Submitted:
24 August 2024
Posted:
27 August 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
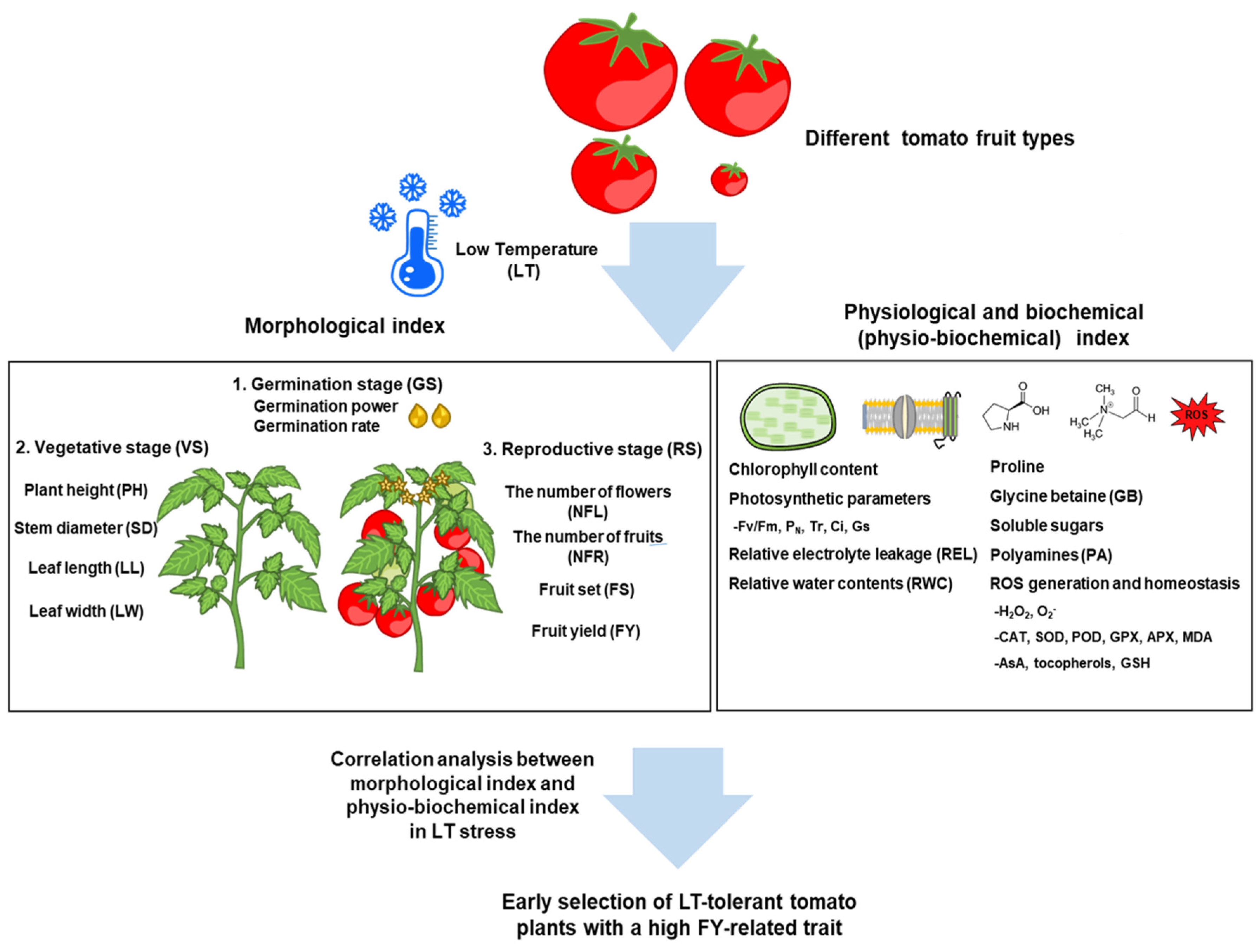
2. Morphological Changes of Tomato Plants in Response to LT
3. Physiological and Biochemical Changes of Tomato Plants to LT
3.1. Physiological Responses
3.1.1. Chlorophyll Contents and Photosynthetic Parameters
3.1.2. Cell Membrane and Relative Electrolyte Leakage (REL)
3.1.3. Relative Water Contents (RWC)
3.2. Biochemical Responses to Low Temperature (LT) Stress
3.2.1. Proline, Soluble Sugars, and Glycine Betaine (GB)
3.2.2. Polyamines (PAs)
3.2.3. The ROS Generation and Regulation by Antioxidant Molecules
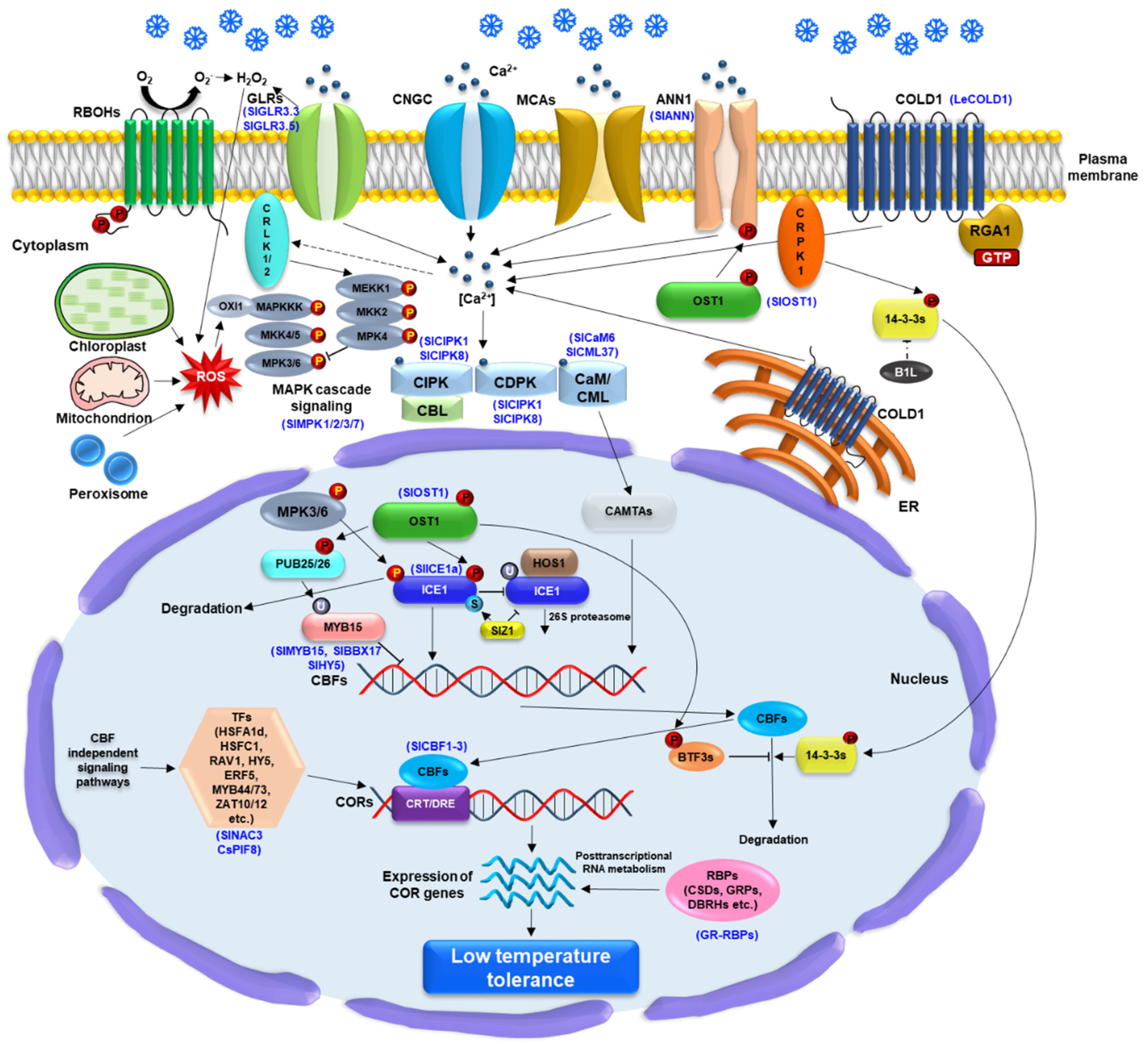
4. Molecular Mechanisms Underlying the LT Response in Tomato Plants
4.1. LT Perception and LT Response
4.2. LT Signaling Pathways via a Calcium Molecule
4.3. LT Signaling Pathways via ROS Molecules
4.4. The LT Signaling Transduction via a CBF Dependent Pathway
4.5. The LT Signaling Transduction via a CBF-Independent Pathway
4.6. The Cellular Roles of RNA-Binding Proteins in LT Response
4.7. Epigenetic Regulation of Fruit Ripening and Abiotic Stress in Tomato Plants
4.7.1. DNA Methylation in Fruit Ripening and Abiotic Stress Response
4.7.2. Histone Modifications in Fruit Ripening and Abiotic Stress Response
4.7.3. Noncoding RNAs in Fruit Ripening and Abiotic Stress Response
4.8. RNA Methylation In Fruit Ripening and Abiotic Stress Response
5. Conclusions and Future Prospects
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Valderas-Martinez, P.; Chiva-Blanch, G.; Casas, R.; Arranz, S.; Martínez-Huélamo, M.; Urpi-Sarda, M.; Torrado, X.; Corella, D.; Lamuela-Raventós, R.M.; Estruch, R. Tomato sauce enriched with olive oil exerts greater effects on cardiovascular disease risk factors than raw tomato and tomato sauce: a randomized trial. Nutrients 2016, 8, 170. [Google Scholar] [CrossRef]
- Vinson, J.A.; Hao, Y.; Su, X.; Zubik, L. Phenol antioxidant quantity and quality in foods: vegetables. J. Agric. Food Chem. 1998, 46, 3630–3634. [Google Scholar] [CrossRef]
- Salehi, B.; Sharifi-Rad, R.; Sharopov, F.; Namiesnik, J.; Roointan, A.; Kamle, M.; Kumar, P.; Martins, N.; Sharifi-Rad, J. Beneficial effects and potential risks of tomato consumption for human health: An overview. Nutrition 2019, 62, 201–208. [Google Scholar] [CrossRef] [PubMed]
- Vallverdú-Queralt, A.; Regueiro, J.; Rinaldi de Alvarenga, J.F.; Torrado, X.; Lamuela-Raventos, R.M. Carotenoid profile of tomato sauces: effect of cooking time and content of extra virgin olive oil. Int. J. Mol. Sci. 2015, 16, 9588–9599. [Google Scholar] [CrossRef] [PubMed]
- Van Ploeg, D.; Heuvelink, E. Influence of sub-optimal temperature on tomato growth and yield: a review. J. HORTIC. SCI. BIOTECH. 2005, 80, 652–659. [Google Scholar] [CrossRef]
- Theocharis, A.; Clément, C.; Barka, E.A. Physiological and molecular changes in plants grown at low temperatures. Planta 2012, 235, 1091–1105. [Google Scholar] [CrossRef]
- Bhandari, R.; Neupane, N.; Adhikari, D.P. Climatic change and its impact on tomato (lycopersicum esculentum l.) production in plain area of Nepal. Environ. Chall. 2021, 4, 100129. [Google Scholar] [CrossRef]
- Gerszberg, A.; Hnatuszko-Konka, K. Tomato tolerance to abiotic stress: a review of most often engineered target sequences. J. Plant Growth Regul. 2017, 83, 175–198. [Google Scholar] [CrossRef]
- Lee, K.; Rajametov, S.N.; Jeong, H.-B.; Cho, M.-C.; Lee, O.-J.; Kim, S.-G.; Yang, E.-Y.; Chae, W.-B. Comprehensive understanding of selecting traits for heat tolerance during vegetative and reproductive growth stages in tomato. Agronomy 2022, 12, 834. [Google Scholar] [CrossRef]
- Kang, Y.; Lee, K.; Hoshikawa, K.; Kang, M.; Jang, S. Molecular bases of heat stress responses in vegetable crops with focusing on heat shock factors and heat shock proteins. Front. Plant Sci. 2022, 13, 837152. [Google Scholar] [CrossRef]
- Song, J.; Shang, L.; Chen, S.; Lu, Y.; Zhang, Y.; Ouyang, B.; Ye, Z.; Zhang, J. Interactions between ShPP2-1, an F-box family gene, and ACR11A regulate cold tolerance of tomato. Hortic. Res. 2021, 8, 148. [Google Scholar] [CrossRef]
- Guan, Y.; Hwarari, D.; Korboe, H.M.; Ahmad, B.; Cao, Y.; Movahedi, A.; Yang, L. Low temperature stress-induced perception and molecular signaling pathways in plants. Environ. Exp. Bot. 2023, 207, 105190. [Google Scholar] [CrossRef]
- Rajametov, S.N.; Lee, K.; Jeong, H.-B.; Cho, M.-C.; Nam, C.-W.; Yang, E.-Y. Physiological traits of thirty-five tomato accessions in response to low temperature. Agriculture 2021, 11, 792. [Google Scholar] [CrossRef]
- Rajametov, S.; Yang, E.Y.; Cho, M.C.; Chae, S.Y.; Kim, J.H.; Nam, C.W.; Chae, W.B. Traits affecting low temperature tolerance in tomato and its application to breeding program. Plant Breed. Biotech. 2019, 7, 350–359. [Google Scholar]
- Yadav, D.; Meena, Y.K.; Bairwa, L.; Singh, U.; Bairwa, S.; Choudhary, M.; Singh, A. Morphological, Physiological and Biochemical Response to Low Temperature Stress in Tomato (Solanum lycopersicum L.): A Review. Int. J. Stress Manag. 2021, 12, 706–712. [Google Scholar] [CrossRef]
- Xiaoa, F.; Yang, Z.; Zhua, L. Low temperature and weak light affect greenhouse tomato growth and fruit quality. J. Plant Sci 2018, 6, 16–24. [Google Scholar]
- Shi, Y.; Ding, Y.; Yang, S. Molecular regulation of CBF signaling in cold acclimation. Trends Plant Sci. 2018, 23, 623–637. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, J.; Sarwar, R.; Zhang, W.; Geng, R.; Zhu, K.-M.; Tan, X.-L. Research progress on the physiological response and molecular mechanism of cold response in plants. Front. Plant Sci. 2024, 15, 1334913. [Google Scholar] [CrossRef]
- Hu, J.; Xu, T.; Kang, H. Crosstalk between RNA m6A modification and epigenetic factors for gene regulation in plants. Plant Commun. 2024. [Google Scholar] [CrossRef]
- Foolad, M.; Lin, G. Relationship between cold tolerance during seed germination and vegetative growth in tomato: analysis of response and correlated response to selection. J. Am. Soc. Hort. Sci. 2001, 126, 216–220. [Google Scholar] [CrossRef]
- Hoek, I.H.; Ten Cate, C.H.H.; Keijzer, C.J.; Schel, J.H.; Dons, H.J. Development of the fifth leaf is indicative for whole plant performance at low temperature in tomato. Ann. Bot. 1993, 72, 367–374. [Google Scholar] [CrossRef]
- Picken, A. A review of pollination and fruit set in the tomato (Lycopersicon esculentum Mill.). J. Hortic. Sci. 1984, 59, 1–13. [Google Scholar] [CrossRef]
- Yang, E.Y.; Rajaemtov, S.N.; Cho, M.C.; Jeong, H.B.; Chae, W.B. Factors Affecting Tolerance to Low Night Temperature Differ by Fruit Types and Sizes in Tomato. Agriculture 2021, 11, 681. [Google Scholar] [CrossRef]
- Ercan, N.; Vural, H. The effects of low temperatures on fruit set of tomatoes. In Proceedings of the II Symposium on Protected Cultivation of Solanacea in Mild Winter Climates 366; 1993; pp. 65–72. [Google Scholar]
- Khan, T.A.; Fariduddin, Q.; Yusuf, M. Lycopersicon esculentum under low temperature stress: an approach toward enhanced antioxidants and yield. Environ. Sci. Pollut. Res. 2015, 22, 14178–14188. [Google Scholar] [CrossRef]
- Mu, J.; Fu, Y.; Liu, B.; Zhang, Y.; Wang, A.; Li, Y.; Zhu, J. SiFBA5, a cold-responsive factor from Saussurea involucrata promotes cold resilience and biomass increase in transgenic tomato plants under cold stress. BMC Plant Biol. 2021, 21, 1–10. [Google Scholar] [CrossRef]
- Zhang, Y.; Peng, Y.; Liu, J.; Yan, J.; Zhu, K.; Sun, X.; Bu, X.; Wang, X.; Ahammed, G.J.; Liu, Y. Tetratricopeptide repeat protein SlREC2 positively regulates cold tolerance in tomato. Plant Physiol. 2023, 192, 648–665. [Google Scholar] [CrossRef]
- Barrero-Sicilia, C.; Silvestre, S.; Haslam, R.P.; Michaelson, L.V. Lipid remodelling: Unravelling the response to cold stress in Arabidopsis and its extremophile relative Eutrema salsugineum. Plant Sci. 2017, 263, 194–200. [Google Scholar] [CrossRef]
- Lamers, J.; Van Der Meer, T.; Testerink, C. How plants sense and respond to stressful environments. Plant Physiol. 2020, 182, 1624–1635. [Google Scholar] [CrossRef]
- de Jong, F.; Munnik, T. Attracted to membranes: lipid-binding domains in plants. Plant Physiol. 2021, 185, 707–723. [Google Scholar] [CrossRef]
- Nadeem, M.; Pham, T.H.; Nieuwenhuis, A.; Ali, W.; Zaeem, M.; Ashiq, W.; Gillani, S.S.M.; Manful, C.; Adigun, O.A.; Galagedara, L. Adaptation strategies of forage soybeans cultivated on acidic soils under cool climate to produce high quality forage. Plant Sci. 2019, 283, 278–289. [Google Scholar] [CrossRef]
- Wu, J.; Nadeem, M.; Galagedara, L.; Thomas, R.; Cheema, M. Recent insights into cell responses to cold stress in plants: Signaling, defence, and potential functions of phosphatidic acid. Environ. Exp. Bot. 2022, 203, 105068. [Google Scholar] [CrossRef]
- Guo, X.; Li, J.; Zhang, L.; Zhang, Z.; He, P.; Wang, W.; Wang, M.; Wang, A.; Zhu, J. Heterotrimeric G-protein α subunit (LeGPA1) confers cold stress tolerance to processing tomato plants (Lycopersicon esculentum Mill). BMC Plant Biol. 2020, 20, 1–16. [Google Scholar] [CrossRef]
- Zhang, L.; Guo, X.; Zhang, Z.; Wang, A.; Zhu, J. Cold-regulated gene LeCOR413PM2 confers cold stress tolerance in tomato plants. Gene 2021, 764, 145097. [Google Scholar] [CrossRef] [PubMed]
- İşeri, Ö.D.; Körpe, D.A.; Sahin, F.I.; Haberal, M. Hydrogen peroxide pretreatment of roots enhanced oxidative stress response of tomato under cold stress. Acta Physiol. Plant 2013, 35, 1905–1913. [Google Scholar] [CrossRef]
- Saleem, M.; Fariduddin, Q.; Janda, T. Multifaceted role of salicylic acid in combating cold stress in plants: a review. J. Plant Growth Regul. 2021, 40, 464–485. [Google Scholar] [CrossRef]
- Meena, Y.; Khurana, D.; Kaur, N.; Singh, K. Towards enhanced low temperature stress tolerance in tomato: An approach. J. Environ. Biol. 2018, 39, 529–535. [Google Scholar] [CrossRef]
- Wang, L.; Wu, B.; Chen, G.; Chen, H.; Peng, Y.; Sohail, H.; Geng, S.; Luo, G.; Xu, D.; Ouyang, B. The essential role of jasmonate signaling in Solanum habrochaites rootstock-mediated cold tolerance in tomato grafts. Hortic. Res. 2023, 10, uhac227. [Google Scholar] [CrossRef]
- Manivannan, P.; Jaleel, C.A.; Kishorekumar, A.; Sankar, B.; Somasundaram, R.; Sridharan, R.; Panneerselvam, R. Changes in antioxidant metabolism of Vigna unguiculata (L.) Walp. by propiconazole under water deficit stress. Colloids Surf. B. Biointerfaces 2007, 57, 69–74. [Google Scholar] [CrossRef] [PubMed]
- Feng, H.-L.; Ma, N.-N.; Meng, X.; Zhang, S.; Wang, J.-R.; Chai, S.; Meng, Q.-W. A novel tomato MYC-type ICE1-like transcription factor, SlICE1a, confers cold, osmotic and salt tolerance in transgenic tobacco. Plant Physiol. Biochem. 2013, 73, 309–320. [Google Scholar] [CrossRef] [PubMed]
- Kushad, M.M.; Yelenosky, G. Evaluation of polyamine and proline levels during low temperature acclimation of citrus. Plant Physiol. 1987, 84, 692–695. [Google Scholar] [CrossRef] [PubMed]
- Xin, Z.; Li, P.H. Relationship between proline and abscisic acid in the induction of chilling tolerance in maize suspension-cultured cells. Plant Physiol. 1993, 103, 607–613. [Google Scholar] [CrossRef] [PubMed]
- Patade, V.Y.; Khatri, D.; Kumari, M.; Grover, A.; Mohan Gupta, S.; Ahmed, Z. Cold tolerance in Osmotin transgenic tomato (Solanum lycopersicum L.) is associated with modulation in transcript abundance of stress responsive genes. SpringerPlus 2013, 2, 117. [Google Scholar] [CrossRef] [PubMed]
- Park, E.-J.; Jeknic, Z.; Chen, T.H. Exogenous application of glycinebetaine increases chilling tolerance in tomato plants. Plant Cell Physiol. 2006, 47, 706–714. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Dai, T.; Liu, Y.; Wang, J.; Wang, Q.; Zhu, W. Effect of exogenous glycine betaine on the germination of tomato seeds under cold stress. Int. J. Mol. Sci. 2022, 23, 10474. [Google Scholar] [CrossRef]
- Dai, T.; Ban, S.; Han, L.; Li, L.; Zhang, Y.; Zhang, Y.; Zhu, W. Effects of exogenous glycine betaine on growth and development of tomato seedlings under cold stress. Front. Plant Sci. 2024, 15, 1332583. [Google Scholar] [CrossRef]
- Wei, D.; Zhang, T.; Wang, B.; Zhang, H.; Ma, M.; Li, S.; Chen, T.H.; Brestic, M.; Liu, Y.; Yang, X. Glycinebetaine mitigates tomato chilling stress by maintaining high-cyclic electron flow rate of photosystem I and stability of photosystem II. Plant Cell Rep. 2022, 41, 1087–1101. [Google Scholar] [CrossRef]
- Shahzad, R.; Ahmed, F.; Wang, Z.; Harlina, P.W.; Nishawy, E.; Ayaad, M.; Manan, A.; Maher, M.; Ewas, M. Comparative analysis of two phytochrome mutants of tomato (Micro-Tom cv.) reveals specific physiological, biochemical, and molecular responses under chilling stress. J. Genet. Eng. Biotechnol. 2020, 18, 77. [Google Scholar] [CrossRef]
- Wani, U.M.; Majeed, S.T.; Raja, V.; Wani, Z.A.; Jan, N.; Andrabi, K.I.; John, R. Ectopic expression of a novel cold-resistance protein 1 from Brassica oleracea promotes tolerance to chilling stress in transgenic tomato. Sci. Rep. 2021, 11, 16574. [Google Scholar] [CrossRef]
- Amini, S.; Maali-Amiri, R.; Kazemi-Shahandashti, S.-S.; Lopez-Gomez, M.; Sadeghzadeh, B.; Sobhani-Najafabadi, A.; Kariman, K. Effect of cold stress on polyamine metabolism and antioxidant responses in chickpea. J. Plant Physiol. 2021, 258, 153387. [Google Scholar] [CrossRef]
- Kocsy, G.; Pál, M.; Soltész, A.; Szalai, G.; Boldizsár, Á.; Kovács, V.; Janda, T. Low temperature and oxidative stress in cereals. Acta Agron. Hung. 2011, 59, 169–189. [Google Scholar] [CrossRef]
- Du, H.; Chen, B.; Li, Q.; Liu, H.; Kurtenbach, R. Conjugated polyamines in root plasma membrane enhanced the tolerance of plum seedling to osmotic stress by stabilizing membrane structure and therefore elevating H+-ATPase activity. Front. Plant Sci. 2022, 12, 812360. [Google Scholar] [CrossRef] [PubMed]
- Borromeo, I.; Domenici, F.; Del Gallo, M.; Forni, C. Role of polyamines in the response to salt stress of tomato. Plants 2023, 12, 1855. [Google Scholar] [CrossRef]
- Zhang, K.; Zhang, H.; Bi, M.; Zhao, X.; Xiang, H.; Yang, F.; Tan, C.; He, Y.; Li, T.; Meng, S. ShWRKY55 enhances the cold resistance of wild tomato LA1777 by regulating the expression of the key gene ShSAMDC2 involved in polyamine synthesis. Environ. Exp. Bot. 2024, 221, 105723. [Google Scholar] [CrossRef]
- Pál, M.; Szalai, G.; Gondor, O.K.; Janda, T. Unfinished story of polyamines: Role of conjugation, transport and light-related regulation in the polyamine metabolism in plants. Plant Sci. 2021, 308, 110923. [Google Scholar] [CrossRef] [PubMed]
- Minocha, R.; Majumdar, R.; Minocha, S.C. Polyamines and abiotic stress in plants: a complex relationship. Front. Plant Sci. 2014, 5, 175. [Google Scholar] [CrossRef] [PubMed]
- Alcázar, R.; Bueno, M.; Tiburcio, A.F. Polyamines: Small amines with large effects on plant abiotic stress tolerance. Cells 2020, 9, 2373. [Google Scholar] [CrossRef]
- Kim, T.E.; Kim, S.K.; Han, T.J.; Lee, J.S.; Chang, S.C. ABA and polyamines act independently in primary leaves of cold-stressed tomato (Lycopersicon esculentum). Physiol. Plant. 2002, 115, 370–376. [Google Scholar] [CrossRef]
- Diao, Q.; Song, Y.; Qi, H. Exogenous spermidine enhances chilling tolerance of tomato (Solanum lycopersicum L.) seedlings via involvement in polyamines metabolism and physiological parameter levels. Acta Physiol. Plant 2015, 37, 1–15. [Google Scholar] [CrossRef]
- Song, Y.; Diao, Q.; Qi, H. Polyamine metabolism and biosynthetic genes expression in tomato (Lycopersicon esculentum Mill.) seedlings during cold acclimation. J. Plant Growth Regul. 2015, 75, 21–32. [Google Scholar] [CrossRef]
- Min, D.; Zhou, J.; Li, J.; Ai, W.; Li, Z.; Zhang, X.; Fu, X.; Zhao, X.; Li, F.; Li, X. SlMYC2 targeted regulation of polyamines biosynthesis contributes to methyl jasmonate-induced chilling tolerance in tomato fruit. Postharvest Biol. Technol. 2021, 174, 111443. [Google Scholar] [CrossRef]
- Ding, F.; Wang, C.; Xu, N.; Wang, M.; Zhang, S. Jasmonic acid-regulated putrescine biosynthesis attenuates cold-induced oxidative stress in tomato plants. Sci. Hortic. 2021, 288, 110373. [Google Scholar] [CrossRef]
- Apel, K.; Hirt, H. Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 2004, 55, 373–399. [Google Scholar] [CrossRef] [PubMed]
- Mansoor, S.; Ali Wani, O.; Lone, J.K.; Manhas, S.; Kour, N.; Alam, P.; Ahmad, A.; Ahmad, P. Reactive oxygen species in plants: from source to sink. Antioxidants 2022, 11, 225. [Google Scholar] [CrossRef] [PubMed]
- Quan, L.J.; Zhang, B.; Shi, W.W.; Li, H.Y. Hydrogen peroxide in plants: a versatile molecule of the reactive oxygen species network. J. Integr. Plant Biol. 2008, 50, 2–18. [Google Scholar] [CrossRef]
- Miller, G.; Suzuki, N.; Ciftci-Yilmaz, S.; Mittler, R. Reactive oxygen species homeostasis and signalling during drought and salinity stresses. Plant, Cell Environ. 2010, 33, 453–467. [Google Scholar] [CrossRef]
- Ashraf, M. Biotechnological approach of improving plant salt tolerance using antioxidants as markers. Biotechnol. Adv. 2009, 27, 84–93. [Google Scholar] [CrossRef]
- Chowdhury, M.E.K.; Choi, B.; Cho, B.-K.; Kim, J.B.; Park, S.U.; Natarajan, S.; Lim, H.-S.; Bae, H. Regulation of 4CL, encoding 4-coumarate: coenzyme A ligase, expression in kenaf under diverse stress conditions. Plant Omics 2013, 6, 254–262. [Google Scholar]
- Mittler, R.; Zandalinas, S.I.; Fichman, Y.; Van Breusegem, F. Reactive oxygen species signalling in plant stress responses. Nat. Rev. Mol. Cell Biol. 2022, 23, 663–679. [Google Scholar] [CrossRef]
- Castro, B.; Citterico, M.; Kimura, S.; Stevens, D.M.; Wrzaczek, M.; Coaker, G. Stress-induced reactive oxygen species compartmentalization, perception and signalling. Nat. Plants 2021, 7, 403–412. [Google Scholar] [CrossRef]
- Shu, D.-F.; Wang, L.-Y.; Duan, M.; Deng, Y.-S.; Meng, Q.-W. Antisense-mediated depletion of tomato chloroplast glutathione reductase enhances susceptibility to chilling stress. Plant Physiol. Biochem. 2011, 49, 1228–1237. [Google Scholar] [CrossRef]
- Iqbal, Z.; Memon, A.G.; Ahmad, A.; Iqbal, M.S. Calcium mediated cold acclimation in plants: underlying signaling and molecular mechanisms. Front. Plant Sci. 2022, 13, 855559. [Google Scholar] [CrossRef] [PubMed]
- Yuan, P.; Yang, T.; Poovaiah, B. Calcium signaling-mediated plant response to cold stress. Int. J. Mol. Sci. 2018, 19, 3896. [Google Scholar] [CrossRef] [PubMed]
- Tian, W.; Wang, C.; Gao, Q.; Li, L.; Luan, S. Calcium spikes, waves and oscillations in plant development and biotic interactions. Nat. Plants 2020, 6, 750–759. [Google Scholar] [CrossRef]
- Chen, X.; Ding, Y.; Yang, Y.; Song, C.; Wang, B.; Yang, S.; Guo, Y.; Gong, Z. Protein kinases in plant responses to drought, salt, and cold stress. J. Integr. Plant Biol. 2021, 63, 53–78. [Google Scholar] [CrossRef] [PubMed]
- Mori, K.; Renhu, N.; Naito, M.; Nakamura, A.; Shiba, H.; Yamamoto, T.; Suzaki, T.; Iida, H.; Miura, K. Ca2+-permeable mechanosensitive channels MCA1 and MCA2 mediate cold-induced cytosolic Ca2+ increase and cold tolerance in Arabidopsis. Sci. Rep. 2018, 8, 550. [Google Scholar] [CrossRef]
- Liu, Q.; Ding, Y.; Shi, Y.; Ma, L.; Wang, Y.; Song, C.; Wilkins, K.A.; Davies, J.M.; Knight, H.; Knight, M.R. The calcium transporter ANNEXIN1 mediates cold-induced calcium signaling and freezing tolerance in plants. EMBO J. 2021, 40, e104559. [Google Scholar] [CrossRef]
- Zhang, L.; Guo, X.; Qin, Y.; Feng, B.; Wu, Y.; He, Y.; Wang, A.; Zhu, J. The chilling tolerance divergence 1 protein confers cold stress tolerance in processing tomato. Plant Physiol. Biochem. 2020, 151, 34–46. [Google Scholar] [CrossRef]
- Ijaz, R.; Ejaz, J.; Gao, S.; Liu, T.; Imtiaz, M.; Ye, Z.; Wang, T. Overexpression of annexin gene AnnSp2, enhances drought and salt tolerance through modulation of ABA synthesis and scavenging ROS in tomato. Sci. Rep. 2017, 7, 12087. [Google Scholar] [CrossRef]
- Chong, L.; Xu, R.; Huang, P.; Guo, P.; Zhu, M.; Du, H.; Sun, X.; Ku, L.; Zhu, J.-K.; Zhu, Y. The tomato OST1–VOZ1 module regulates drought-mediated flowering. Plant Cell 2022, 34, 2001–2018. [Google Scholar] [CrossRef]
- Nawaz, Z.; Kakar, K.U.; Ullah, R.; Yu, S.; Zhang, J.; Shu, Q.-Y.; Ren, X.-l. Genome-wide identification, evolution and expression analysis of cyclic nucleotide-gated channels in tobacco (Nicotiana tabacum L.). Genomics 2019, 111, 142–158. [Google Scholar] [CrossRef]
- Wang, L.; Li, M.; Liu, Z.; Dai, L.; Zhang, M.; Wang, L.; Zhao, J.; Liu, M. Genome-wide identification of CNGC genes in Chinese jujube (Ziziphus jujuba Mill.) and ZjCNGC2 mediated signalling cascades in response to cold stress. BMC Genomics 2020, 21, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Cui, Y.; Lu, S.; Li, Z.; Cheng, J.; Hu, P.; Zhu, T.; Wang, X.; Jin, M.; Wang, X.; Li, L. CYCLIC NUCLEOTIDE-GATED ION CHANNELs 14 and 16 promote tolerance to heat and chilling in rice. Plant Physiol. 2020, 183, 1794–1808. [Google Scholar] [CrossRef]
- Wang, J.; Ren, Y.; Liu, X.; Luo, S.; Zhang, X.; Liu, X.; Lin, Q.; Zhu, S.; Wan, H.; Yang, Y. Transcriptional activation and phosphorylation of OsCNGC9 confer enhanced chilling tolerance in rice. Mol. Plant 2021, 14, 315–329. [Google Scholar] [CrossRef] [PubMed]
- Tang, M.; Xu, C.; Cao, H.; Shi, Y.; Chen, J.; Chai, Y.; Li, Z. Tomato calmodulin-like protein SlCML37 is a calcium (Ca2+) sensor that interacts with proteasome maturation factor SlUMP1 and plays a role in tomato fruit chilling stress tolerance. J. Plant Physiol. 2021, 258, 153373. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhou, X.n.; Liu, S.; Yu, A.; Yang, C.; Chen, X.; Liu, J.; Wang, A. Identification and functional analysis of tomato CIPK gene family. Int. J. Mol. Sci. 2019, 21, 110. [Google Scholar] [CrossRef] [PubMed]
- Lin, R.; Song, J.; Tang, M.; Wang, L.; Yu, J.; Zhou, Y. CALMODULIN6 negatively regulates cold tolerance by attenuating ICE1-dependent stress responses in tomato. Plant Physiol. 2023, 193, 2105–2121. [Google Scholar] [CrossRef]
- Yang, T.; Shad Ali, G.; Yang, L.; Du, L.; Reddy, A.; Poovaiah, B. Calcium/calmodulin-regulated receptor-like kinase CRLK1 interacts with MEKK1 in plants. Plant Signal. Behav. 2010, 5, 991–994. [Google Scholar] [CrossRef]
- Lee, B.-h.; Lee, H.; Xiong, L.; Zhu, J.-K. A mitochondrial complex I defect impairs cold-regulated nuclear gene expression. Plant Cell 2002, 14, 1235–1251. [Google Scholar] [CrossRef]
- Dong, C.-H.; Zolman, B.K.; Bartel, B.; Lee, B.-h.; Stevenson, B.; Agarwal, M.; Zhu, J.-K. Disruption of Arabidopsis CHY1 reveals an important role of metabolic status in plant cold stress signaling. Mol. Plant 2009, 2, 59–72. [Google Scholar] [CrossRef]
- Li, H.; Jiang, X.; Lv, X.; Ahammed, G.J.; Guo, Z.; Qi, Z.; Yu, J.; Zhou, Y. Tomato GLR3. 3 and GLR3. 5 mediate cold acclimation-induced chilling tolerance by regulating apoplastic H2O2 production and redox homeostasis. Plant, Cell Environ. 2019, 42, 3326–3339. [Google Scholar] [CrossRef]
- Ahmad, F.; Singh, A.; Kamal, A. Salicylic acid–mediated defense mechanisms to abiotic stress tolerance. In Plant signaling molecules; Elsevier: 2019, pp. 355-369.
- Gudesblat, G.E.; Iusem, N.D.; Morris, P.C. Guard cell-specific inhibition of Arabidopsis MPK3 expression causes abnormal stomatal responses to abscisic acid and hydrogen peroxide. New Phytol. 2007, 173, 713–721. [Google Scholar] [CrossRef] [PubMed]
- Yuasa, T.; Ichimura, K.; Mizoguchi, T.; Shinozaki, K. Oxidative stress activates ATMPK6, an Arabidopsis homologue of MAP kinase. Plant Cell Physiol. 2001, 42, 1012–1016. [Google Scholar] [CrossRef]
- Yu, L.; Yan, J.; Yang, Y.; He, L.; Zhu, W. Enhanced tolerance to chilling stress in tomato by overexpression of a mitogen-activated protein kinase, SlMPK7. Plant Mol. Biol. Rep. 2016, 34, 76–88. [Google Scholar] [CrossRef]
- Wang, L.; Zhao, R.; Zheng, Y.; Chen, L.; Li, R.; Ma, J.; Hong, X.; Ma, P.; Sheng, J.; Shen, L. SlMAPK1/2/3 and antioxidant enzymes are associated with H2O2-induced chilling tolerance in tomato plants. J. Agric. Food Chem. 2017, 65, 6812–6820. [Google Scholar] [CrossRef]
- Chinnusamy, V.; Zhu, J.-K.; Sunkar, R. Gene regulation during cold stress acclimation in plants. Plant stress tolerance: Methods and protocols 2010, 39–55. [Google Scholar]
- Jaglo-Ottosen, K.R.; Gilmour, S.J.; Zarka, D.G.; Schabenberger, O.; Thomashow, M.F. Arabidopsis CBF1 overexpression induces COR genes and enhances freezing tolerance. Science 1998, 280, 104–106. [Google Scholar] [CrossRef]
- Qin, F.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Achievements and challenges in understanding plant abiotic stress responses and tolerance. Plant Cell Physiol. 2011, 52, 1569–1582. [Google Scholar] [CrossRef] [PubMed]
- Jiang, B.; Shi, Y.; Zhang, X.; Xin, X.; Qi, L.; Guo, H.; Li, J.; Yang, S. PIF3 is a negative regulator of the CBF pathway and freezing tolerance in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 2017, 114, E6695–E6702. [Google Scholar] [CrossRef]
- Zhao, C.; Zhu, J.-K. The broad roles of CBF genes: from development to abiotic stress. Plant Signal. Behav. 2016, 11, e1215794. [Google Scholar] [CrossRef]
- Wang, P.; Cui, X.; Zhao, C.; Shi, L.; Zhang, G.; Sun, F.; Cao, X.; Yuan, L.; Xie, Q.; Xu, X. COR27 and COR28 encode nighttime repressors integrating Arabidopsis circadian clock and cold response. J. Integr. Plant Biol. 2017, 59, 78–85. [Google Scholar] [CrossRef]
- Novillo, F.; Alonso, J.M.; Ecker, J.R.; Salinas, J. CBF2/DREB1C is a negative regulator of CBF1/DREB1B and CBF3/DREB1A expression and plays a central role in stress tolerance in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 2004, 101, 3985–3990. [Google Scholar] [CrossRef]
- Wang, Y.; Jiang, C.-J.; Li, Y.-Y.; Wei, C.-L.; Deng, W.-W. CsICE1 and CsCBF1: two transcription factors involved in cold responses in Camellia sinensis. Plant Cell Rep. 2012, 31, 27–34. [Google Scholar] [CrossRef]
- Wang, L.; Nick, P. Cold sensing in grapevine—which signals are upstream of the microtubular “thermometer”. Plant, Cell Environ. 2017, 40, 2844–2857. [Google Scholar] [CrossRef] [PubMed]
- Verma, S.; Negi, N.P.; Narwal, P.; Kumari, P.; Kisku, A.V.; Gahlot, P.; Mittal, N.; Kumar, D. Calcium signaling in coordinating plant development, circadian oscillations and environmental stress responses in plants. Environ. Exp. Bot. 2022, 201, 104935. [Google Scholar] [CrossRef]
- Kim, Y.; Park, S.; Gilmour, S.J.; Thomashow, M.F. Roles of CAMTA transcription factors and salicylic acid in configuring the low-temperature transcriptome and freezing tolerance of A rabidopsis. Plant J. 2013, 75, 364–376. [Google Scholar] [CrossRef]
- Kidokoro, S.; Yoneda, K.; Takasaki, H.; Takahashi, F.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Different cold-signaling pathways function in the responses to rapid and gradual decreases in temperature. Plant Cell 2017, 29, 760–774. [Google Scholar] [CrossRef]
- Li, R.; Zhang, L.; Wang, L.; Chen, L.; Zhao, R.; Sheng, J.; Shen, L. Reduction of tomato-plant chilling tolerance by CRISPR–Cas9-mediated SlCBF1 mutagenesis. J. Agric. Food Chem. 2018, 66, 9042–9051. [Google Scholar] [CrossRef] [PubMed]
- Yu, W.; Shen, L.; Sheng, J. Synergistic effects of SlCBF1 and ethylene signaling on the maintenance of tomatoes quality during long-term cold storage. Postharvest Biol. Technol. 2024, 217, 113090. [Google Scholar] [CrossRef]
- Zhang, L.; Jiang, X.; Liu, Q.; Ahammed, G.J.; Lin, R.; Wang, L.; Shao, S.; Yu, J.; Zhou, Y. The HY5 and MYB15 transcription factors positively regulate cold tolerance in tomato via the CBF pathway. Plant, Cell Environ. 2020, 43, 2712–2726. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Jia, Y.; Shi, Y.; Zhang, X.; Song, C.; Gong, Z.; Yang, S. OST 1-mediated BTF 3L phosphorylation positively regulates CBF s during plant cold responses. EMBO J. 2018, 37, e98228. [Google Scholar] [CrossRef]
- Zarka, D.G.; Vogel, J.T.; Cook, D.; Thomashow, M.F. Cold induction of Arabidopsis CBF genes involves multiple ICE (inducer of CBF expression) promoter elements and a cold-regulatory circuit that is desensitized by low temperature. Plant Physiol. 2003, 133, 910–918. [Google Scholar] [CrossRef]
- Thomashow, M.F.; Torii, K.U. SCREAMing twist on the role of ICE1 in freezing tolerance. Plant Cell 2020, 32, 816–819. [Google Scholar] [CrossRef] [PubMed]
- Lee, B.-h.; Henderson, D.A.; Zhu, J.-K. The Arabidopsis cold-responsive transcriptome and its regulation by ICE1. Plant Cell 2005, 17, 3155–3175. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Li, H.; Zhang, X.; Xie, Q.; Gong, Z.; Yang, S. OST1 kinase modulates freezing tolerance by enhancing ICE1 stability in Arabidopsis. Dev. Cell 2015, 32, 278–289. [Google Scholar] [CrossRef] [PubMed]
- Dong, C.-H.; Agarwal, M.; Zhang, Y.; Xie, Q.; Zhu, J.-K. The negative regulator of plant cold responses, HOS1, is a RING E3 ligase that mediates the ubiquitination and degradation of ICE1. Proc. Natl. Acad. Sci. U.S.A. 2006, 103, 8281–8286. [Google Scholar] [CrossRef]
- Jung, J.-H.; Park, C.-M. HOS1-mediated activation of FLC via chromatin remodeling under cold stress. Plant Signal. Behav. 2013, 8, e27342. [Google Scholar] [CrossRef]
- Kim, S.H.; Kim, H.S.; Bahk, S.; An, J.; Yoo, Y.; Kim, J.-Y.; Chung, W.S. Phosphorylation of the transcriptional repressor MYB15 by mitogen-activated protein kinase 6 is required for freezing tolerance in Arabidopsis. Nucleic Acids Res. 2017, 45, 6613–6627. [Google Scholar] [CrossRef]
- Wang, X.; Zhang, X.; Song, C.-P.; Gong, Z.; Yang, S.; Ding, Y. PUB25 and PUB26 dynamically modulate ICE1 stability via differential ubiquitination during cold stress in Arabidopsis. Plant Cell 2023, 35, 3585–3603. [Google Scholar] [CrossRef]
- Chen, T.; Zhang, W.; Yang, G.; Chen, J.-H.; Chen, B.-X.; Sun, R.; Zhang, H.; An, L.-Z. TRANSTHYRETIN-LIKE and BYPASS1-LIKE co-regulate growth and cold tolerance in Arabidopsis. BMC Plant Biol. 2020, 20, 1–11. [Google Scholar] [CrossRef]
- Teige, M.; Scheikl, E.; Eulgem, T.; Doczi, R.; Ichimura, K.; Shinozaki, K.; Dangl, J.L.; Hirt, H. The MKK2 pathway mediates cold and salt stress signaling in Arabidopsis. Mol. Cell 2004, 15, 141–152. [Google Scholar] [CrossRef]
- Li, H.; Ding, Y.; Shi, Y.; Zhang, X.; Zhang, S.; Gong, Z.; Yang, S. MPK3-and MPK6-mediated ICE1 phosphorylation negatively regulates ICE1 stability and freezing tolerance in Arabidopsis. Dev. Cell 2017, 43, 630–642. [Google Scholar] [CrossRef] [PubMed]
- Song, J.; Lin, R.; Tang, M.; Wang, L.; Fan, P.; Xia, X.; Yu, J.; Zhou, Y. SlMPK1-and SlMPK2-mediated SlBBX17 phosphorylation positively regulates CBF-dependent cold tolerance in tomato. New Phytol. 2023, 239, 1887–1902. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Zhang, Z.; Xie, S.; Si, T.; Li, Y.; Zhu, J.-K. Mutational evidence for the critical role of CBF transcription factors in cold acclimation in Arabidopsis. Plant Physiol. 2016, 171, 2744–2759. [Google Scholar] [CrossRef] [PubMed]
- Xin, Z.; Browse, J. Eskimo1 mutants of Arabidopsis are constitutively freezing-tolerant. Proc. Natl. Acad. Sci. U.S.A. 1998, 95, 7799–7804. [Google Scholar] [CrossRef]
- Liu, H.; Zhang, Y.; Lu, S.; Chen, H.; Wu, J.; Zhu, X.; Zou, B.; Hua, J. HsfA1d promotes hypocotyl elongation under chilling via enhancing expression of ribosomal protein genes in Arabidopsis. New Phytol. 2021, 231, 646–660. [Google Scholar] [CrossRef]
- Jiao, S.-Z.; Guo, C.; Yao, W.-K.; Zhang, N.-B.; Zhang, J.-Y.; Xu, W.-R. An Amur grape VaHsfC1 is involved in multiple abiotic stresses. Sci. Hortic. 2022, 295, 110785. [Google Scholar] [CrossRef]
- Zhu, J.; Shi, H.; Lee, B.-h.; Damsz, B.; Cheng, S.; Stirm, V.; Zhu, J.-K.; Hasegawa, P.M.; Bressan, R.A. An Arabidopsis homeodomain transcription factor gene, HOS9, mediates cold tolerance through a CBF-independent pathway. Proc. Natl. Acad. Sci. U.S.A. 2004, 101, 9873–9878. [Google Scholar] [CrossRef]
- Vogel, J.T.; Zarka, D.G.; Van Buskirk, H.A.; Fowler, S.G.; Thomashow, M.F. Roles of the CBF2 and ZAT12 transcription factors in configuring the low temperature transcriptome of Arabidopsis. Plant J. 2005, 41, 195–211. [Google Scholar] [CrossRef]
- Catalá, R.; Medina, J.; Salinas, J. Integration of low temperature and light signaling during cold acclimation response in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 2011, 108, 16475–16480. [Google Scholar] [CrossRef]
- Park, S.; Lee, C.M.; Doherty, C.J.; Gilmour, S.J.; Kim, Y.; Thomashow, M.F. Regulation of the Arabidopsis CBF regulon by a complex low-temperature regulatory network. Plant J. 2015, 82, 193–207. [Google Scholar] [CrossRef]
- He, Z.; Zhao, T.; Yin, Z.; Liu, J.; Cheng, Y.; Xu, J. The phytochrome-interacting transcription factor CsPIF8 contributes to cold tolerance in citrus by regulating superoxide dismutase expression. Plant Sci. 2020, 298, 110584. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Ma, X.; Chen, Y.; Wang, C.; Xia, Z.; Liu, Z.; Gao, L.; Zhang, W. SlNAC3 suppresses cold tolerance in tomatoes by enhancing ethylene biosynthesis. Plant, Cell Environ. 2024, 47, 3132–3146. [Google Scholar] [CrossRef]
- Lee, K.; Kang, H. Emerging roles of RNA-binding proteins in plant growth, development, and stress responses. Mol. Cells 2016, 39, 179–185. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.; Kang, H. Roles of organellar RNA-binding proteins in plant growth, development, and abiotic stress responses. Int. J. Mol. Sci. 2020, 21, 4548. [Google Scholar] [CrossRef] [PubMed]
- Muthusamy, M.; Kim, J.-H.; Kim, J.A.; Lee, S.-I. Plant RNA binding proteins as critical modulators in drought, high salinity, heat, and cold stress responses: an updated overview. Int. J. Mol. Sci. 2021, 22, 6731. [Google Scholar] [CrossRef]
- Cheng, K.; Zhang, C.; Lu, Y.; Li, J.; Tang, H.; Ma, L.; Zhu, H. The Glycine-Rich RNA-Binding Protein Is a Vital Post-Transcriptional Regulator in Crops. Plants 2023, 12, 3504. [Google Scholar] [CrossRef]
- Yang, Y.; Zhu, G.; Li, R.; Yan, S.; Fu, D.; Zhu, B.; Tian, H.; Luo, Y.; Zhu, H. The RNA editing factor SlORRM4 is required for normal fruit ripening in tomato. Plant Physiol. 2017, 175, 1690–1702. [Google Scholar] [CrossRef]
- Yang, Y.; Liu, X.; Wang, K.; Li, J.; Zhu, G.; Ren, S.; Deng, Z.; Zhu, B.; Fu, D.; Qu, G. Molecular and functional diversity of organelle RNA editing mediated by RNA recognition motif-containing protein ORRM4 in tomato. New Phytol. 2020, 228, 570–585. [Google Scholar] [CrossRef]
- Li, X.; Yang, Y.; Zeng, N.; Qu, G.; Fu, D.; Zhu, B.; Luo, Y.; Ostersetzer-Biran, O.; Zhu, H. Glycine-rich RNA-binding cofactor RZ1AL is associated with tomato ripening and development. Hortic. Res. 2022, 9, uhac134. [Google Scholar] [CrossRef]
- Ruggieri, G.M.; Triassi, A.; Alvarez, C.E.; Gola, A.; Wiggenhauser, J.; Budde, C.O.; Lara, M.V.; Drincovich, M.F.; Müller, G.L. Overexpression of glycine-rich RNA-binding protein in tomato renders fruits with higher protein content after cold storage. Biol. Plant. 2018, 62, 501–510. [Google Scholar] [CrossRef]
- Zhang, Y.; Mo, Y.; Li, J.; Liu, L.; Gao, Y.; Zhang, Y.; Huang, Y.; Ren, L.; Zhu, H.; Jiang, X. Divergence in regulatory mechanisms of GR-RBP genes in different plants under abiotic stress. Sci. Rep. 2024, 14, 8743. [Google Scholar] [CrossRef]
- Tang, D.; Gallusci, P.; Lang, Z. Fruit development and epigenetic modifications. New Phytol. 2020, 228, 839–844. [Google Scholar] [CrossRef] [PubMed]
- Ming, Y.; Jiang, L.; Ji, D. Epigenetic regulation in tomato fruit ripening. Front. Plant Sci. 2023, 14, 1269090. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Wu, X.; Liu, H.; Zhang, M.; Liao, W. DNA methylation in tomato fruit ripening. Physiol. Plant. 2022, 174, e13627. [Google Scholar] [CrossRef]
- Zhong, S.; Fei, Z.; Chen, Y.-R.; Zheng, Y.; Huang, M.; Vrebalov, J.; McQuinn, R.; Gapper, N.; Liu, B.; Xiang, J. Single-base resolution methylomes of tomato fruit development reveal epigenome modifications associated with ripening. Nat. Biotechnol. 2013, 31, 154–159. [Google Scholar] [CrossRef]
- Lang, Z.; Wang, Y.; Tang, K.; Tang, D.; Datsenka, T.; Cheng, J.; Zhang, Y.; Handa, A.K.; Zhu, J.-K. Critical roles of DNA demethylation in the activation of ripening-induced genes and inhibition of ripening-repressed genes in tomato fruit. Proc. Natl. Acad. Sci. U.S.A. 2017, 114, E4511–E4519. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Tieman, D.M.; Jiao, C.; Xu, Y.; Chen, K.; Fei, Z.; Giovannoni, J.J.; Klee, H.J. Chilling-induced tomato flavor loss is associated with altered volatile synthesis and transient changes in DNA methylation. Proc. Natl. Acad. Sci. U.S.A. 2016, 113, 12580–12585. [Google Scholar] [CrossRef]
- Zhang, C.; Duan, W.; Chen, K.; Zhang, B. Transcriptome and methylome analysis reveals effects of ripening on and off the vine on flavor quality of tomato fruit. Postharvest Biol. Technol. 2020, 162, 111096. [Google Scholar] [CrossRef]
- Zhou, J.; Zhou, S.; Chen, B.; Sangsoy, K.; Luengwilai, K.; Albornoz, K.; Beckles, D.M. Integrative analysis of the methylome and transcriptome of tomato fruit (Solanum lycopersicum L.) induced by postharvest handling. Hortic. Res. 2024, 11, uhae095. [Google Scholar] [CrossRef]
- Liang, Q.; Deng, H.; Li, Y.; Liu, Z.; Shu, P.; Fu, R.; Zhang, Y.; Pirrello, J.; Zhang, Y.; Grierson, D. Like Heterochromatin Protein 1b represses fruit ripening via regulating the H3K27me3 levels in ripening-related genes in tomato. New Phytol. 2020, 227, 485–497. [Google Scholar] [CrossRef]
- Li, Z.; Jiang, G.; Liu, X.; Ding, X.; Zhang, D.; Wang, X.; Zhou, Y.; Yan, H.; Li, T.; Wu, K. Histone demethylase SlJMJ6 promotes fruit ripening by removing H3K27 methylation of ripening-related genes in tomato. New Phytol. 2020, 227, 1138–1156. [Google Scholar] [CrossRef]
- Li, Z.; Zeng, J.; Zhou, Y.; Ding, X.; Jiang, G.; Wu, K.; Jiang, Y.; Duan, X. Histone H3K27 demethylase SlJMJ3 modulates fruit ripening in tomato. Plant Physiol. 2024, kiae233. [Google Scholar] [CrossRef]
- Ding, X.; Liu, X.; Jiang, G.; Li, Z.; Song, Y.; Zhang, D.; Jiang, Y.; Duan, X. SlJMJ7 orchestrates tomato fruit ripening via crosstalk between H3K4me3 and DML2-mediated DNA demethylation. New Phytol. 2022, 233, 1202–1219. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.-E.; Hu, Z.; Zhu, M.; Li, F.; Zhu, Z.; Lu, Y.; Chen, G. The tomato histone deacetylase SlHDA1 contributes to the repression of fruit ripening and carotenoid accumulation. Sci. Rep. 2017, 7, 7930. [Google Scholar] [CrossRef]
- Guo, J.-E.; Hu, Z.; Yu, X.; Li, A.; Li, F.; Wang, Y.; Tian, S.; Chen, G. A histone deacetylase gene, SlHDA3, acts as a negative regulator of fruit ripening and carotenoid accumulation. Plant Cell Rep. 2018, 37, 125–135. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.-E. Histone deacetylase gene SlHDT1 regulates tomato fruit ripening by affecting carotenoid accumulation and ethylene biosynthesis. Plant Sci. 2022, 318, 111235. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.-E.; Wang, H.; Yang, Y.; Li, J.; Zhu, Z. Histone deacetylase gene SlHDA3 is involved in drought and salt response in tomato. J. Plant Growth Regul. 2023, 99, 359–372. [Google Scholar] [CrossRef]
- Guo, J.-E.; Wang, H. Histone deacetylase gene SlHDA1 regulates drought and salt tolerance in tomato (Solanum lycopersicum). Sci. Hortic. 2023, 313, 111899. [Google Scholar] [CrossRef]
- Ma, L.; Mu, J.; Grierson, D.; Wang, Y.; Gao, L.; Zhao, X.; Zhu, B.; Luo, Y.; Shi, K.; Wang, Q. Noncoding RNAs: functional regulatory factors in tomato fruit ripening. Theor. Appl. Genet. 2020, 133, 1753–1762. [Google Scholar] [CrossRef]
- Lin, D.; Zhu, X.; Qi, B.; Gao, Z.; Tian, P.; Li, Z.; Lin, Z.; Zhang, Y.; Huang, T. SlMIR164A regulates fruit ripening and quality by controlling SlNAM2 and SlNAM3 in tomato. Plant Biotechnol. J. 2022, 20, 1456–1469. [Google Scholar] [CrossRef]
- Dong, Y.; Tang, M.; Huang, Z.; Song, J.; Xu, J.; Ahammed, G.J.; Yu, J.; Zhou, Y. The miR164a-NAM3 module confers cold tolerance by inducing ethylene production in tomato. Plant J. 2022, 111, 440–456. [Google Scholar] [CrossRef]
- Li, Y.; Liu, Y.; Gao, Z.; Wang, F.; Xu, T.; Qi, M.; Liu, Y.; Li, T. MicroRNA162 regulates stomatal conductance in response to low night temperature stress via abscisic acid signaling pathway in tomato. Front. Plant Sci. 2023, 14, 1045112. [Google Scholar] [CrossRef] [PubMed]
- Tan, J.; Zhou, Z.; Niu, Y.; Sun, X.; Deng, Z. Identification and functional characterization of tomato circRNAs derived from genes involved in fruit pigment accumulation. Sci. Rep. 2017, 7, 8594. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Zhao, W.; Gao, L.; Zhao, L. Genome-wide profiling of long non-coding RNAs from tomato and a comparison with mRNAs associated with the regulation of fruit ripening. BMC Plant Biol. 2018, 18, 1–10. [Google Scholar] [CrossRef]
- Wang, Y.; Gao, L.; Zhu, B.; Zhu, H.; Luo, Y.; Wang, Q.; Zuo, J. Integrative analysis of long non-coding RNA acting as ceRNAs involved in chilling injury in tomato fruit. Gene 2018, 667, 25–33. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Cai, J.; Xu, T.; Kang, H. Epitranscriptomic mRNA modifications governing plant stress responses: underlying mechanism and potential application. Plant Biotechnol. J. 2022, 20, 2245–2257. [Google Scholar] [CrossRef]
- Tang, J.; Chen, S.; Jia, G. Detection, regulation, and functions of RNA N6-methyladenosine modification in plants. Plant Commun. 2023, 4, 100546. [Google Scholar] [CrossRef]
- Hu, J.; Cai, J.; Umme, A.; Chen, Y.; Xu, T.; Kang, H. Unique features of mRNA m6A methylomes during expansion of tomato (Solanum lycopersicum) fruits. Plant Physiol. 2022, 188, 2215–2227. [Google Scholar] [CrossRef]
- Shen, H.; Luo, B.; Wang, Y.; Li, J.; Hu, Z.; Xie, Q.; Wu, T.; Chen, G. Genome-wide identification, classification and expression analysis of m6A gene family in Solanum lycopersicum. Int. J. Mol. Sci. 2022, 23, 4522. [Google Scholar] [CrossRef]
- Zhou, L.; Tian, S.; Qin, G. RNA methylomes reveal the m 6 A-mediated regulation of DNA demethylase gene SlDML2 in tomato fruit ripening. Genome Biol. 2019, 20, 1–23. [Google Scholar]
- Yin, S.; Ao, Q.; Qiu, T.; Tan, C.; Tu, Y.; Kuang, T.; Yang, Y. Tomato SlYTH1 encoding a putative RNA m6A reader affects plant growth and fruit shape. Plant Sci. 2022, 323, 111417. [Google Scholar] [CrossRef] [PubMed]
- Bai, C.; Fang, M.; Zhai, B.; Ma, L.; Fu, A.; Gao, L.; Kou, X.; Meng, D.; Wang, Q.; Zheng, S. Regulations of m6A methylation on tomato fruit chilling injury. Hortic. Plant J. 2021, 7, 434–442. [Google Scholar] [CrossRef]
- Yang, D.; Xu, H.; Liu, Y.; Li, M.; Ali, M.; Xu, X.; Lu, G. RNA N6-methyladenosine responds to low-temperature stress in tomato anthers. Front. Plant Sci. 2021, 12, 687826. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).




