Submitted:
26 August 2024
Posted:
26 August 2024
You are already at the latest version
Abstract
Keywords:
Introduction
miRNAs, ROS and Melanoma
Oxidative stress Signalling Pathways and miRNA Expression in Melanoma
MITF Signalling Pathway
HIF-1α Signalling Pathway
MAPK Signalling Pathway
PI3K- AKT Signalling Pathway
Wnt Signalling Pathway
NRF2/KEAP1 Wnt Signalling Pathway
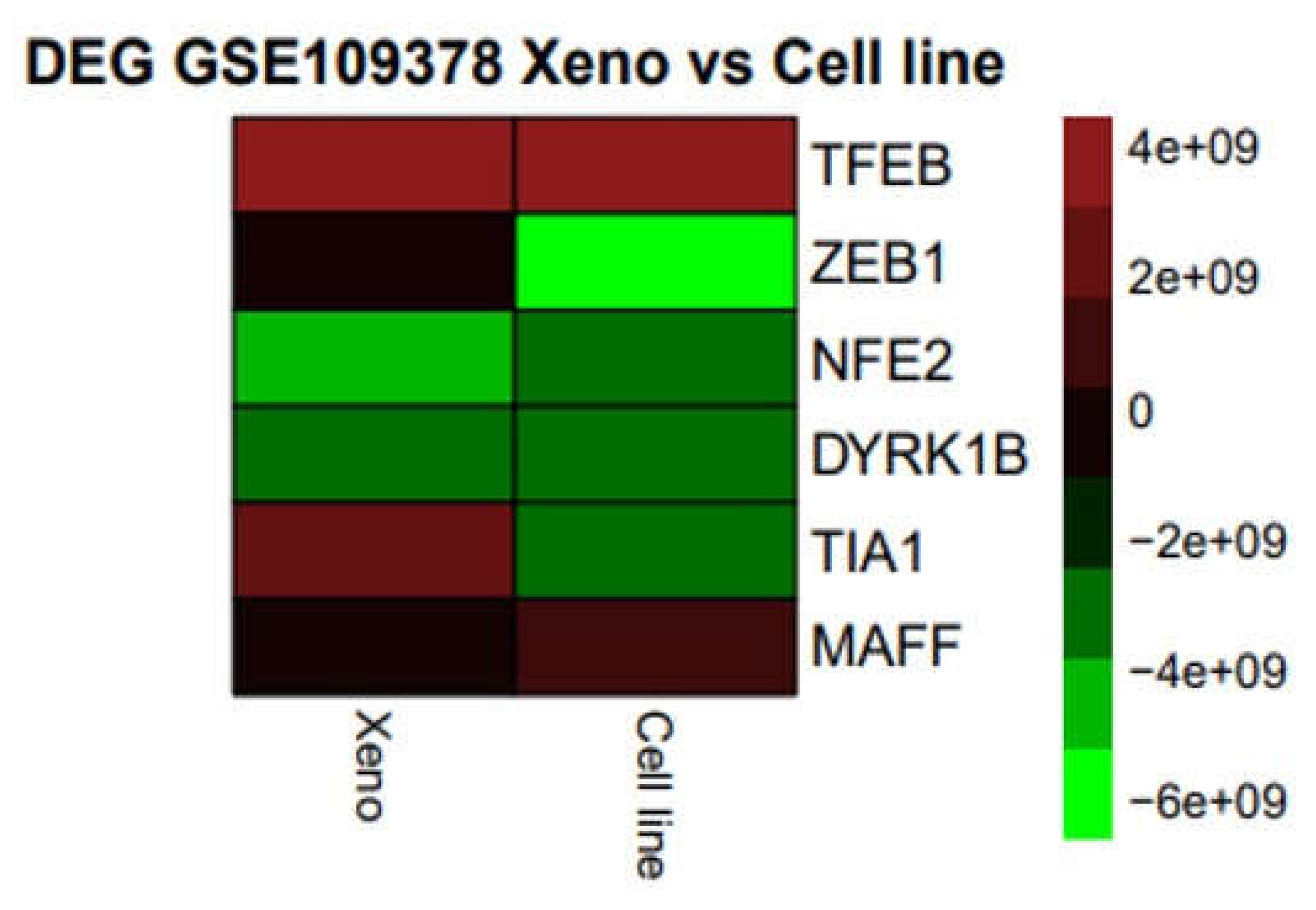
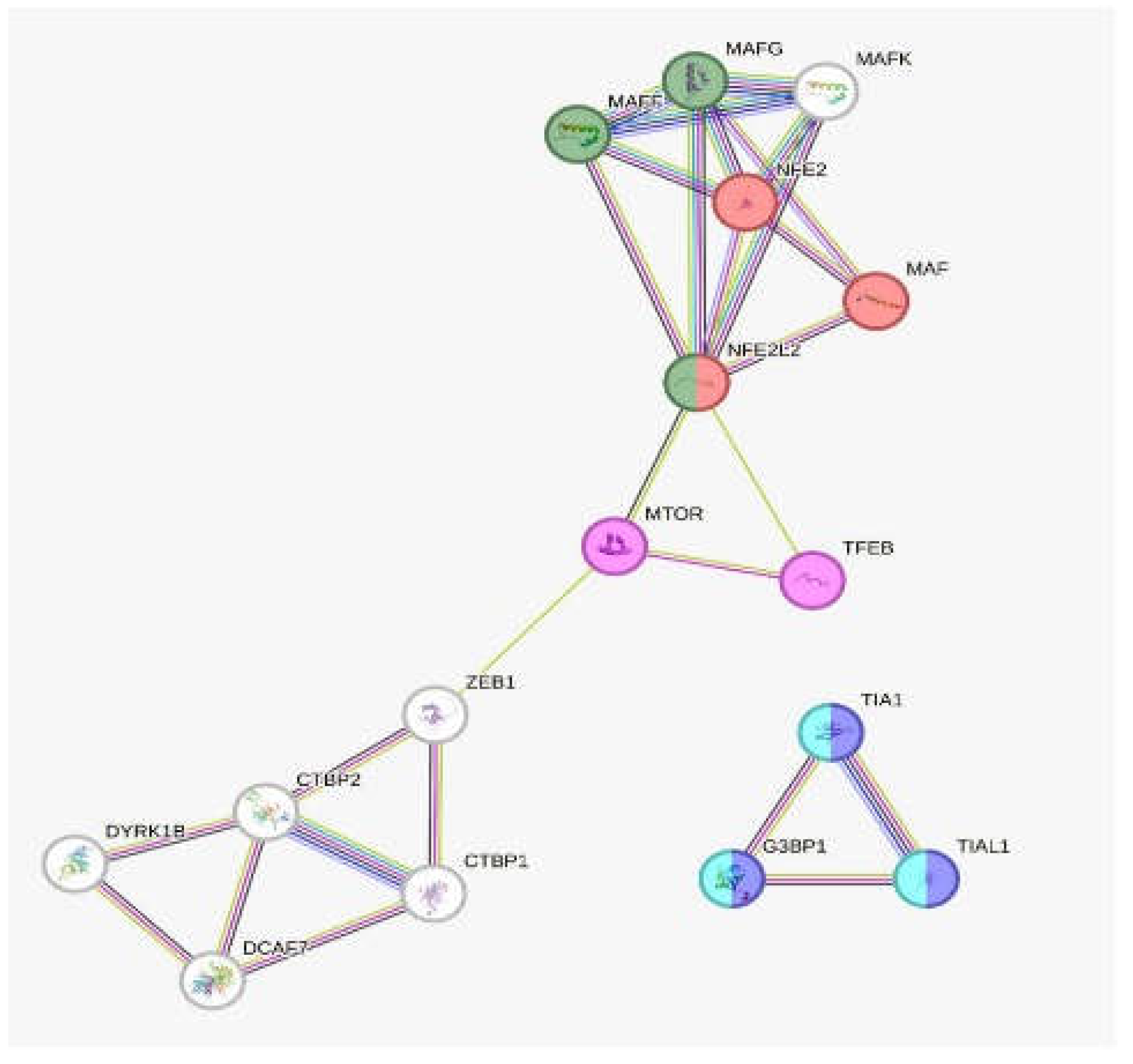
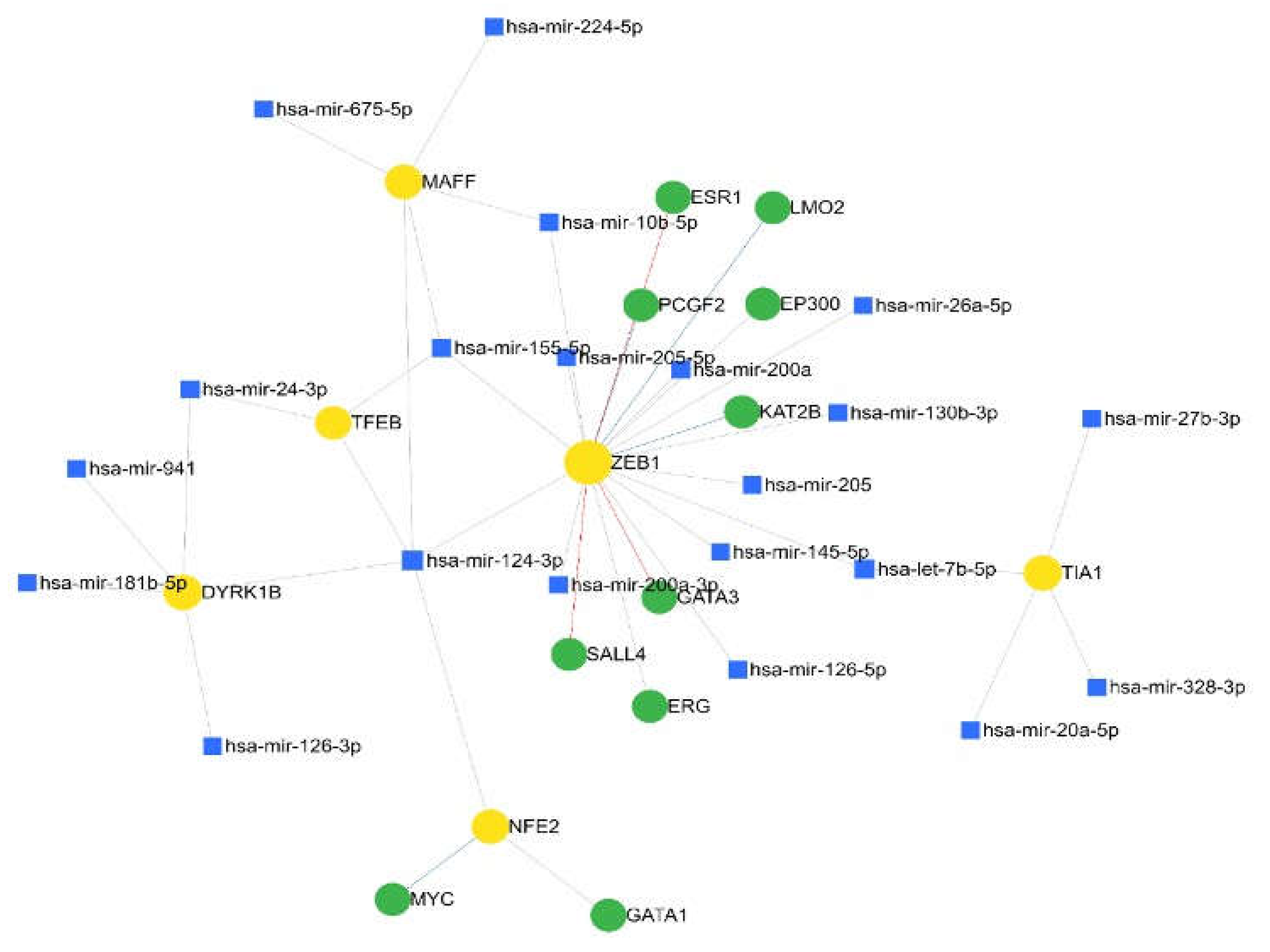
Expression Networks and Regulation under Oxidative Stress-Induced Melanoma
Conclusion
Acknowledgments
Conflicts of Interest
References
- Piskounova E, Agathocleous M, Murphy MM, Hu Z, Huddlestun SE, Zhao Z, et al. Oxidative stress inhibits distant metastasis by human melanoma cells. Nature 2015;527:186–91. [CrossRef]
- Ralph SJ, Rodríguez-Enríquez S, Neuzil J, Moreno-Sánchez R. Bioenergetic pathways in tumor mitochondria as targets for cancer therapy and the importance of the ROS-induced apoptotic trigger. Mol Aspects Med 2010;31:29–59. [CrossRef]
- Venza I, Venza M, Visalli M, Lentini G, Teti D, d’Alcontres FS. ROS as Regulators of Cellular Processes in Melanoma. Oxid Med Cell Longev 2021;2021:1–19. [CrossRef]
- Pozzobon FC, Acosta ÁE, Castillo JS. Cáncer de piel en Colombia: cifras del Instituto Nacional de Cancerología. Revista de La Asociación Colombiana de Dermatología y Cirugía Dermatológica 2018;26. [CrossRef]
- IARC, Internacional Agency for Research on cancer. Cancer Today 2020. https://gco.iarc.fr/today/online-analysis-table?v=2020&mode=cancer&mode_population=continents&population=900&populations=904&key=asr&sex=0&cancer=39&type=0&statistic=5&prevalence=0&population_group=0&ages_group%5B%5D=0&ages_group%5B%5D=17&group_cancer=1&include_nmsc=1&include_nmsc_other=1#collapse-group-0-1 (accessed on 28 June 2023).
- Tormo, R. Estudio de los perfiles de expresión de micrornas en líneas celulares de cancer de mama triple negativo tratadas con doxorubicina: implicación de la familia mir-449 2017.
- Garzon R, Calin GA, Croce CM. MicroRNAs in Cancer. Annu Rev Med 2009;60:167–79. [CrossRef]
- Yang C, Yan Z, Hu F, Wei W, Sun Z, Xu W. Silencing of microRNA-517a induces oxidative stress injury in melanoma cells via inactivation of the JNK signaling pathway by upregulating CDKN1C. Cancer Cell Int 2020;20:32. [CrossRef]
- Schadendorf D, van Akkooi ACJ, Berking C, Griewank KG, Gutzmer R, Hauschild A, et al. Melanoma. The Lancet 2018;392:971–84. [CrossRef]
- Bennett, DC. Genetics of melanoma progression: the rise and fall of cell senescence. Pigment Cell Melanoma Res 2016;29:122–40. [CrossRef]
- Akbani R, Akdemir KC, Aksoy BA, Albert M, Ally A, Amin SB, et al. Genomic Classification of Cutaneous Melanoma. Cell 2015;161:1681–96. [CrossRef]
- Hodis E, Watson IR, Kryukov GV, Arold ST, Imielinski M, Theurillat J-P, et al. A Landscape of Driver Mutations in Melanoma. Cell 2012;150:251–63. [CrossRef]
- Shain AH, Yeh I, Kovalyshyn I, Sriharan A, Talevich E, Gagnon A, et al. The Genetic Evolution of Melanoma from Precursor Lesions. New England Journal of Medicine 2015;373:1926–36. [CrossRef]
- Liu S, Howell PM, Riker AI. Up-Regulation of miR-182 Expression after Epigenetic Modulation of Human Melanoma Cells. Ann Surg Oncol 2013;20:1745–52. [CrossRef]
- Glud M, Gniadecki R. MicroRNAs in the pathogenesis of malignant melanoma. Journal of the European Academy of Dermatology and Venereology 2013;27:142–50. [CrossRef]
- Kunz, M. MicroRNAs in Melanoma Biology, 2013, p. 103–20. [CrossRef]
- Mione M, Bosserhoff A. Micro RNA s in melanocyte and melanoma biology. Pigment Cell Melanoma Res 2015;28:340–54. [CrossRef]
- Ameres SL, Zamore PD. Diversifying microRNA sequence and function. Nat Rev Mol Cell Biol 2013;14:475–88. [CrossRef]
- Ha M, Kim VN. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol 2014;15:509–24. [CrossRef]
- Pritchard CC, Cheng HH, Tewari M. MicroRNA profiling: approaches and considerations. Nat Rev Genet 2012;13:358–69. [CrossRef]
- Price C, Chen J. MicroRNAs in cancer biology and therapy: Current status and perspectives. Genes Dis 2014;1:53–63. [CrossRef]
- Wang D, Qiu C, Zhang H, Wang J, Cui Q, Yin Y. Human MicroRNA Oncogenes and Tumor Suppressors Show Significantly Different Biological Patterns: From Functions to Targets. PLoS One 2010;5:e13067. [CrossRef]
- Luna Buitrago D, Lovering RC, Caporali A. Insights into Online microRNA Bioinformatics Tools. Noncoding RNA 2023;9:18. [CrossRef]
- Chao J, Guo Y, Li P, Chao L. Role of Kallistatin Treatment in Aging and Cancer by Modulating miR-34a and miR-21 Expression. Oxid Med Cell Longev 2017;2017:1–7. [CrossRef]
- Zhang X, Ng W-L, Wang P, Tian L, Werner E, Wang H, et al. MicroRNA-21 Modulates the Levels of Reactive Oxygen Species by Targeting SOD3 and TNF α. Cancer Res 2012;72:4707–13. [CrossRef]
- Yadav P, Sharma P, Sundaram S, Venkatraman G, Bera AK, Karunagaran D. SLC7A11/ xCT is a target of miR-5096 and its restoration partially rescues miR-5096-mediated ferroptosis and anti-tumor effects in human breast cancer cells. Cancer Lett 2021;522:211–24. [CrossRef]
- Li S-Z, Hu Y-Y, Zhao J, Zhao Y-B, Sun J-D, Yang Y, et al. MicroRNA-34a induces apoptosis in the human glioma cell line, A172, through enhanced ROS production and NOX2 expression. Biochem Biophys Res Commun 2014;444:6–12. [CrossRef]
- Díaz Arce, D. Revista cubana de investigaciones biomédicas. vol. 23. Editorial Ciencias Médicas; 2004.
- Andreucci E, Ruzzolini J, Bianchini F, Versienti G, Biagioni A, Lulli M, et al. miR-214-Enriched Extracellular Vesicles Released by Acid-Adapted Melanoma Cells Promote Inflammatory Macrophage-Dependent Tumor Trans-Endothelial Migration. Cancers (Basel) 2022;14:5090. [CrossRef]
- Zhou J, Xu D, Xie H, Tang J, Liu R, Li J, et al. miR-33a functions as a tumor suppressor in melanoma by targeting HIF-1α. Cancer Biol Ther 2015;16:846–55. [CrossRef]
- CHEN Y, CAO K, WANG S, CHEN J, HE B, HE G, et al. MicroRNA-138 suppresses proliferation, invasion and glycolysis in malignant melanoma cells by targeting HIF-1α. Exp Ther Med 2016;11:2513–8. [CrossRef]
- Boyle GM, Woods SL, Bonazzi VF, Stark MS, Hacker E, Aoude LG, et al. Melanoma cell invasiveness is regulated by miR-211 suppression of the BRN2 transcription factor. Pigment Cell Melanoma Res 2011;24:525–37. [CrossRef]
- Völler D, Ott C, Bosserhoff A. MicroRNAs in malignant melanoma. Clin Biochem 2013;46:909–17. [CrossRef]
- Noguchi S, Kumazaki M, Yasui Y, Mori T, Yamada N, Akao Y. MicroRNA-203 Regulates Melanosome Transport and Tyrosinase Expression in Melanoma Cells by Targeting Kinesin Superfamily Protein 5b. Journal of Investigative Dermatology 2014;134:461–9. [CrossRef]
- Shirvani H, Ghanavi J, Aliabadi A, Mousavinasab F, Talebi M, Majidpoor J, et al. MiR-211 plays a dual role in cancer development: From tumor suppressor to tumor enhancer. Cell Signal 2023;101:110504. [CrossRef]
- Donadelli M, Dando I, Fiorini C, Palmieri M. Regulation of miR-23b expression and its dual role on ROS production and tumour development. Cancer Lett 2014;349:107–13. [CrossRef]
- Kozak J, Jonak K, Maciejewski R. The function of miR-200 family in oxidative stress response evoked in cancer chemotherapy and radiotherapy. Biomedicine & Pharmacotherapy 2020;125:110037. [CrossRef]
- Cui Y, She K, Tian D, Zhang P, Xin X. miR-146a Inhibits Proliferation and Enhances Chemosensitivity in Epithelial Ovarian Cancer via Reduction of SOD2. Oncology Research Featuring Preclinical and Clinical Cancer Therapeutics 2016;23:275–82. [CrossRef]
- Pflaum J, Schlosser S, Müller M. p53 Family and Cellular Stress Responses in Cancer. Front Oncol 2014;4. [CrossRef]
- Dar AA, Majid S, Rittsteuer C, de Semir D, Bezrookove V, Tong S, et al. The Role of miR-18b in MDM2-p53 Pathway Signaling and Melanoma Progression. JNCI: Journal of the National Cancer Institute 2013;105:433–42. [CrossRef]
- Hanahan, D. Hallmarks of Cancer: New Dimensions. Cancer Discov 2022;12:31–46. [CrossRef]
- Irvine M, Stewart A, Pedersen B, Boyd S, Kefford R, Rizos H. Oncogenic PI3K/AKT promotes the step-wise evolution of combination BRAF/MEK inhibitor resistance in melanoma. Oncogenesis 2018;7:72. [CrossRef]
- Hambright HG, Meng P, Kumar AP, Ghosh R. Inhibition of PI3K/AKT/mTOR axis disrupts oxidative stress-mediated survival of melanoma cells. Oncotarget 2015;6:7195–208. [CrossRef]
- Fraga A, Ribeiro R, Medeiros R. Hipoxia tumoral: Papel del factor inducible por hipoxia. Actas Urol Esp 2009;33. [CrossRef]
- Joshi S, Singh AR, Durden DL. MDM2 Regulates Hypoxic Hypoxia-inducible Factor 1α Stability in an E3 Ligase, Proteasome, and PTEN-Phosphatidylinositol 3-Kinase-AKT-dependent Manner. Journal of Biological Chemistry 2014;289:22785–97. [CrossRef]
- Malekan M, Ebrahimzadeh MA, Sheida F. The role of Hypoxia-Inducible Factor-1alpha and its signaling in melanoma. Biomedicine & Pharmacotherapy 2021;141:111873. [CrossRef]
- Carpenter EL, Becker AL, Indra AK. NRF2 and Key Transcriptional Targets in Melanoma Redox Manipulation. Cancers (Basel) 2022;14:1531. [CrossRef]
- Xue G, Romano E, Massi D, Mandalà M. Wnt/β-catenin signaling in melanoma: Preclinical rationale and novel therapeutic insights. Cancer Treat Rev 2016;49:1–12. [CrossRef]
- Varrone F, Caputo E. The miRNAs Role in Melanoma and in Its Resistance to Therapy. Int J Mol Sci 2020;21:878. [CrossRef]
- Lister JA, Capper A, Zeng Z, Mathers ME, Richardson J, Paranthaman K, et al. A Conditional Zebrafish MITF Mutation Reveals MITF Levels Are Critical for Melanoma Promotion vs. Regression In Vivo. Journal of Investigative Dermatology 2014;134:133–40. [CrossRef]
- Hartman ML, Czyz M. MITF in melanoma: mechanisms behind its expression and activity. Cellular and Molecular Life Sciences 2015;72:1249–60. [CrossRef]
- Park HY, Kosmadaki M, Yaar M, Gilchrest BA. Cellular mechanisms regulating human melanogenesis. Cellular and Molecular Life Sciences 2009;66:1493–506. [CrossRef]
- Thompson JF, Scolyer RA, Kefford RF. Cutaneous melanoma. The Lancet 2005;365:687–701. [CrossRef]
- Levy C, Khaled M, Robinson KC, Veguilla RA, Chen P-H, Yokoyama S, et al. Lineage-Specific Transcriptional Regulation of DICER by MITF in Melanocytes. Cell 2010;141:994–1005. [CrossRef]
- Hsiao JJ, Fisher DE. The roles of microphthalmia-associated transcription factor and pigmentation in melanoma. Arch Biochem Biophys 2014;563:28–34. [CrossRef]
- Bruneel K, Verstappe J, Vandamme N, Berx G. Intrinsic Balance between ZEB Family Members Is Important for Melanocyte Homeostasis and Melanoma Progression. Cancers (Basel) 2020;12:2248. [CrossRef]
- Denecker G, Vandamme N, Akay Ö, Koludrovic D, Taminau J, Lemeire K, et al. Identification of a ZEB2-MITF-ZEB1 transcriptional network that controls melanogenesis and melanoma progression. Cell Death Differ 2014;21:1250–61. [CrossRef]
- TargetScanHuman 8.0 n.d. https://www.targetscan.org/vert_80/ (accessed 29 March 2024).
- Segura MF, Hanniford D, Menendez S, Reavie L, Zou X, Alvarez-Diaz S, et al. Aberrant miR-182 expression promotes melanoma metastasis by repressing FOXO3 and microphthalmia-associated transcription factor. Proceedings of the National Academy of Sciences 2009;106:1814–9. [CrossRef]
- Bemis LT, Chen R, Amato CM, Classen EH, Robinson SE, Coffey DG, et al. MicroRNA-137 Targets Microphthalmia-Associated Transcription Factor in Melanoma Cell Lines. Cancer Res 2008;68:1362–8. [CrossRef]
- Mazar J, DeYoung K, Khaitan D, Meister E, Almodovar A, Goydos J, et al. The Regulation of miRNA-211 Expression and Its Role in Melanoma Cell Invasiveness. PLoS One 2010;5:e13779. [CrossRef]
- Margue C, Philippidou D, Reinsbach SE, Schmitt M, Behrmann I, Kreis S. New Target Genes of MITF-Induced microRNA-211 Contribute to Melanoma Cell Invasion. PLoS One 2013;8:e73473. [CrossRef]
- De Luca T, Pelosi A, Trisciuoglio D, D’Aguanno S, Desideri M, Farini V, et al. miR-211 and MITF modulation by Bcl-2 protein in melanoma cells. Mol Carcinog 2016;55:2304–12. [CrossRef]
- Mills CN, Joshi SS, Niles RM. Expression and function of hypoxia inducible factor-1 alpha in human melanoma under non-hypoxic conditions. Mol Cancer 2009;8:104. [CrossRef]
- Serocki M, Bartoszewska S, Janaszak-Jasiecka A, Ochocka RJ, Collawn JF, Bartoszewski R. miRNAs regulate the HIF switch during hypoxia: a novel therapeutic target. Angiogenesis 2018;21:183–202. [CrossRef]
- Seok J-K, Lee SH, Kim MJ, Lee Y-M. MicroRNA-382 induced by HIF-1α is an angiogenic miR targeting the tumor suppressor phosphatase and tensin homolog. Nucleic Acids Res 2014;42:8062–72. [CrossRef]
- Bhatt K, Wei Q, Pabla N, Dong G, Mi Q-S, Liang M, et al. MicroRNA-687 Induced by Hypoxia-Inducible Factor-1 Targets Phosphatase and Tensin Homolog in Renal Ischemia-Reperfusion Injury. Journal of the American Society of Nephrology 2015;26:1588–96. [CrossRef]
- Feige E, Yokoyama S, Levy C, Khaled M, Igras V, Lin RJ, et al. Hypoxia-induced transcriptional repression of the melanoma-associated oncogene MITF. Proceedings of the National Academy of Sciences 2011;108. [CrossRef]
- Buscà R, Berra E, Gaggioli C, Khaled M, Bille K, Marchetti B, et al. Hypoxia-inducible factor 1α is a new target of microphthalmia-associated transcription factor (MITF) in melanoma cells. J Cell Biol 2005;170:49–59. [CrossRef]
- Shen G, Li X, Jia Y, Piazza GA, Xi Y. Hypoxia-regulated microRNAs in human cancer. Acta Pharmacol Sin 2013;34:336–41. [CrossRef]
- Qiu H, Chen F, Chen M. MicroRNA-138 negatively regulates the hypoxia-inducible factor 1α to suppress melanoma growth and metastasis. Biol Open 2019;8. [CrossRef]
- Špaková I, Rabajdová M, Mičková H, Graier WF, Mareková M. Effect of hypoxia factors gene silencing on ROS production and metabolic status of A375 malignant melanoma cells. Sci Rep 2021;11:10325. [CrossRef]
- Ascierto PA, Kirkwood JM, Grob J-J, Simeone E, Grimaldi AM, Maio M, et al. The role of BRAF V600 mutation in melanoma. J Transl Med 2012;10:85. [CrossRef]
- Poulikakos PI, Zhang C, Bollag G, Shokat KM, Rosen N. RAF inhibitors transactivate RAF dimers and ERK signalling in cells with wild-type BRAF. Nature 2010;464:427–30. [CrossRef]
- McKay MM, Morrison DK. Integrating signals from RTKs to ERK/MAPK. Oncogene 2007;26:3113–21. [CrossRef]
- Held L, Eigentler TK, Meier F, Held M, Röcken M, Garbe C, et al. Oncogenetics of melanoma: basis for molecular diagnostics and therapy. JDDG: Journal Der Deutschen Dermatologischen Gesellschaft 2011;9:510–6. [CrossRef]
- Wang J, Shen WH, Jin YJ, Brandt-Rauf PW, Yin Y. A Molecular Link between E2F-1 and the MAPK Cascade. Journal of Biological Chemistry 2007;282:18521–31. [CrossRef]
- Cheng L, Lopez-Beltran A, Massari F, MacLennan GT, Montironi R. Molecular testing for BRAF mutations to inform melanoma treatment decisions: a move toward precision medicine. Modern Pathology 2018;31:24–38. [CrossRef]
- Colombino M, Capone M, Lissia A, Cossu A, Rubino C, De Giorgi V, et al. BRAF/NRAS Mutation Frequencies Among Primary Tumors and Metastases in Patients With Melanoma. Journal of Clinical Oncology 2012;30:2522–9. [CrossRef]
- Son Y, Cheong Y-K, Kim N-H, Chung H-T, Kang DG, Pae H-O. Mitogen-Activated Protein Kinases and Reactive Oxygen Species: How Can ROS Activate MAPK Pathways? J Signal Transduct 2011;2011:1–6. [CrossRef]
- Matsuzawa A, Ichijo H. Redox control of cell fate by MAP kinase: physiological roles of ASK1-MAP kinase pathway in stress signaling. Biochimica et Biophysica Acta (BBA) - General Subjects 2008;1780:1325–36. [CrossRef]
- Poenitzsch Strong AM, Setaluri V, Spiegelman VS. microRNA-340 as a modulator of RAS–RAF–MAPK signaling in melanoma. Arch Biochem Biophys 2014;563:118–24. [CrossRef]
- Vera O, Bok I, Jasani N, Nakamura K, Xu X, Mecozzi N, et al. A MAPK/miR-29 Axis Suppresses Melanoma by Targeting MAFG and MYBL2. Cancers (Basel) 2021;13:1408. [CrossRef]
- Madhunapantula S, V. , Robertson GP. The PTEN-AKT3 signaling cascade as a therapeutic target in melanoma. Pigment Cell Melanoma Res 2009;22:400–19. [CrossRef]
- Leslie, NR. Redox regulation of PI 3-kinase signalling via inactivation of PTEN. EMBO J 2003;22:5501–10. [CrossRef]
- Liu P, Hu Y, Ma L, Du M, Xia L, Hu Z. miR-425 inhibits melanoma metastasis through repression of PI3K-Akt pathway by targeting IGF-1. Biomedicine & Pharmacotherapy 2015;75:51–7. [CrossRef]
- Yang B, Wu Y, Chen Y, Li Y, Wang J, Cha X, et al. MiR-5195-3p targets the PCBP2/PI3K/AKT pathway to inhibit melanoma cell proliferation and migration. Heliyon 2023;9:e19227. [CrossRef]
- Atkinson JM, Rank KB, Zeng Y, Capen A, Yadav V, Manro JR, et al. Activating the Wnt/β-Catenin Pathway for the Treatment of Melanoma – Application of LY2090314, a Novel Selective Inhibitor of Glycogen Synthase Kinase-3. PLoS One 2015;10:e0125028. [CrossRef]
- Mao J, Wang J, Liu B, Pan W, Farr GH, Flynn C, et al. Low-Density Lipoprotein Receptor-Related Protein-5 Binds to Axin and Regulates the Canonical Wnt Signaling Pathway. Mol Cell 2001;7:801–9. [CrossRef]
- Wang J-J, Li Z-F, Li X-J, Han Z, Zhang L, Liu Z-J. Effects of microRNA-136 on melanoma cell proliferation, apoptosis, and epithelial–mesenchymal transition by targetting PMEL through the Wnt signaling pathway. Biosci Rep 2017;37. [CrossRef]
- Huo J, Zhang Y, Li R, Wang Y, Wu J, Zhang D. Upregulated MicroRNA-25 Mediates the Migration of Melanoma Cells by Targeting DKK3 through the WNT/β-Catenin Pathway. Int J Mol Sci 2016;17:1124. [CrossRef]
- L Shi, J-W Huo, S-S Chen, J-X Xue, W-Y Gao, X-Y Li, et al. MicroRNA-22 targets FMNL2 to inhibit melanoma progression via the regulation of the Wnt/β-catenin signaling pathway and epithelial-mesenchymal transition. Eur Rev Med Pharmacol Sci 2019.
- Wu Y, Antony S, Meitzler JL, Doroshow JH. Molecular mechanisms underlying chronic inflammation-associated cancers. Cancer Lett 2014;345:164–73. [CrossRef]
- Malakoutikhah Z, Mohajeri Z, Dana N, Haghjooy Javanmard S. The dual role of Nrf2 in melanoma: a systematic review. BMC Mol Cell Biol 2023;24:5. [CrossRef]
- Raghunath A, Sundarraj K, Arfuso F, Sethi G, Perumal E. Dysregulation of Nrf2 in Hepatocellular Carcinoma: Role in Cancer Progression and Chemoresistance. Cancers (Basel) 2018;10:481. [CrossRef]
- Jessen C, Kreß JKC, Baluapuri A, Hufnagel A, Schmitz W, Kneitz S, et al. The transcription factor NRF2 enhances melanoma malignancy by blocking differentiation and inducing COX2 expression. Oncogene 2020;39:6841–55. [CrossRef]
- De Backer J, Lin A, Berghe W Vanden, Bogaerts A, Hoogewijs D. Cytoglobin inhibits non-thermal plasma-induced apoptosis in melanoma cells through regulation of the NRF2-mediated antioxidant response. Redox Biol 2022;55. [CrossRef]
- Wu S, Lu H, Bai Y. Nrf2 in cancers: A double-edged sword. Cancer Med 2019;8:2252–67. [CrossRef]
- Taguchi K, Yamamoto M. The KEAP1–NRF2 System in Cancer. Front Oncol 2017;7. [CrossRef]
- Shah NM, Rushworth SA, Murray MY, Bowles KM, MacEwan DJ. Understanding the role of NRF2-regulated miRNAs in human malignancies. Oncotarget 2013;4:1130–42. [CrossRef]
- Liu Y-Y, Slotine J-J, Barabási A-L. Controllability of complex networks. Nature 2011;473:167–73. [CrossRef]
- MacNeil LT, Walhout AJM. Gene regulatory networks and the role of robustness and stochasticity in the control of gene expression. Genome Res 2011;21:645–57. [CrossRef]
- Home - GEO - NCBI, n.d. https://www.ncbi.nlm.nih.gov/geo/ (accessed on 28 March 2024).
- Sahoo A, Sahoo SK, Joshi P, Lee B, Perera RJ. MicroRNA-211 Loss Promotes Metabolic Vulnerability and BRAF Inhibitor Sensitivity in Melanoma. Journal of Investigative Dermatology 2019;139:167–76. [CrossRef]
- Efron B, Tibshirani R, Storey JD, Tusher V. Empirical Bayes Analysis of a Microarray Experiment. J Am Stat Assoc 2001;96:1151–60. [CrossRef]
- Schwender H, Ickstadt K. Empirical Bayes analysis of single nucleotide polymorphisms. BMC Bioinformatics 2008;9:1–15. [CrossRef]
- Grant GR, Manduchi E, Stoeckert CJ. Analysis and Management of Microarray Gene Expression Data. Curr Protoc Mol Biol 2007;77. [CrossRef]
- Waku T, Nakada S, Masuda H, Sumi H, Wada A, Hirose S, et al. The CNC-family transcription factor Nrf3 coordinates the melanogenesis cascade through macropinocytosis and autophagy regulation. Cell Rep 2023;42:111906. [CrossRef]
- Ballesteros-Álvarez J, Dilshat R, Fock V, Möller K, Karl L, Larue L, et al. MITF and TFEB cross-regulation in melanoma cells. PLoS One 2020;15:e0238546. [CrossRef]
- Möller K, Sigurbjornsdottir S, Arnthorsson AO, Pogenberg V, Dilshat R, Fock V, et al. MITF has a central role in regulating starvation-induced autophagy in melanoma. Sci Rep 2019;9:1055. [CrossRef]
- Ariano C, Costanza F, Akman M, Riganti C, Corà D, Casanova E, et al. TFEB inhibition induces melanoma shut-down by blocking the cell cycle and rewiring metabolism. Cell Death Dis 2023;14:314. [CrossRef]
- Richard G, Dalle S, Monet M, Ligier M, Boespflug A, Pommier RM, et al. ZEB 1-mediated melanoma cell plasticity enhances resistance to MAPK inhibitors. EMBO Mol Med 2016;8:1143–61. [CrossRef]
- Durand S, Tang Y, Pommier RM, Benboubker V, Grimont M, Boivin F, et al. ZEB1 controls a lineage-specific transcriptional program essential for melanoma cell state transitions. BioRxiv 2023:2023.02.10.526467. [CrossRef]
- Gasiorek JJ, Blank V. Regulation and function of the NFE2 transcription factor in hematopoietic and non-hematopoietic cells. Cellular and Molecular Life Sciences 2015;72:2323–35. [CrossRef]
- Motohashi H, Kimura M, Fujita R, Inoue A, Pan X, Takayama M, et al. NF-E2 domination over Nrf2 promotes ROS accumulation and megakaryocytic maturation. Blood 2010;115:677–86. [CrossRef]
- Ashford AL, Dunkley TPJ, Cockerill M, Rowlinson RA, Baak LM, Gallo R, et al. Identification of DYRK1B as a substrate of ERK1/2 and characterisation of the kinase activity of DYRK1B mutants from cancer and metabolic syndrome. Cellular and Molecular Life Sciences 2016;73:883–900. [CrossRef]
- Hamada J, Shoda K, Masuda K, Fujita Y, Naruto T, Kohmoto T, et al. Tumor-promoting function and prognostic significance of the RNA-binding protein T-cell intracellular antigen-1 in esophageal squamous cell carcinoma. Oncotarget 2016;7:17111–28. [CrossRef]
- Garaigorta U, Heim MH, Boyd B, Wieland S, Chisari F V. Hepatitis C Virus (HCV) Induces Formation of Stress Granules Whose Proteins Regulate HCV RNA Replication and Virus Assembly and Egress. J Virol 2012;86:11043–56. [CrossRef]
- Takayama K, Suzuki T, Fujimura T, Takahashi S, Inoue S. Association of USP10 with G3BP2 Inhibits p53 Signaling and Contributes to Poor Outcome in Prostate Cancer. Molecular Cancer Research 2018;16:846–56. [CrossRef]
- Legrand N, Dixon DA, Sobolewski C. Stress granules in colorectal cancer: Current knowledge and potential therapeutic applications. World J Gastroenterol 2020;26:5223–47. [CrossRef]
- Choi S, Sa M, Cho N, Kim KK, Park S-H. Rbfox2 dissociation from stress granules suppresses cancer progression. Exp Mol Med 2019;51:1–12. [CrossRef]
- Glenewinkel F, Cohen MJ, King CR, Kaspar S, Bamberg-Lemper S, Mymryk JS, et al. The adaptor protein DCAF7 mediates the interaction of the adenovirus E1A oncoprotein with the protein kinases DYRK1A and HIPK2. Sci Rep 2016;6:28241. [CrossRef]
- Deng H, Liu J, Deng Y, Han G, Shellman YG, Robinson SE, et al. CtBP1 Is Expressed in Melanoma and Represses the Transcription of p16INK4a and Brca1. Journal of Investigative Dermatology 2013;133:1294–301. [CrossRef]
- Pajares M, Rojo AI, Arias E, Díaz-Carretero A, Cuervo AM, Cuadrado A. Transcription factor NFE2L2/NRF2 modulates chaperone-mediated autophagy through the regulation of LAMP2A. Autophagy 2018;14:1310–22. [CrossRef]
- Sagwal SK, Bekeschus S. ROS Pleiotropy in Melanoma and Local Therapy with Physical Modalities. Oxid Med Cell Longev 2021;2021:1–21. [CrossRef]
- Wu H-T, Zhong H-T, Li G-W, Shen J-X, Ye Q-Q, Zhang M-L, et al. Oncogenic functions of the EMT-related transcription factor ZEB1 in breast cancer. J Transl Med 2020;18:51. [CrossRef]
- Wei S, Zhong L, Wang X, Zhang W. Low expression of GATA3 promotes cell proliferation and metastasis in gastric cancer. Cancer Manag Res 2017;Volume 9:769–80. [CrossRef]
- Chen T, Tsang JYS, Su X, Li P, Sun W, Wong ILK, et al. SALL4 promotes tumor progression in breast cancer by targeting EMT. Mol Carcinog 2020;59:1209–26. [CrossRef]
- Han X, Duan X, Liu Z, Long Y, Liu C, Zhou J, et al. ZEB1 directly inhibits GPX4 transcription contributing to ROS accumulation in breast cancer cells. Breast Cancer Res Treat 2021;188:329–42. [CrossRef]
- Han X, Long Y, Duan X, Liu Z, Hu X, Zhou J, et al. ZEB1 induces ROS generation through directly promoting MCT4 transcription to facilitate breast cancer. Exp Cell Res 2022;412:113044. [CrossRef]
- Plaschka M, Benboubker V, Grimont M, Berthet J, Tonon L, Lopez J, et al. ZEB1 transcription factor promotes immune escape in melanoma. J Immunother Cancer 2022;10:e003484. [CrossRef]
- Wu M, Deng X, Zhong Y, Hu L, Zhang X, Liang Y, et al. MafF Is Regulated via the circ-ITCH/miR-224-5p Axis and Acts as a Tumor Suppressor in Hepatocellular Carcinoma. Oncology Research Featuring Preclinical and Clinical Cancer Therapeutics 2020;28:299–309. [CrossRef]
- Caporali S, Amaro A, Levati L, Alvino E, Lacal PM, Mastroeni S, et al. miR-126-3p down-regulation contributes to dabrafenib acquired resistance in melanoma by up-regulating ADAM9 and VEGF-A. Journal of Experimental & Clinical Cancer Research 2019;38:272. [CrossRef]
- Itoh T, Fukatani K, Nakashima A, Suzuki K. MicroRNA-141-3p and microRNA-200a-3p regulate α-melanocyte stimulating hormone-stimulated melanogenesis by directly targeting microphthalmia-associated transcription factor. Sci Rep 2020;10:2149. [CrossRef]
- Li A, Wu N, Sun J. E2F1-induced microRNA-224-5p expression is associated with hepatocellular carcinoma cell migration, invasion and epithelial-mesenchymal transition via MREG. Oncol Lett 2022;23:82. [CrossRef]
- Rana S, Valbuena GN, Curry E, Bevan CL, Keun HC. MicroRNAs as biomarkers for prostate cancer prognosis: a systematic review and a systematic reanalysis of public data. Br J Cancer 2022;126:502–13. [CrossRef]
- Kang H, Rho JG, Kim C, Tak H, Lee H, Ji E, et al. The miR-24-3p/p130Cas: a novel axis regulating the migration and invasion of cancer cells. Sci Rep 2017;7:44847. [CrossRef]
- Zhou X, Yan T, Huang C, Xu Z, Wang L, Jiang E, et al. Melanoma cell-secreted exosomal miR-155-5p induce proangiogenic switch of cancer-associated fibroblasts via SOCS1/JAK2/STAT3 signaling pathway. Journal of Experimental & Clinical Cancer Research 2018;37:242. [CrossRef]
- Tian Y, Zeng J, Yang Z. MicroRNA-27b inhibits the development of melanoma by targeting MYC. Oncol Lett 2021;21:370. [CrossRef]
- Stope M, Ahrend H, Daeschlein G, Grove E, Paditz M, Mustea A, et al. MicroRNA-20a-3p and microRNA-20a-5p exhibit anti-proliferative activities in a melanoma in vitro model. SDRP Journal of Cellular and Molecular Physiology 2019;3:1–10. [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




