1. Introduction
Colorectal cancer (CRC) has emerged as a significant public health challenge globally. Its rising incidence and mortality rates mark it as an imperative health issue that cannot be overlooked [
1]. Epidemiological data indicate substantial variability in CRC incidence across countries and regions. The overall rate is up to 13%-15%, showing a trend of annual increase [
2]. In particular, CRC has become one of the most common malignant tumors in Western developed countries. Factors such as lifestyle changes, dietary shifts, and an aging population contribute to the prevalence of CRC [
3]. Therefore, comprehensively understanding its pathogenesis and exploring effective prevention strategies are of profound significance for reducing the incidence and mortality of CRC and improving public health.
Substantial advancement has been made in research regarding CRC and intestinal microbes. Extensive studies have shown that intestinal dysbacteriosis is a pivotal risk factor for CRC. The enrichment or depletion of specific bacterial populations is closely linked to disease risk. For example, an increase in Fusobacterium nucleatum and Bacteroides fragilis may elevate the risk of CRC [
4,
5]. Moreover, metabolites produced by intestinal microbes, such as short-chain fatty acids and bile acids, as well as those produced by specific amino acids (like tryptophan), e.g., trans-3-indole propionic acid, IDA, play crucial roles in the pathogenesis of CRC [
6]. These metabolites promote or inhibit the progression of CRC by affecting cell signaling pathways and ferroptosis [
7]. A complex immune-microbial metabolic axis has been identified between intestinal microbes and the host, jointly regulating the host's immune metabolism and functions [
8]. Intestinal dysbacteriosis may disrupt immune homeostasis and induce the onset of CRC [
9]. Meanwhile, interventions in intestinal microbes can influence the host's metabolic pathways, potentially providing effective strategies for CRC treatment [
10]. These findings offer essential evidence for the prevention, diagnosis, and treatment of CRC. Furthermore, intestinal microbes have been reported to be closely involved in various diseases, including CRC [
11]. However, the causal relationships between intestinal microbes and CRC remain obscure.
Mendelian randomization (MR) was employed in this study. It is a powerful tool for exploring causal relationships between environmental exposures and diseases. Data from genome-wide association studies (GWAS) were utilized to analyze the potential links between intestinal microbes and CRC, delving into their possible causal relationships. The results of this research provide new perspectives for the prevention and treatment of CRC and may promote the formulation of healthcare policies and the rational allocation of healthcare resources. Identifying the causal associations between intestinal microbes and CRC can enhance the understanding of CRC's pathogenesis, thereby spurring the development of effective prevention and treatment strategies. Moreover, this study offers valuable insights for research on other similar diseases.
2. Material and Method
2.1. Study Design
This study focused on the causal relationships between intestinal microbes and CRC and its subtypes (such as colon cancer, rectal cancer, and rectal anastomosis cancer). Utilizing publicly available GWAS data and a European CRC cohort 16S rRNA dataset, the relationship between intestinal flora traits and CRC across diverse pathological sites and stages was explored through two-sample univariate MR and MR-Bayesian Model Averaging (MR-BMA) methods [
12]. The MR-BMA method is particularly suitable for identifying causal factors in high-correlation data, such as intestinal microbes associated with CRC [
13]. Moreover, the NetMoss method was employed to construct a multi-population cohort and validate the causal relationships between intestinal microbes and CRC. The overall study design aimed to deepen the understanding of the potential role of intestinal microbes in the development of CRC.
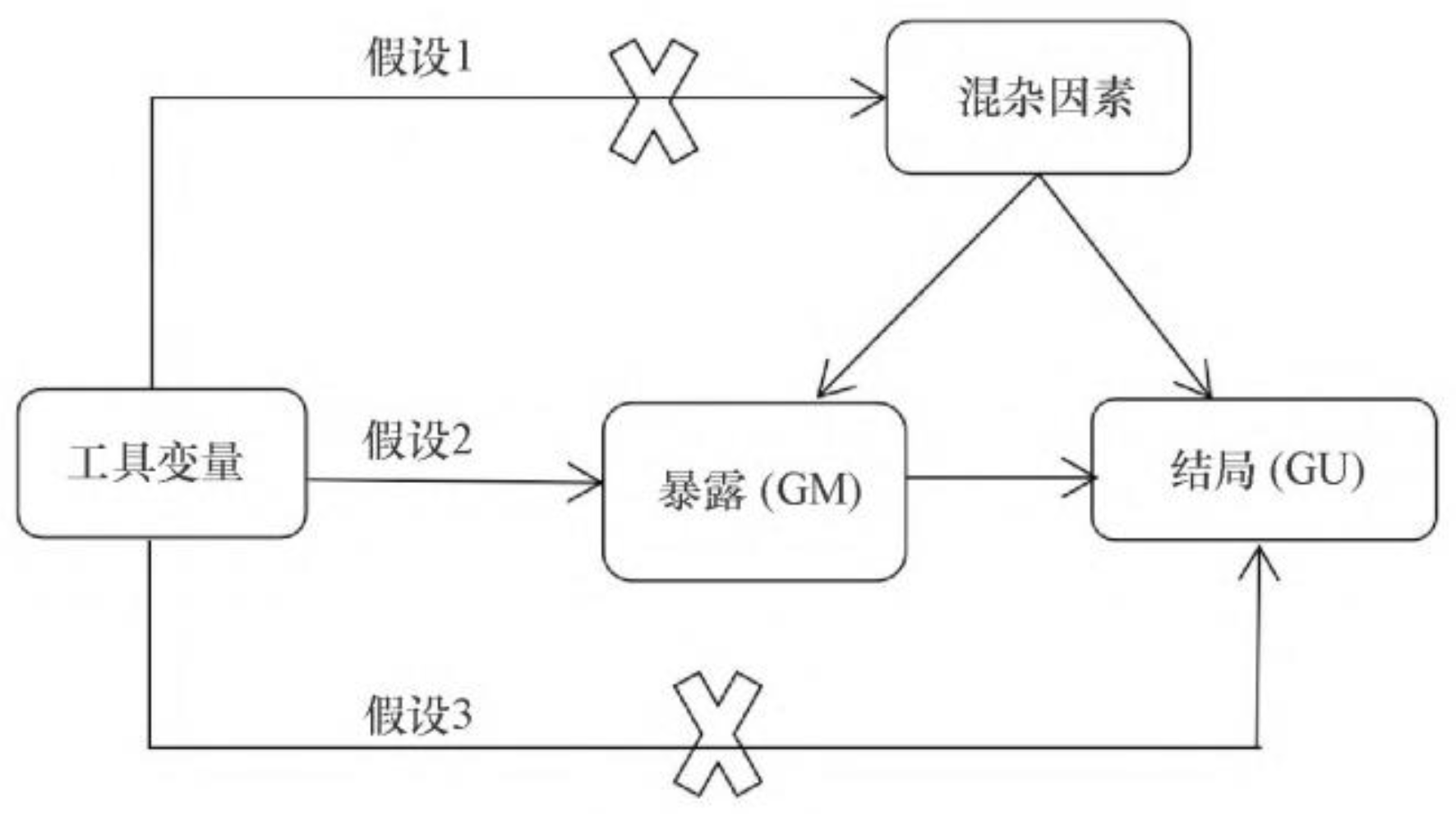
Figure 1.
The MR Model
1.
Figure 1.
The MR Model
1.
2.2. Data Source
2.2.1. Intestinal Microbe Dataset
GWAS data from the Dutch LifeLines cohort were collected, involving 207 intestinal microbial taxa and 205 functional pathways. LifeLines is a large-scale prospective study with complete fecal and phenotypic information from 7,738 participants, covering 5,584,686 SNPs. Based on further metagenomic sequencing data, an in-depth analysis of microbial composition was conducted and subjected to rigorous quality control.
2.2.2. CRC Dataset
GWAS summary data from the Finnish Biobank (FinnGen) were adopted, encompassing CRC across various pathological locations and stages. The specific data include CRC (CRC, IV=221,814), colon malignancy (CC, IV=220,595), rectal malignancy (CR, IV=219,870), benign colorectal tumors (BCR, IV=228,104), benign colon tumors (BC, IV=not mentioned), and benign rectal tumors (BR, IV=220,900). The FinnGen project aims to investigate the genomes of 500,000 Finns and data from the National Health Registry, currently covering 224,737 genotypic and phenotypic participants. The study identified genome-wide significant associations at 2,491 independent loci based on 1,932 clinical endpoints defined by the National Health Registry. Researchers applied mixed-effects logistic regression to adjust for age, sex, ten principal components, and genotype batch to ensure the accuracy of results.
Table 1.
Detailed Information of GWAS Data.
Table 1.
Detailed Information of GWAS Data.
| Type |
Phenotype |
Sample Size |
GWAS ID |
Institution |
Race |
| CRC |
colorectal malignant tumor |
221,814 |
fmn-b-c3 |
FinnGen |
European |
| |
colon malignancy |
220,595 |
fmn-b-c3 |
FinnGen |
European |
| |
rectal malignancy |
219,870 |
fmn-b-c3 |
FinnGen |
European |
| |
benign colorectal tumors |
228,104 |
finn-b-CD2 |
FinnGen |
European |
| |
benign colon tumors |
218,792 |
finn-b-CD2 |
FinnGen |
European |
| |
benign rectal tumors |
220,900 |
finn-b-CD2 |
FinnGen |
European |
| Intestinal microbes |
Microbial taxa |
7,738 |
NA |
NA |
European |
| |
Microbial functional pathways |
7,738 |
NA |
NA |
European |
| Other diseases |
Crohn's disease |
211,107, 211,268 |
finn-b-CHRONLARGE, finn-b-CHRONSMALL |
FinnGen |
European |
| |
Ulcerative colitis |
214,620 |
fmn-b-KLL_ULCER |
FinnGen |
European |
| |
Irritable bowel syndrome |
187,028 |
fmn-b-KLL_IBS |
FinnGen |
European |
| |
Non-alcoholic fatty liver disease |
218,792 |
fmn-b-NAFLD |
FinnGen |
European |
| |
Type 2 diabetes |
215,654 |
fmn-b-E4_DM2 |
FinnGen |
European |
| Dietary habits |
Alcohol consumption status |
360,726 |
ukb-d-2 |
NA |
European |
| |
Bread intake |
452,236 |
ukb-b-11348 |
MRC-IEU |
European |
| |
Grain type: Biscuits |
299,898 |
ukb-d-14682 |
NA |
European |
| |
Lamb/Lamb Intake |
460,006 |
ukb-b-14179 |
MRC-IEU |
European |
| |
Ferritin |
23,986 |
ieu-a-1050 |
GIS |
European |
| |
Liver intake |
64,944 |
ukb-b-6373 |
MRC-IEU |
European |
| |
The type of milk used |
360,806 |
ukb-d-14186 |
NA |
European |
| |
Minerals and other dietary supplements |
336,314 |
ukb-a-495 |
Neale Lab |
European |
| |
Single layer pastry intake |
64,949 |
ukb-b-2024, ukb-b-11189 |
MRC-IEU |
European |
| |
Vitamin and trace elements |
335,591, 460,351 |
ukb-a-464, ukb-b-15175 |
Neale Lab, MRC-IEU |
European |
2.3. Instrumental Variable (IV) Screening
A series of stringent quality control measures were implemented to screen effective SNPs, thus accurately assessing the causal relationships between intestinal microbes and specific health outcomes. First, a low-significance threshold (lxl, T5) was set to preliminarily screen SNPs that were highly correlated with microbial traits. Subsequently, PLINK software was used to exclude SNPs with linkage disequilibrium (r < 0.01, focal distance = 10,000 kb), as well as those with a minor allele frequency (MAF) less than 0.01 and non-derived allele frequency palindromic SNPs [
14]. Furthermore, F-statistics were calculated to quantify the strength of IVs according to MR assumptions, eliminating weak IVs with F-statistics less than ten [
15]. Additionally, each SNP in the PhenoScanner GWAS database was evaluated for direct association with the outcomes to decline confounding factors. Those found to be directly related were removed to meet the assumptions of MR analysis. Ultimately, SNPs satisfying the conditions were selected for the MR analysis [
16].
2.4. Univariate MR Analysis
The univariate MR analysis was employed with selected high-quality SNPs as IVs. Various MR estimation methods, including inverse variance weighting (IVW), Wald ratio, and MR Egger, were used to assess the potential causal relationships between intestinal flora traits and CRC across different pathological locations and stages. Each method calculated corresponding statistics and P-values to identify microbial traits with significant or potential associations. Additionally, the MR-BMA method was applied to rank the causal importance of potential biomarkers, further confirming the causal relationships between key microbial taxa and CRC.
2.5. Two-Sample MR Analysis
MR analysis software (TwoSampleMR) was used to verify the specificity of microbial biomarkers closely related to the development of CRC. First, GWAS summary data on six non-CRC diseases (including Crohn's disease of the colon, Crohn's disease of the small intestine, ulcerative colitis, irritable bowel syndrome, non-alcoholic fatty liver disease, and type 2 diabetes) were obtained from the FinnGen database. Subsequently, detailed MR analysis was conducted using these data along with previously identified microbial traits associated with CRC. Parameters were first set, including but not limited to the type of MR estimates (e.g., IVW), methods for adjusting effect sizes, and criteria for determining significance levels. Corrected P-values were then utilized to assess statistical significance, with a threshold set at P < 0.2, aiming to clearly identify microbial biomarkers specifically related to the development of CRC [
17].
Figure 2.
The MR Analysis Framework.
Figure 2.
The MR Analysis Framework.
3. Result
3.1. IV
Appropriate IVs are crucial for exploring the potential causal relationships between intestinal microbes and CRC across diverse pathological locations and stages. This study conducted rigorous screening of a large number of genetic variants to ensure they could serve as effective IVs representing intestinal flora traits. Finally, 3,310 SNPs were retained, which were closely related to 412 microbial traits, encompassing 207 microbial taxa and 205 functional pathways. Each microbial trait had at least one SNP as an IV, with a maximum of up to 19. The F-values for SNPs were calculated to assess their strength. All F-statistics are greater than 10, eliminating the possibility of weak IVs. However, further analysis revealed that rs2450114 had a significant association with benign colorectal tumors (BCR) and benign rectal tumors (BR) (P < lxl(T5)). It was excluded from the subsequent MR analysis.
3.2. Univariate MR Analysis
After selecting appropriate IVs, various univariate MR methods were applied to explore the potential causal relationships between intestinal microbial markers and CRC across different pathological locations and stages. Through IVW (M), Wald ratio, and MR Egger, multiple groups of intestinal flora diversity were identified with significant associations with CRC and its subtypes (CRC, colon cancer, rectal cancer, rectosigmoid colon cancer [BCR], unspecified rectal cancers [BC], and unspecified rectal cancers [BR]) (P < 0.05). Notably, 12 and 20 microbial traits having significant causal relationships with CRC and its subtypes were discovered (IVW(M) < 0.2), respectively, revealing differences in microbial markers between the colon and rectum.
A total of 12 key microbial traits were identified in CRC, including but not limited to Clostridium, Bacteroides, and Bifidobacterium. These floras significantly influenced the occurrence and progression of CRC through distinct metabolic activities and interactions with the host. For colon cancer, major associated floras were concentrated in Ruminococcus and Lactobacillus, which may play a key regulatory role in the local environment of the colon. Regarding rectal cancer and its subtypes, more complex distribution patterns were observed, such as significant associations between benign colorectal tumors and Akkermansia and potential links between unspecified rectal cancers and benign rectal tumors with Prevotella and Fusobacterium. These findings provide valuable insights into the mechanisms underlying CRC.
The univariate MR analysis identified 19 to 29 intestinal microbial taxa that had potential causal relationships with CRC, colon cancer, rectal cancer, rectosigmoid colon cancer, unspecified rectal cancers, and BR. Among them, Clostridium, whose metabolites influence intestinal immunity and inflammation, was related to the overall risk of CRC; Bacteroides, promoting intestinal health through polysaccharide degradation, was significantly associated with the subtypes of colon cancer; Bifidobacterium, a probiotic that regulates intestinal flora balance, showed potential benefits for various CRC subtypes. Additionally, Ruminococcus exhibited prominent characteristics in rectal cancer and rectosigmoid colon cancer, affecting the local environment through cellulose degradation; Lactobacillus was commonly linked to the subtypes of CRC, enhancing intestinal barrier function.
In order to probe into the causal relationships between these microbes and disease outcomes, a potential biomarker ranking method based on MR-BMA was utilized, with a prior probability of 0.1 as a benchmark for causal importance ranking. Upon stringent screening and pruning, 14 to 23 microbial taxa were analyzed and measured by 87 to 157 genetic variants as IVs. Preliminary MR-BMA analysis revealed significant effects of specific SNPs (e.g., rs12736307 and rs76321722) on colon cancer and rectal cancer outcomes. Through further adjustment for potential confounders (such as age, sex, and lifestyle), more robust and reliable results were obtained. The MR-BMA analysis indicated that the previously identified key SNPs maintained significant associations with CRC subtypes, with their effect sizes and directions remaining substantially unchanged.
Table 2.
MR-BMA Ranking of MIP and MACE for the Microbial Biomarkers of CRC.
Table 2.
MR-BMA Ranking of MIP and MACE for the Microbial Biomarkers of CRC.
| Microbial Biomarker |
MIP |
MACE |
Pval |
| Pseudoflavonifractor |
0.906 |
0.257 |
0.020 |
| Streptococcus thermophilus |
0.562 |
-0.106 |
0.020 |
| Coprococcus catus |
0.433 |
-0.114 |
0.010 |
| Parabacteroides distasonis |
0.076 |
0.011 |
0.347 |
| Erysipelotrichaceae |
0.065 |
-0.010 |
0.535 |
| Sutterella wadsworthensis |
0.056 |
-0.009 |
0.802 |
| Streptococcus |
0.055 |
-0.003 |
0.455 |
| Roseburia inulinivorans |
0.029 |
0.002 |
1.000 |
| Flavonifractor |
0.028 |
0.002 |
0.960 |
| Gammaproteobacteria |
0.026 |
-0.001 |
1.000 |
Table 3.
MR-BMA Ranking of MIP and MACE for Colonic Malignant Tumors.
Table 3.
MR-BMA Ranking of MIP and MACE for Colonic Malignant Tumors.
| Microbial Biomarker |
MIP |
MACE |
Pval |
| Gammaproteobacteria |
0.204 |
0.049 |
0.089 |
| Eubacterium ventriosum |
0.169 |
-0.025 |
0.030 |
| Holdemania unclassified |
0.148 |
-0.020 |
0.059 |
| Sutterella wadsworthensis |
0.136 |
-0.024 |
0.158 |
| Prevotella copri |
0.088 |
0.012 |
0.188 |
| Roseburia inulinivorans |
0.082 |
0.012 |
0.198 |
| Betaproteobacteria |
0.078 |
-0.010 |
0.366 |
| Paraprevotella xylaniphila |
0.071 |
-0.007 |
0.188 |
| Sutterellaceae |
0.071 |
-0.008 |
0.495 |
| Parabacteroides goldsteinii |
0.064 |
0.005 |
0.079 |
3.3. Two-Sample MR Analysis
Through the results of two-sample univariate MR analysis and potential biomarker ranking, several key microorganisms were identified to have potential relationships with CRC and its subtypes. For colon cancer, emopMw, as a unique microbial strain, exhibited significant protective effects by modulating the intestinal microenvironment, promoting beneficial bacterial growth, and inhibiting the proliferation of harmful bacteria [
18]. Its protective effects primarily stem from reducing inflammatory responses and oxidative stress in the intestine, thereby maintaining the health of colon cells [
19].
In the subgroup of rectal cancer, members of the Erysipelotrichaceae family are particularly notable. By enhancing intestinal barrier function and reducing local inflammatory responses, they protect rectal health [
20]. This protective effect is not only evident in the prevention of CRC but also significantly lowers the risk of inflammatory bowel disease (IBS), further validating their extensive benefits for intestinal health.
In contrast, for CRC subtypes, including rectosigmoid colon cancer (BCR) and basal cell carcinoma (BC), microbial species such as Orato were identified as potential risk factors. These microbes may increase the risk of specific CRC subtypes by promoting inflammation, disrupting the intestinal barrier, or producing harmful metabolites [
21].
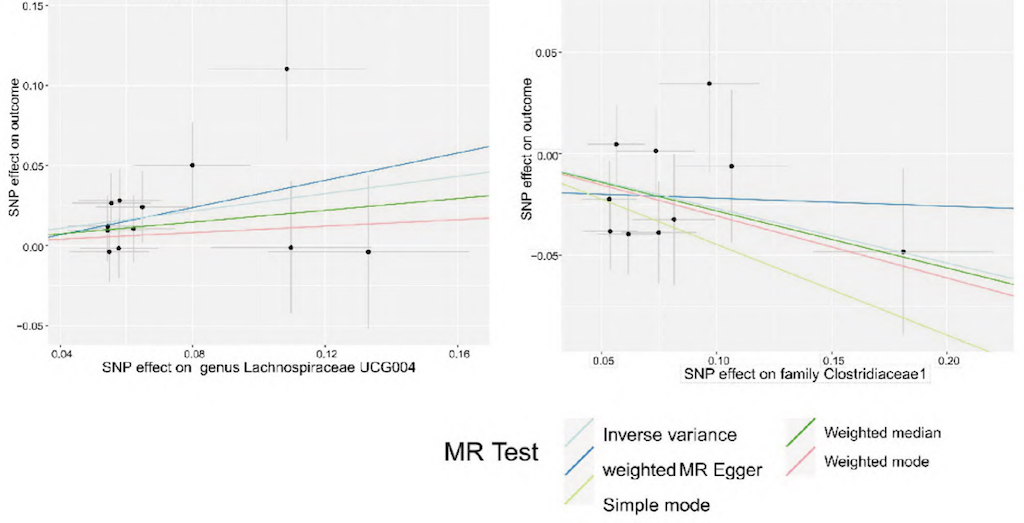
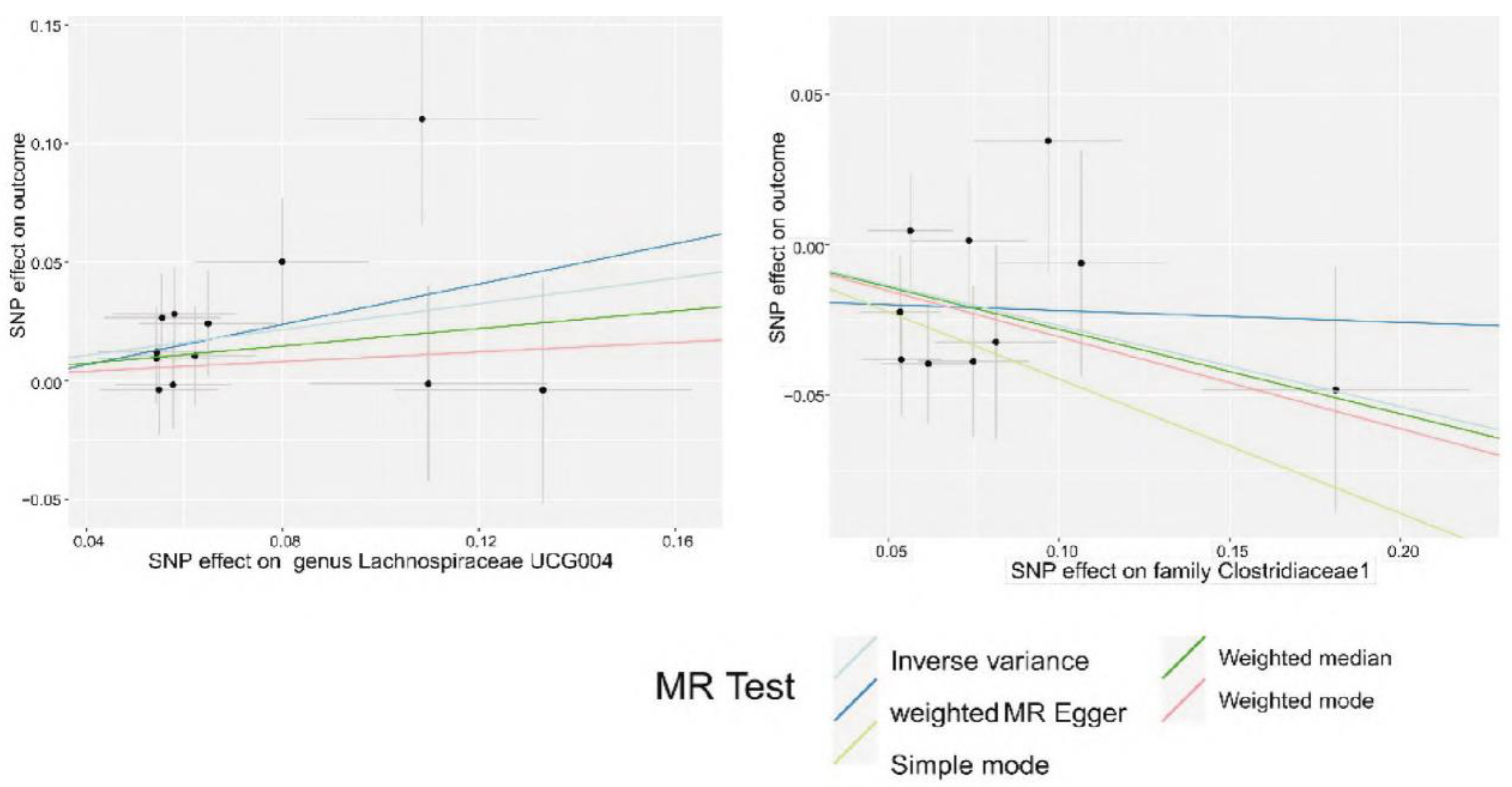
Figure 3.
The scatter plot of five MR models of intestinal microbes causally related to CRC.
Figure 3.
The scatter plot of five MR models of intestinal microbes causally related to CRC.
The two-sample univariate MR analysis was conducted to further explore whether these microbes could serve as specific biomarkers for CRC. The results showed that Erysipelotrichaceae significantly reduced the risk of IBS (OR = 0.897, 95% CI = 0.844-0.954, P = 0.0006), indicating its potentially positive role in preventing IBS. Additionally, Streptococcus thermophilus, Eubacterium ventriosum, Streptococcus, Coprococcus sp. strain ART55-1, Eubacterium siraeum, Bacteroides ovatus, and Bifidobacterium ado/cew were identified as specific biomarkers associated with CRC. These findings provide novel perspectives and potential targets for the diagnosis and treatment of CRC.
4. Discussion
While prior MR studies have investigated the causal relationships between intestinal microbiota and CRC, this study presents a more exhaustive analysis. It not only examined the causal link between intestinal microbes and CRC but also delved into specific associations with the disease's pathological location and stage. It identified and ranked causal microbial biomarkers across various cancer subsites and stages. Leveraging univariate MR and MR-BMA analyses. These findings were validated through multiple population cohort datasets.
Through rigorous screening and analysis, this study elucidated the potential causal relationships between intestinal microbial biomarkers and CRC and its subtypes, providing a valuable reference for CRC pathogenesis. Multiple microbial traits significantly associated with CRC were confirmed, demonstrating associations not only with CRC overall but also with specific subtypes, such as colon cancer and rectal cancer.
The identified key microbes, such as EmopMw and Erysipelotrichaceae, exhibited notable potential protective effects. Importantly, these strains have been reported to modulate intestinal immunity and promote intestinal barrier health, aligning with the protective effects observed in CRC [
22]. Notably, this study is pioneering in establishing a direct link between these strains and CRC subtypes and quantifying their causal effects on CRC outcomes, a feat not accomplished by previous research. Meanwhile, some microbial species, such as Orato et al., were found to be associated with an increased risk of specific CRC subtypes, contradicting some findings. This discrepancy underscores the complexity and heterogeneity of microbial communities in CRC development [
23,
24]. Contributing factors may include variations in study design, sample selection, analytical methods, and population characteristics, highlighting the necessity for further exploration of the intricate network of microbe-host interactions [
25].
Additionally, the MR-BMA analysis revealed the involvement of SNPs in CRC and its subtypes, offering new avenues for genetic research and laying a theoretical foundation for personalized CRC treatment. However, research in this domain remains in its infancy, necessitating large-scale interdisciplinary studies to validate these findings and investigate the specific mechanisms underlying microbe-gene-environment interactions in CRC development.
Funding
I have full access to all of the data in this study and I take full responsibility for the integrity of the data and the accuracy of the data analyses. And there was no Funding.
Institutional Review Board Statement
Ethics, Consent to Participate, and Consent to Publish declarations: not applicable.
References
- Zhao, W.; Yin, Z.; Wang, Y.; Li, W. Interpretation of 2024 American Cancer Statistics Report and Comparison of Cancer Epidemiology between China and the U.S. Cancer Research on Prevention and Treatment, 1–12.
- Tang, M.; Wang, X.; Li, X. . Research Progress on Statins in the Prevention and Treatment of Colorectal Cancer. The Journal of Practical Medicine 2024, 40, 2041–2046. [Google Scholar]
- Wang, G.; Zhang, G.; Tang, J.; Zhang, Y.; Yang, Y.; Wang, J. The Impact of SOX4 on Immune Evasion and Cell Migration in Colorectal Cancer Cells by Targeting miR-17 Expression Levels. Laboratory Medicine and Clinic 2024, 21, 1825–1830. [Google Scholar]
- Yi, D.; Tian, X. Screening Results of Colorectal Cancer in Fecal DNA Positive Cases in the Wuchang Area of Wuhan from 2021 to 2022. Journal of Public Health and Preventive Medicine 2024, 35, 103–106. [Google Scholar]
- Liu, Z.; Zhang, B.; Li, S. Application Effects of Bifidobacterium Tri-Active Capsules in Postoperative Chemotherapy for Colorectal Cancer. Chinese Journal of Clinical Rational Drug Use 2024, 17, 8–11. [Google Scholar]
- He, R.; Zhang, Y.; Li, P.; Song, G.; Li, A.; Wang, Y. Progress in Imaging Assessment of Perineural Invasion in Rectal Cancer. Chinese Journal of Digestion and Medical Imageology (Electronic Edition) 2024, 14, 378–381. [Google Scholar]
- Chen, L.; Lu, N.; Chen, H. Cardiovascular Disease Risk Factors Associated with Prognosis in Esophageal and Colorectal Cancer Patients after Surgery. Chinese Journal of Hypertension (Bilingual Edition), 1–7.
- Zhao, J. Study on the Effects of Different Enteral Nutritional Preparations on Nutritional Status and Gastrointestinal Function in Colorectal Cancer Patients after Radical Surgery. The Journal of Medical Theory and Practice 2024, 37, 2228–2230. [Google Scholar]
- Hou, W.; Li, Y.; Bi, Y.; Han, L.; Ren, X. Research Progress on Lipid Metabolism Reprogramming in Colorectal Cancer. Chinese Journal of Gerontology 2024, 44, 3323–3327. [Google Scholar]
- Wang, W.; Xu, S. Research Progress on Correlation between intestinal flora and Colorectal Cancer. Journal of Anhui Medical University 2024, 59, 1285–1289. [Google Scholar] [CrossRef]
- Liu, Y.; Xu, H.; Ji, G. Bile Acids Driving Gastrointestinal Inflammation-Cancer Transformation and Traditional Chinese Medicine Prevention Strategies. Bulletin of National Natural Science Foundation of China 2024, 38, 461–468. [Google Scholar]
- Han, W.; Fan, Z.; Li, Y.; Miao, L. Changes in Intestinal Flora in Colorectal Cancer Patients and Their Relationship with Serum Inflammatory Factors and Pathological Staging. Clinical Journal of Medical Officer 2024, 52, 641–643. [Google Scholar]
- Xu, J.; Han, L.; Guo, K. Mendelian Randomization Analysis of the Causal Relationship between intestinal flora and Systemic Sclerosis. Progress in Microbiology and Immunology, 1–8.
- Guo, Y.; Cui, Y. Exploring the Causal Relationship between intestinal flora and Insomnia Based on Mendelian Randomization. Chinese General Practice, 1–8.
- Liang, J.; Yang, S.; Wang, H. Exploring the Causal Relationship between Intestinal Flora and Childhood Asthma Based on Mendelian Randomization. West China Medical Journal 2024, 39, 553–560. [Google Scholar]
- Guo, Y.; Cui, Y.; Kong, Y. Causal Relationship between Intestinal Flora and Cluster Headache: A Two-Sample Mendelian Randomization Analysis. Practical Journal of Cardiac Cerebral Pneumal and Vascular Disease 2024, 32, 43–48. [Google Scholar]
- Ma, J.; Zhou, Y.; Chen, W. Two-Sample Bidirectional Mendelian Randomization Analysis of Causal Relationship between Gastroesophageal Reflux Disease and Migraine. Journal of Xi'an Jiaotong University (Medical Edition) 2024, 45, 262–270. [Google Scholar]
- Chai, J.; Li, S.; Li, W. intestinal flora and Drug-Related Avascular Necrosis: A Two-Sample Bidirectional Mendelian Randomization Analysis. Chinese Journal of Tissue Engineering Research 2024, 28, 4325–4331. [Google Scholar]
- Wu, M.; An, Z.; He, Y. Exploring the Causal Relationship between intestinal Microbes and Non-Alcoholic Fatty Liver Disease Using Mendelian Randomization. Acta Universitatis Medicinalis Anhui 2024, 59, 236–242. [Google Scholar]
- Wang, X.; Chai, R. The Causal Relationship between Intestinal Flora and Allergic Diseases: Insights from Bidirectional Two-Sample Mendelian Randomization Analysis. Chinese Journal of Allergy & Clinical Immunology 2023, 17, 579. [Google Scholar]
- Yu, L.; Zhang, M. Progress in the Application of Mendelian Randomization Analysis in Diabetic Retinopathy. Chronic Pathematology Journal 2023, 24, 1790–1793. [Google Scholar]
- Wei, Y.; Lan, Y.; Huang, X. A Study on the Association between Intestinal Flora and Frailty Based on Two-Sample Mendelian Randomization Analysis. Guangxi Medical Journal 2023, 45, 2880–2885. [Google Scholar]
- Dong, X.; Xue, J.; Liang, X. The Application of Mendelian Randomization in the Study of Risk Factors for Primary Carcinoma of Liver. Infectious Disease Information 2023, 36, 473–476. [Google Scholar]
- Ren, Q.; He, W. Two-Sample Mendelian Randomization Analysis of Causal Relationship between Intestinal Flora and Lung Cancer. Chinese Journal of Clinical Thoracic and Cardiovascular Surgery 2023, 30, 1618–1627. [Google Scholar]
- Wang, C.; Sun, X.; Chen, W. The Relationship between Type 2 Diabetes and Sepsis: A Two-Sample Mendelian Randomization Study Based on GWAS Database. Chinese Journal of Critical Care Medicine 2023, 43, 794–799. [Google Scholar]
- Wang, D. Mendelian Randomization Study of intestinal flora and Metabolic Cardiovascular Disease. Central South University, 2023. [Google Scholar]
- Lü, B. Systematic Genetic Studies of Intestinal Flora and Associated Diseases. Huazhong Agricultural University, 2020. [Google Scholar]
| 1 |
[Peng Liuqing. Comparison and Applications of Mendelian Randomization Methods for Correcting Weak Instrument Bias[D]. Shanxi Medical University, 2023.] |
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).