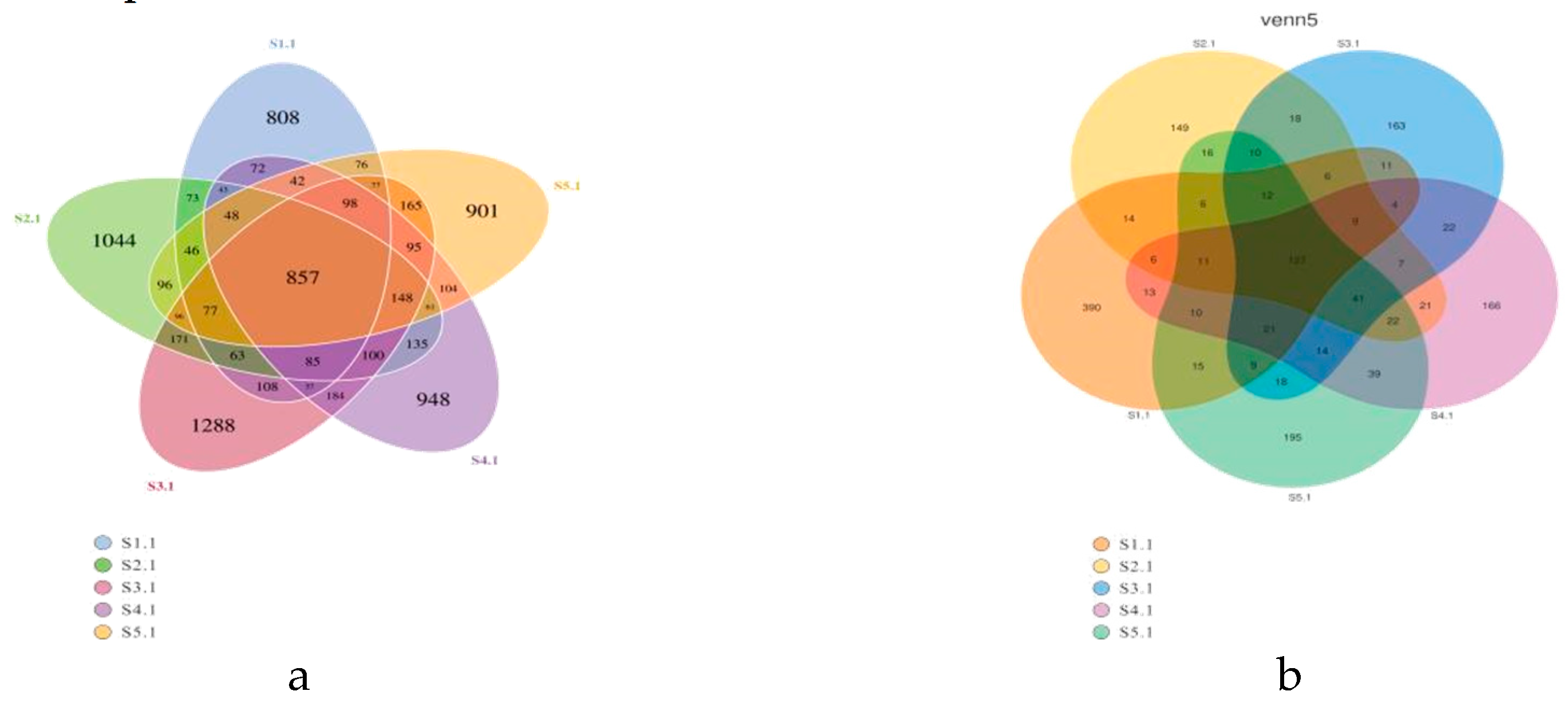
21]. Based on the analysis results of OTUs, there are eight main bacterial phyla in the rhizosphere microbial community (relative abundance greater than 1%), among which
Actinobacteriota and
Proteobacteria are the dominant phyla, especially the relative abundance of
Proteobacteria accounts for more than 40% of all phyla.
Proteobacteria are enriched in the rhizosphere soil of alfalfa[
22], strawberry[
23], potato[
24] and asparagus[
25], indicating that
Proteobacteria can adapt to the rhizosphere environment of various plants and are the main components of rhizosphere bacteria. However, the rhizosphere soils of different plants showed species characteristics for microbial diversity. The rhizosphere soils of the plant grew in mounds, semi-dunes, slopes of wet saline soil, gravel soil, sand dunes, grassland and under pine forests at an altitude of 900 m-2,400 m, with pH ranging from 6.68 to 7.06, mostly in neutral and slightly acidic soil. Based on this special growth environment,
Actinobacteriota,
Proteobacteria,
Acidobacteriota,
Verrucomicrobiota, and
Gemmatimonadota are the main dominant phyla of
H. arenarium (L.) Moench.. Though, soil fungi is fewer than bacteria, it is critical to maintaining ecosystems[
26]. There are four major fungal phyla (relative abundance >1%) in the rhizosphere microbial community of
H. arenarium (L.) Moench., among which,
Ascomycota and
Mortierellomycota,
Basidiomycota are dominant phyla, and especially
Ascomycota is the absolute dominant phyla, with the highest relative abundance accounting for more than 70% of all the phyla, which is is consistent with the study of Li Qingshan et al.[
27].
Ascomycota is the main decomposers of difficult organic matter such as plant litters and lignin in soil. Ren Neifan et al. isolated a strain of
Penicillium cinerea BIBA-G563 from the root of healthy Ruthenica Ruthenica, which belongs to
Ascomycota and has a control effect of 75.4% on the root rot of Ruthenica Ruthenica[
28].
H. arenarium (L.) Moench. is a root-breeding plant, and its root system provides a large number of attachment points for rhizosphere microorganisms, thus increasing the abundance and diversity of rhizosphere microorganisms and facilitating the enrichment of dominant flora.
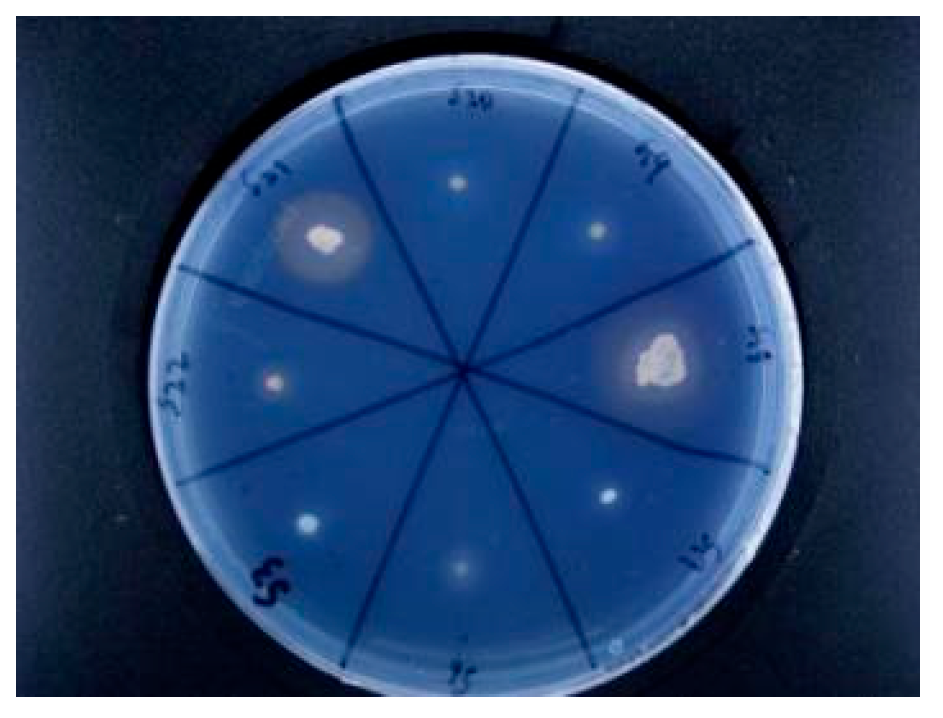
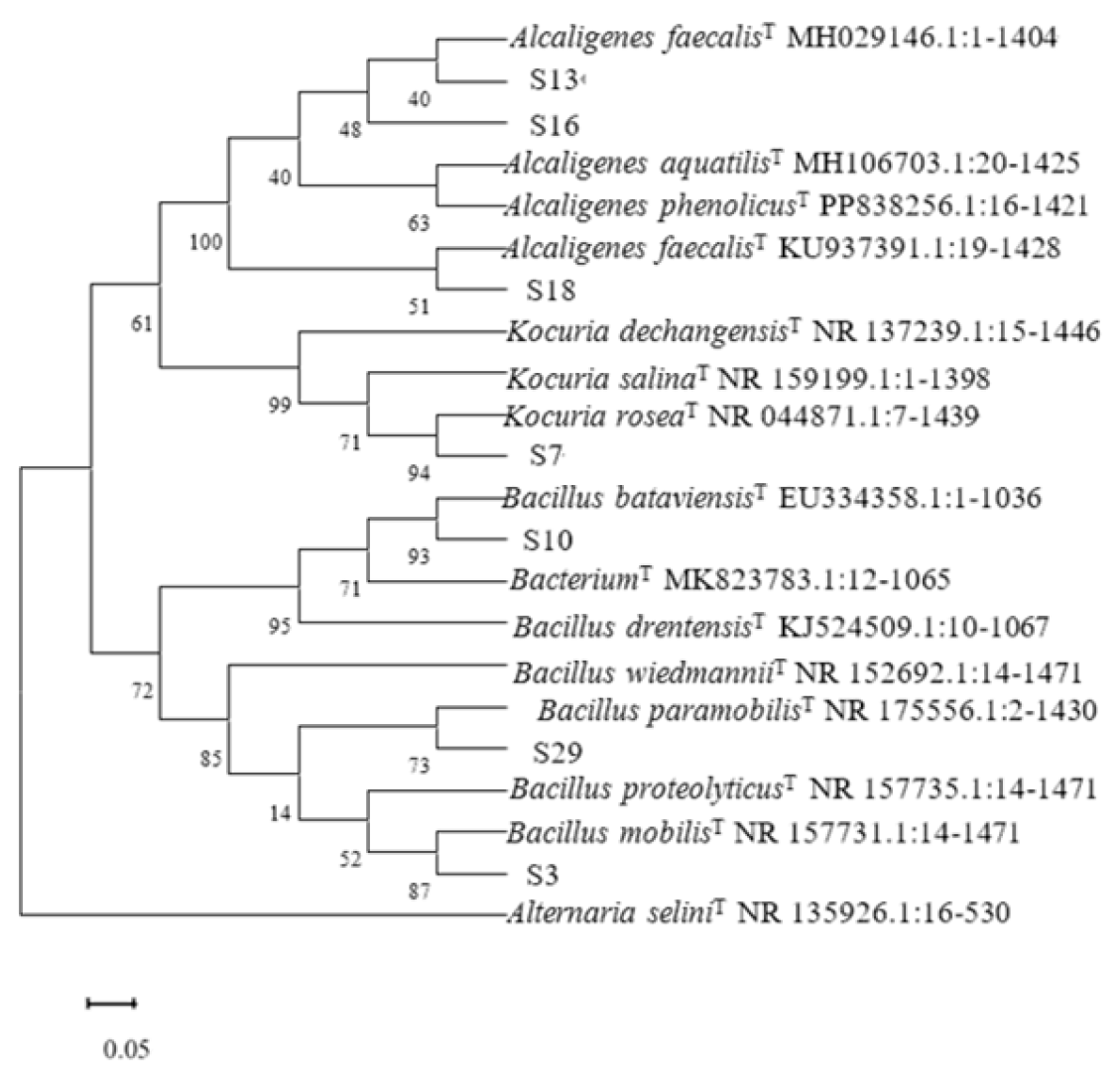
Rhizosphere growth-promoting bacteria promote plant nutrient absorption through biological nitrogen fixation, phosphorus solubilization, potassium solubilization, secretion of plant hormones and production of iron carriers. In this study, nine growth-promoting bacteria were screened, which belonged to the genus
Bacillus under
Firmicutes, the genus
Alcaligenes under
Proteobacteria, and the genus
Kocuria under
Actinomyces, of which five belong to the genus
Bacillus, three belong to the genus
Alcaligenes, and one belong to the genus
Kocuria. Most of the growth-promoting bacteria isolated by previous authors were
Bacillus and
Pseudomonas, S16 has the strongest ability to detoxify phosphorus and produce iron carriers and belongs to
Alcaligenes; S10 has the strongest ability to fix nitrogen and belongs to
Bacillus. Nitrogen-fixing bacteria reduce nitrogen to ammonia to increase the N content in the soil; phosphorus-solubilizing bacteria convert insoluble phosphorus in the soil into phosphorus that can be easily absorbed by plants to increase the effective phosphorus content in the soil; iron-carrier-producing bacterial strains promote iron nutrition in plants through the production and utilization of iron carriers[
29,
30,
31]. In the later stage of this study, the optimal growth-promoting bacteria will be screened by combining the strain with the growth-promoting experiment, which will provide an experimental basis for the development and promotion of microbial fertilizer for the mulberry, provide a research basis for the protection of wild resources of the mulberry and the development and utilization of rhizosphere microorganisms, and provide a strong support for the future application of the mulberry in food and medicine.
In the later stage of this study, the optimal growth-promoting bacteria will be screened by combining the strain with the growth-promoting experiment, which will provide an experimental basis for the development and promotion of microbial fertilizer for the H. arenarium (L.) Moench., provide a research basis for the protection of wild resources of the H. arenarium (L.) Moench. and the development and utilization of rhizosphere microorganisms, and provide a strong support for the future application of the H. arenarium (L.) Moench..