Submitted:
29 August 2024
Posted:
30 August 2024
Read the latest preprint version here
Abstract
Keywords:
Background
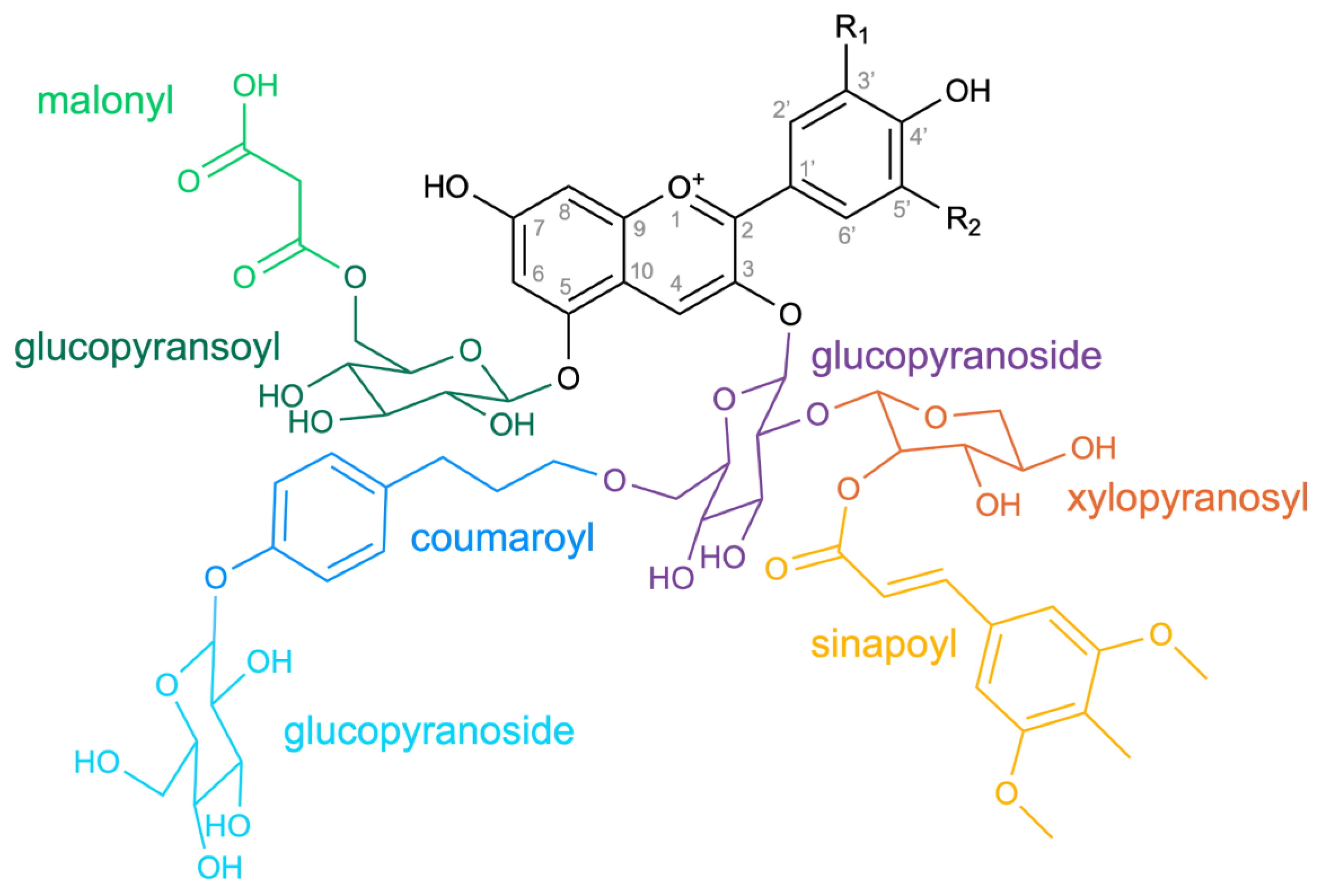
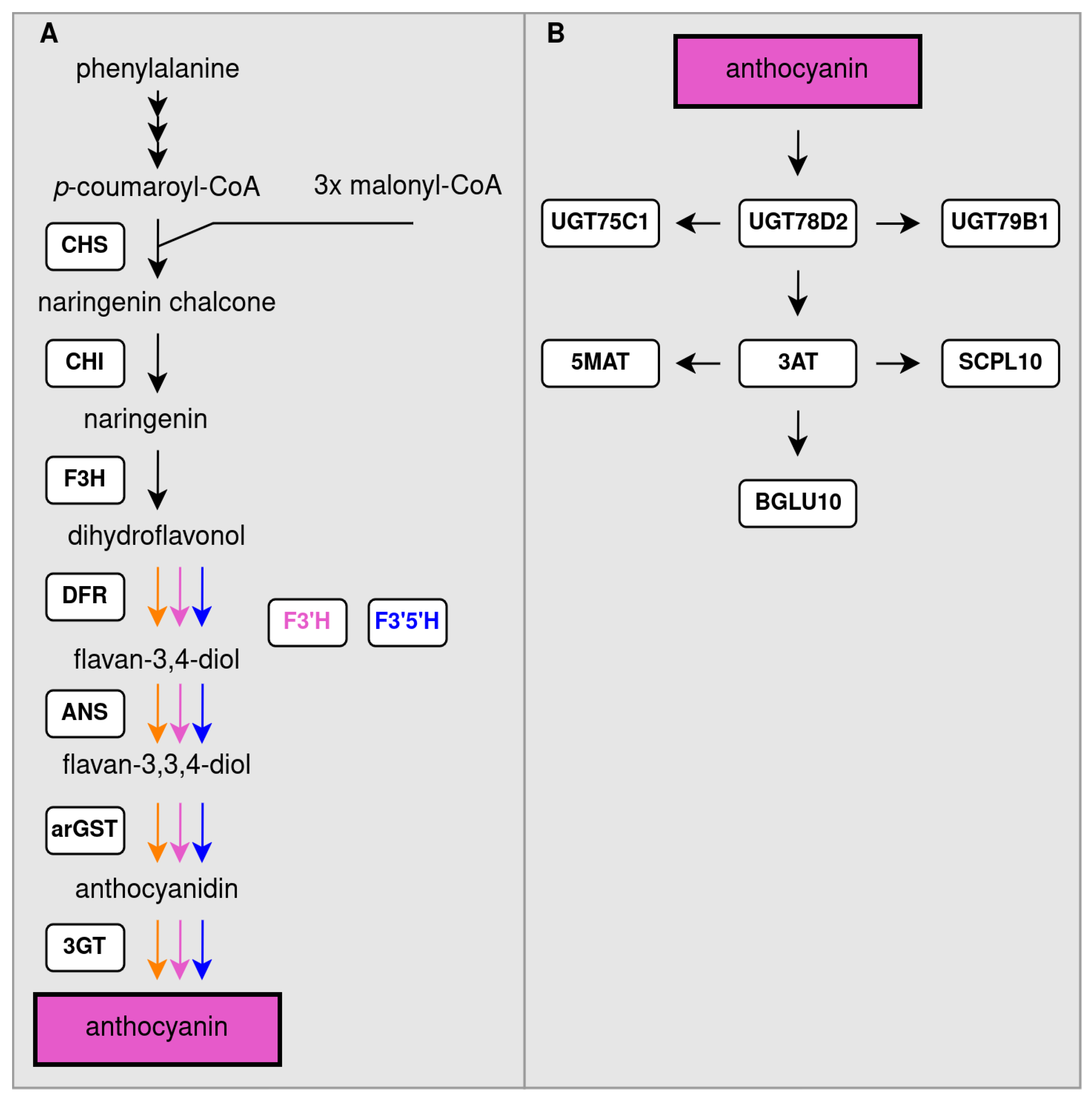
Biosynthesis of a Diverse Set of Anthocyanins
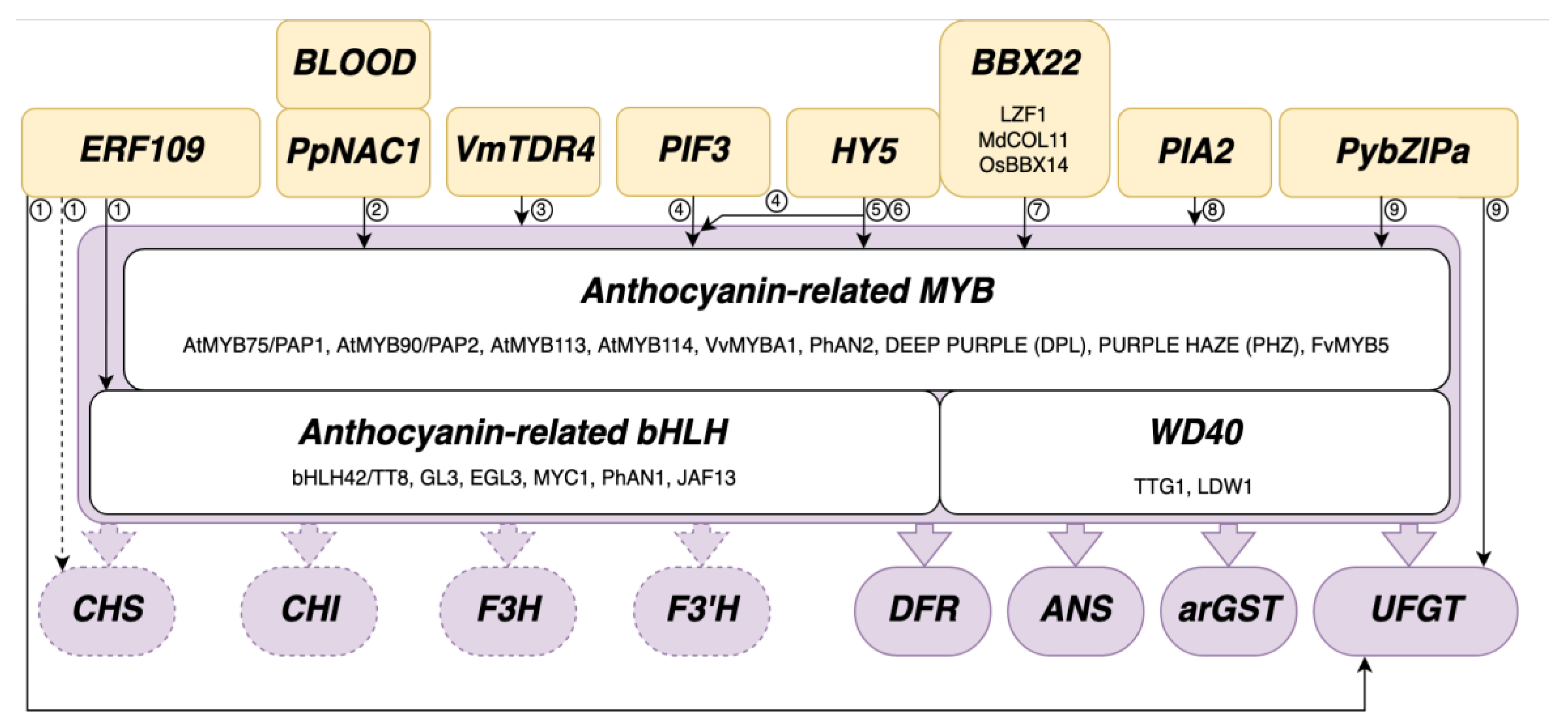
Transcriptional Regulation of Anthocyanin Biosynthesis
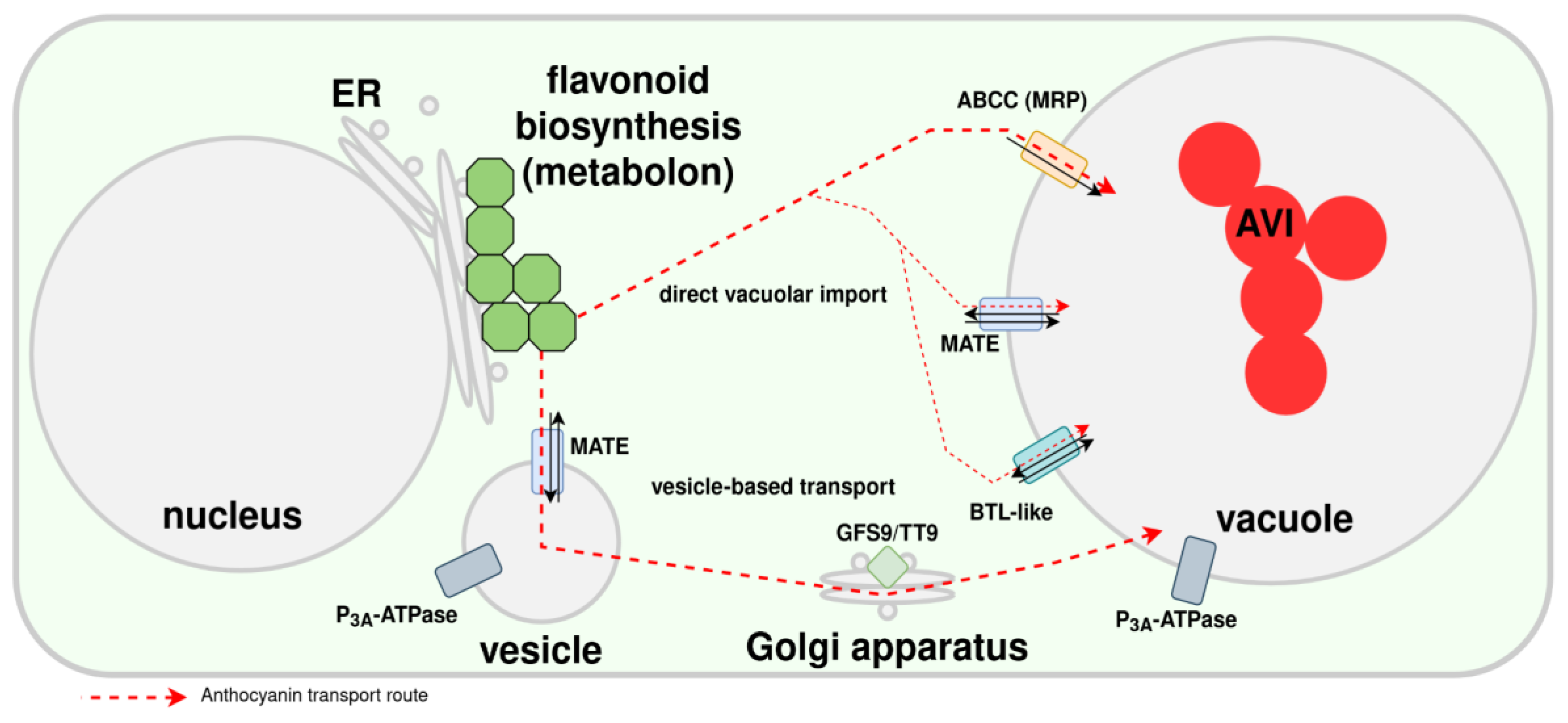
Transport of Anthocyanins
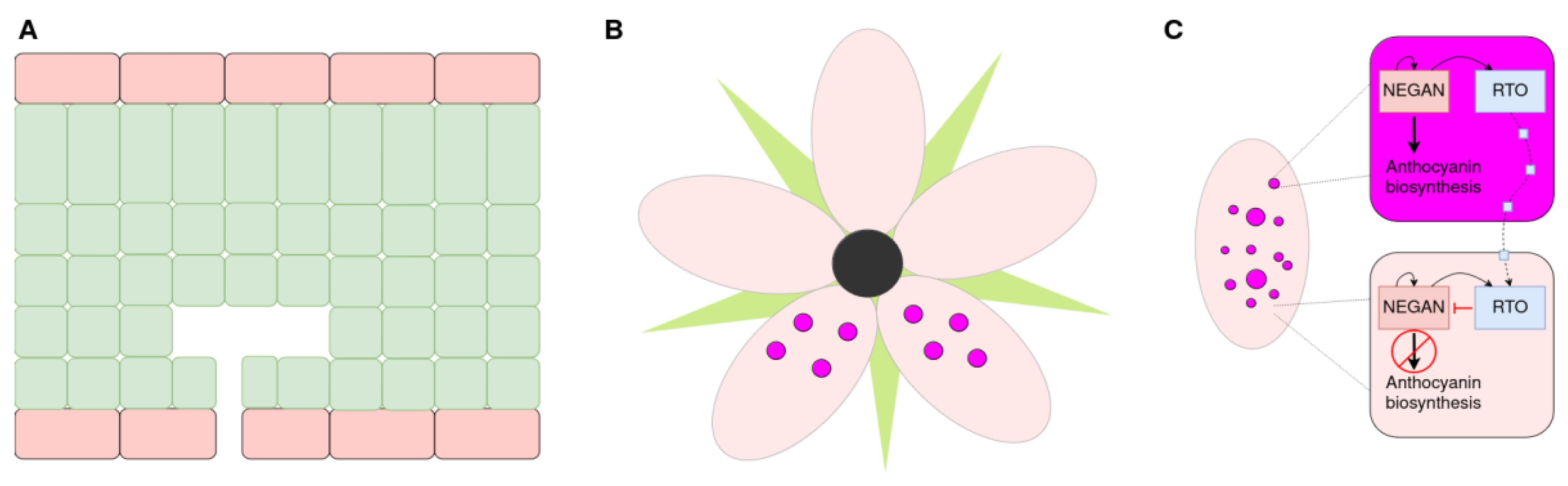
Cell-Specific Accumulation of Anthocyanins and Pigmentation Patterns
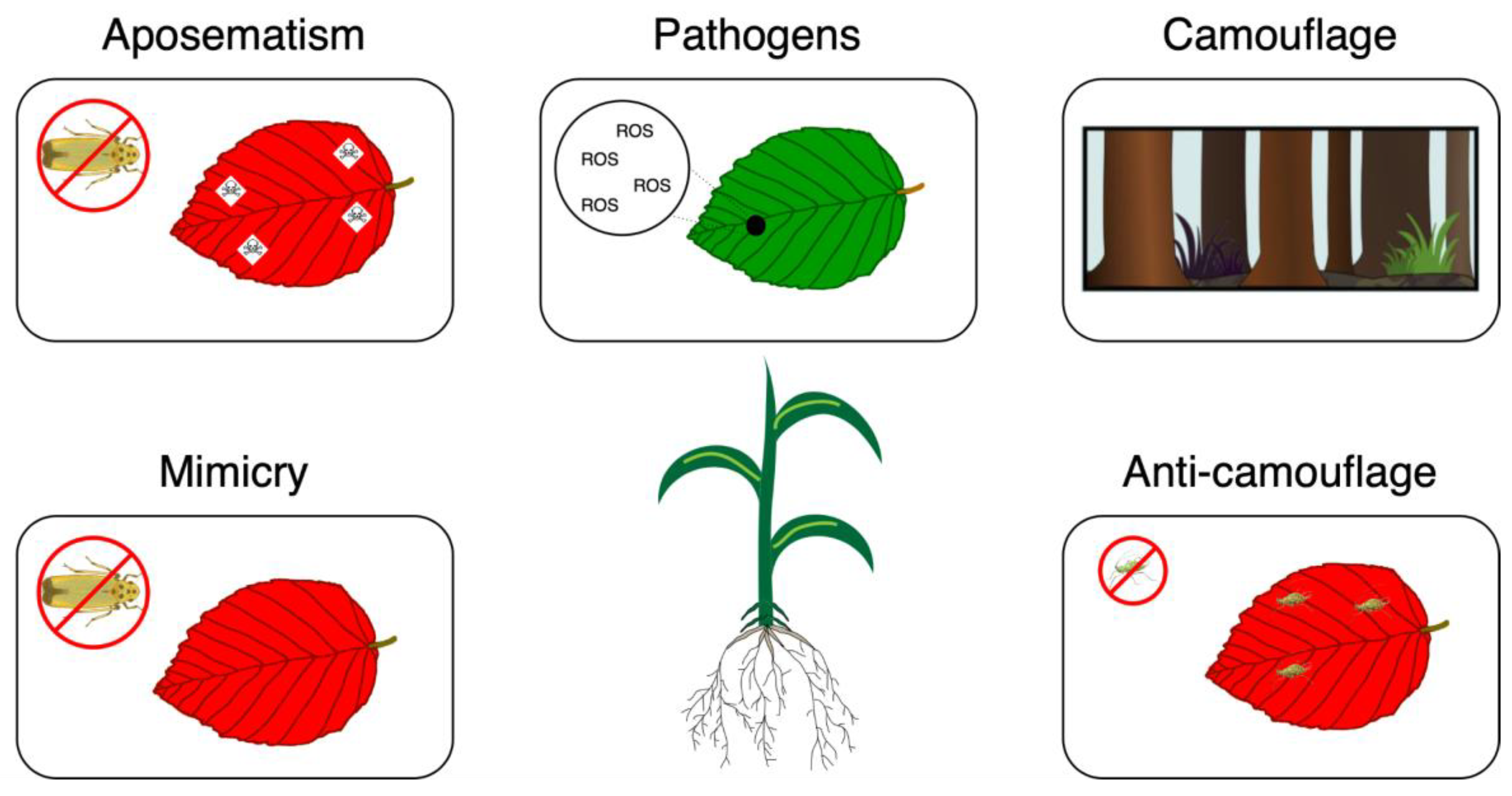
Ecological Functions of Anthocyanins
Protective Functions of Anthocyanins in Photosynthetically Active Plant Organs
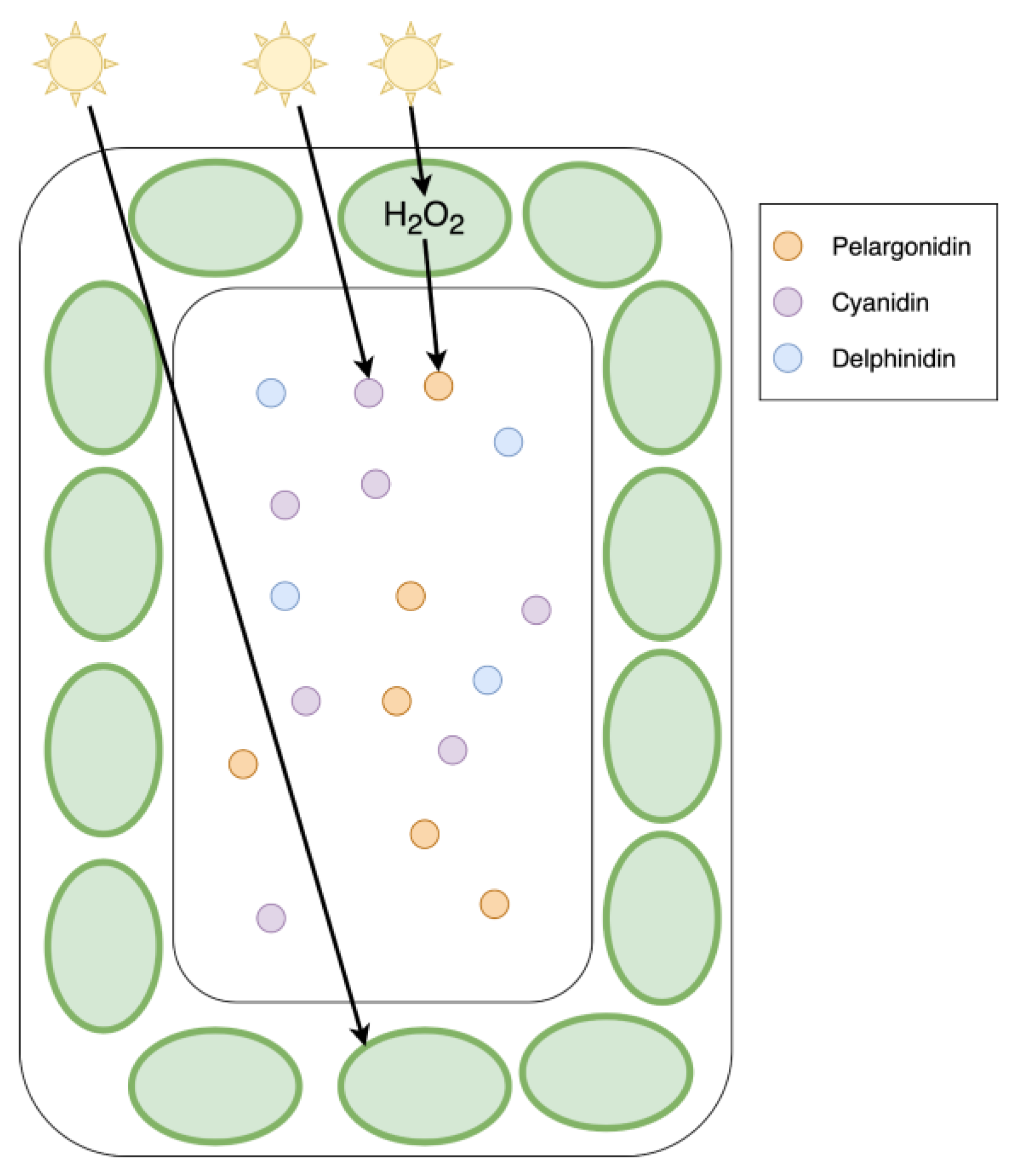
Importance of Anthocyanins in Drought and Salt Stress Response
Cold Stress Response
Anthocyanin Accumulation as Sign of Nutritional Imbalance
Pollinator Attraction
Seed Disperser Attraction
Herbivore Repellence and Pathogen Resistances
Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Abid MA, Wei Y, Meng Z, Wang Y, Ye Y, Wang Y, He H, Zhou Q, Li Y, Wang P, et al.2022. Increasing floral visitation and hybrid seed production mediated by beauty mark in Gossypium hirsutum. Plant Biotechnology Journal 20: 1274–1284. [CrossRef]
- Agati G, Brunetti C; M, Di Ferdinando; F, Ferrini; S, Pollastri; M, Tattini. Functional roles of flavonoids in photoprotection: new evidence, lessons from the past. Plant physiology and biochemistry: PPB 2013, 72, 35–45. [Google Scholar] [CrossRef]
- Aharoni A, De Vos CHR; M, Wein; Z, Sun; R, Greco; A, Kroon; JNM, Mol; AP, O’Connell. The strawberry FaMYB1 transcription factor suppresses anthocyanin and flavonol accumulation in transgenic tobacco. The Plant Journal 2001, 28, 319–332. [Google Scholar] [CrossRef] [PubMed]
- Alabd A, Ahmad M; X, Zhang; Y, Gao; L, Peng; L, Zhang; J, Ni; S, Bai; Y, Teng. Light-responsive transcription factor PpWRKY44 induces anthocyanin accumulation by regulating PpMYB10 expression in pear. Horticulture Research 9: uhac199, 2022. [Google Scholar] [CrossRef]
- Albert NW, Davies KM; DH, Lewis; H, Zhang; M, Montefiori; C, Brendolise; MR, Boase; H, Ngo; PE, Jameson; KE, Schwinn. A conserved network of transcriptional activators and repressors regulates anthocyanin pigmentation in eudicots. The Plant Cell 2014a, 26, 962–980. [Google Scholar] [CrossRef] [PubMed]
- Albert NW, Davies KM; KE, Schwinn. Gene regulation networks generate diverse pigmentation patterns in plants. Plant Signaling & Behavior 2014b, 9, e29526. [Google Scholar] [CrossRef]
- Albert NW, Iorizzo M, Mengist MF, Montanari S, Zalapa J, Maule A, Edger PP, Yocca AE, Platts AE, Pucker B, et al.2023. Vaccinium as a comparative system for understanding of complex flavonoid accumulation profiles and regulation in fruit. Plant Physiology 192: 1696–1710. [CrossRef]
- Albert NW, Lewis DH; H, Zhang; LJ, Irving; PE, Jameson; KM, Davies. Light-induced vegetative anthocyanin pigmentation in Petunia. Journal of Experimental Botany 2009, 60, 2191–2202. [Google Scholar] [CrossRef]
- An J-P, Qu F-J; J-F, Yao; X-N, Wang; C-X, You; X-F, Wang; Y-J, Hao. The bZIP transcription factor MdHY5 regulates anthocyanin accumulation and nitrate assimilation in apple. Horticulture Research 2017, 4, 17023. [Google Scholar] [CrossRef]
- An J-P, Zhang X-W; C-X, You; S-Q, Bi; X-F, Wang; Y-J, Hao. MdWRKY40 promotes wounding-induced anthocyanin biosynthesis in association with MdMYB1 and undergoes MdBT2-mediated degradation. New Phytologist 2019, 224, 380–395. [Google Scholar] [CrossRef]
- Andersen TB, Hansen NB; T, Laursen; C, Weitzel; HT, Simonsen. Evolution of NADPH-cytochrome P450 oxidoreductases (POR) in Apiales – POR 1 is missing. Molecular Phylogenetics and Evolution 2016, 98, 21–28. [Google Scholar] [CrossRef]
- Andersen ØM, Jordheim M. The Anthocyanins. In: Flavonoids: Chemistry, Biochemistry and Applications; 2006. [Google Scholar]
- Appelhagen I, Nordholt N; T, Seidel; K, Spelt; R, Koes; F, Quattrochio; M, Sagasser; B, Weisshaar. TRANSPARENT TESTA 13 is a tonoplast P3A-ATPase required for vacuolar deposition of proanthocyanidins in Arabidopsis thaliana seeds. The Plant Journal 2015, 82, 840–849. [Google Scholar] [CrossRef]
- Araguirang GE, Richter AS. Activation of anthocyanin biosynthesis in high light – what is the initial signal? New Phytologist 2022, 236, 2037–2043. [Google Scholar] [CrossRef]
- Archetti M. 2009. Phylogenetic analysis reveals a scattered distribution of autumn colours. Annals of Botany 103: 703–713. [CrossRef]
- Archetti M, Döring TF, Hagen SB, Hughes NM, Leather SR, Lee DW, Lev-Yadun S, Manetas Y, Ougham HJ, Schaberg PG, et al.2009. Unravelling the evolution of autumn colours: an interdisciplinary approach. Trends in Ecology & Evolution 24: 166–173. [CrossRef]
- Armbruster WS. 2002. Can indirect selection and genetic context contribute to trait diversification? A transition-probability study of blossom-colour evolution in two genera. Journal of Evolutionary Biology 15: 468–486. [CrossRef]
- Azuma A, Kobayashi S; H, Yakushui; M, Yamada; N, Mitani; A, Sato. VvmybA1 genotype determines grape skin color. VITIS - Journal of Grapevine Research 2007, 46, 154–154. [Google Scholar] [CrossRef]
- Bai S, Saito T; C, Honda; Y, Hatsuyama; A, Ito; T, Moriguchi. An apple B-box protein, MdCOL11, is involved in UV-B- and temperature-induced anthocyanin biosynthesis. Planta 2014, 240, 1051–1062. [Google Scholar] [CrossRef] [PubMed]
- Bai S, Tao R, Tang Y, Yin L, Ma Y, Ni J, Yan X, Yang Q, Wu Z, Zeng Y, et al.2019a. BBX16, a B-box protein, positively regulates light-induced anthocyanin accumulation by activating MYB10 in red pear. Plant Biotechnology Journal 17: 1985–1997. [CrossRef]
- Bai S, Tao R; L, Yin; J, Ni; Q, Yang; X, Yan; F, Yang; X, Guo; H, Li; Y, Teng. Two B-box proteins, PpBBX18 and PpBBX21, antagonistically regulate anthocyanin biosynthesis via competitive association with Pyrus pyrifolia ELONGATED HYPOCOTYL 5 in the peel of pear fruit. The Plant Journal: For Cell and Molecular Biology 2019b, 100, 1208–1223. [Google Scholar] [CrossRef]
- Ban Z, Qin H; AJ, Mitchell; B, Liu; F, Zhang; J-K, Weng; RA, Dixon; G, Wang. Noncatalytic chalcone isomerase-fold proteins in Humulus lupulus are auxiliary components in prenylated flavonoid biosynthesis. Proceedings of the National Academy of Sciences of the United States of America 2018, 115, E5223–E5232. [Google Scholar] [CrossRef]
- Barbehenn RV, Constabel CP. Tannins in plant-herbivore interactions. Phytochemistry 2011, 72, 1551–1565. [Google Scholar] [CrossRef] [PubMed]
- Barker A, Pilbeam D. Handbook of plant nutrition. 2015. [Google Scholar]
- Behrens CE, Smith KE; CV, Iancu; J, Choe; JV, Dean. Transport of Anthocyanins and other Flavonoids by the Arabidopsis ATP-Binding Cassette Transporter AtABCC2. Scientific Reports 2019, 9, 437. [Google Scholar] [CrossRef]
- Bienert GP, Schjoerring JK; TP, Jahn. Membrane transport of hydrogen peroxide. Biochimica Et Biophysica Acta 2006, 1758, 994–1003. [Google Scholar] [CrossRef]
- Bloor SJ, Abrahams S. The structure of the major anthocyanin in Arabidopsis thaliana. Phytochemistry 2002, 59, 343–346. [Google Scholar] [CrossRef]
- Borevitz JO, Xia Y; J, Blount; RA, Dixon; C, Lamb. Activation Tagging Identifies a Conserved MYB Regulator of Phenylpropanoid Biosynthesis. The Plant Cell 2000, 12, 2383–2393. [Google Scholar] [CrossRef]
- Broeckling BE, Watson RA; B, Steinwand; DR, Bush. Intronic Sequence Regulates Sugar-Dependent Expression of Arabidopsis thaliana Production of Anthocyanin Pigment-1/MYB75. PLOS ONE 2016, 11, e0156673. [Google Scholar] [CrossRef]
- Broucke E, Dang TTV; Y, Li; S, Hulsmans; J, Van Leene; G, De Jaeger; I, Hwang; E, Wim V den; F, Rolland. SnRK1 inhibits anthocyanin biosynthesis through both transcriptional regulation and direct phosphorylation and dissociation of the MYB/bHLH/TTG1 MBW complex. The Plant Journal 2023, 115, 1193–1213. [Google Scholar] [CrossRef] [PubMed]
- Brugliera F, Holton TA; TW, Stevenson; E, Farcy; C-Y, Lu; EC, Cornish. Isolation and characterization of a cDNA clone corresponding to the Rt locus of Petunia hybrida. The Plant Journal 1994, 5, 81–92. [Google Scholar] [CrossRef] [PubMed]
- Burger J, Edwards GE. Photosynthetic Efficiency, and Photodamage by UV and Visible Radiation, in Red versus Green Leaf Coleus Varieties. Plant and Cell Physiology 1996, 37, 395–399. [Google Scholar] [CrossRef]
- Busche M, Acatay C; S, Martens; B, Weisshaar; R, Stracke. Functional Characterisation of Banana (Musa spp.) 2-Oxoglutarate-Dependent Dioxygenases Involved in Flavonoid Biosynthesis. Frontiers in Plant Science 12, 2021. [Google Scholar] [CrossRef]
- Busche M, Pucker B; B, Weisshaar; R, Stracke. Three R2R3-MYB transcription factors from banana (Musa acuminata) activate structural anthocyanin biosynthesis genes as part of an MBW complex. BMC Research Notes 2023, 16, 103. [Google Scholar] [CrossRef]
- Cardi T, Murovec J, Bakhsh A, Boniecka J, Bruegmann T, Bull SE, Eeckhaut T, Fladung M, Galovic V, Linkiewicz A, et al.2023. CRISPR/Cas-mediated plant genome editing: outstanding challenges a decade after implementation. Trends in Plant Science 28: 1144–1165. [CrossRef]
- Cavallini E, Matus JT; L, Finezzo; S, Zenoni; R, Loyola; F, Guzzo; R, Schlechter; A, Ageorges; P, Arce-Johnson; GB, Tornielli. The Phenylpropanoid Pathway Is Controlled at Different Branches by a Set of R2R3-MYB C2 Repressors in Grapevine. Plant Physiology 2015, 167, 1448–1470. [Google Scholar] [CrossRef]
- Cavallini E, Zenoni S; L, Finezzo; F, Guzzo; A, Zamboni; L, Avesani; GB, Tornielli. Functional Diversification of Grapevine MYB5a and MYB5b in the Control of Flavonoid Biosynthesis in a Petunia Anthocyanin Regulatory Mutant. Plant and Cell Physiology 2014, 55, 517–534. [Google Scholar] [CrossRef]
- Chalker-Scott L. 1999. Environmental Significance of Anthocyanins in Plant Stress Responses. Photochemistry and Photobiology 70: 1–9. [CrossRef]
- Chandler VL, Radicella JP; TP, Robbins; J, Chen; D, Turks. Two regulatory genes of the maize anthocyanin pathway are homologous: isolation of B utilizing R genomic sequences. The Plant Cell 1989, 1, 1175–1183. [Google Scholar] [CrossRef]
- Chang CJ, Li Y-H, Chen L-T, Chen W-C, Hsieh W-P, Shin J, Jane W-N, Chou S-J, Choi G, Hu J-M, et al.2008. LZF1, a HY5-regulated transcriptional factor, functions in Arabidopsis de-etiolation. The Plant Journal: For Cell and Molecular Biology 54: 205–219. [CrossRef]
- Chen Y, Huang Z; L, Tang. Invisible red in young leaves: Anthocyanin likely plays a defensive role in some other way beyond visual warning. Flora 2021, 280, 151833. [Google Scholar] [CrossRef]
- Chen Q, Man C; D, Li; H, Tan; Y, Xie; J, Huang. Arogenate Dehydratase Isoforms Differentially Regulate Anthocyanin Biosynthesis in Arabidopsis thaliana. Molecular Plant 2016, 9, 1609–1619. [Google Scholar] [CrossRef]
- Chialva C, Blein T; M, Crespi; D, Lijavetzky. Insights into long non-coding RNA regulation of anthocyanin carrot root pigmentation. Scientific Reports 2021, 11, 4093. [Google Scholar] [CrossRef]
- Chopy M, Cavallini-Speisser Q; P, Chambrier; P, Morel; J, Just; V, Hugouvieux; S, Rodrigues Bento; C, Zubieta; M, Vandenbussche; M, Monniaux. Cell layer–specific expression of the homeotic MADS-box transcription factor PhDEF contributes to modular petal morphogenesis in petunia. The Plant Cell 2024, 36, 324–345. [Google Scholar] [CrossRef] [PubMed]
- Choudhary N, Pucker B. Conserved amino acid residues and gene expression patterns associated with the substrate preferences of the competing enzymes FLS and DFR. : 2023.11.05.565693; 2023, 2024. [Google Scholar] [CrossRef]
- Christie PJ, Alfenito MR; V, Walbot. Impact of low-temperature stress on general phenylpropanoid and anthocyanin pathways: Enhancement of transcript abundance and anthocyanin pigmentation in maize seedlings. Planta 1994, 194, 541–549. [Google Scholar] [CrossRef]
- Cone KC, Burr FA; B, Burr. Molecular analysis of the maize anthocyanin regulatory locus C1. Proceedings of the National Academy of Sciences of the United States of America 1986, 83, 9631–9635. [Google Scholar] [CrossRef]
- Cook AD, Atsatt PR; CA, Simon. Doves and Dove Weed: Multiple Defenses against Avian Predation. BioScience 1971, 21, 277–281. [Google Scholar] [CrossRef]
- Costa-Arbulú C, Gianoli E; WL, Gonzáles; HM, Niemeyer. Feeding by the aphid Sipha flava produces a reddish spot on leaves of Sorghum halepense: an induced defense? Journal of Chemical Ecology 2001, 27, 273–283. [Google Scholar] [CrossRef]
- Davies KM, Albert NW; KE, Schwinn; KM, Davies; NW, Albert; KE, Schwinn. From landing lights to mimicry: the molecular regulation of flower colouration and mechanisms for pigmentation patterning. Functional Plant Biology 2012, 39, 619–638. [Google Scholar] [CrossRef] [PubMed]
- Del Valle JC, Alcalde-Eon C; MT, Escribano-Bailón; ML, Buide; JB, Whittall; E, Narbona. Stability of petal color polymorphism: the significance of anthocyanin accumulation in photosynthetic tissues. BMC Plant Biology 2019, 19, 496. [Google Scholar] [CrossRef]
- Delph LF, Lively CM. THE EVOLUTION OF FLORAL COLOR CHANGE: POLLINATOR ATTRACTION VERSUS PHYSIOLOGICAL CONSTRAINTS IN FUCHSIA EXCORTICATA. Evolution; International Journal of Organic Evolution 1989, 43, 1252–1262. [Google Scholar] [CrossRef]
- Deng G-M, Zhang S, Yang Q-S, Gao H-J, Sheng O, Bi F-C, Li C-Y, Dong T, Yi G-J, He W-D, et al.2021. MaMYB4, an R2R3-MYB Repressor Transcription Factor, Negatively Regulates the Biosynthesis of Anthocyanin in Banana. Frontiers in Plant Science 11.
- D’Hooghe P, Escamez S; J, Trouverie; J-C, Avice. Sulphur limitation provokes physiological and leaf proteome changes in oilseed rape that lead to perturbation of sulphur, carbon and oxidative metabolisms. BMC Plant Biology 2013, 13, 23. [Google Scholar] [CrossRef]
- Ding B, Patterson EL, Holalu SV, Li J, Johnson GA, Stanley LE, Greenlee AB, Peng F, Bradshaw HD, Blinov ML, et al.2020. Two MYB Proteins in a Self-Organizing Activator-Inhibitor System Produce Spotted Pigmentation Patterns. Current biology: CB 30: 802-814.e8. [CrossRef]
- Dodd IC, Critchley C; GS, Woodall; GR, Stewart. Photoinhibition in differently coloured juvenile leaves of Syzygium species. Journal of Experimental Botany 1998, 49, 1437–1445. [Google Scholar] [CrossRef]
- Dubos C, Le Gourrierec J; A, Baudry; G, Huep; E, Lanet; I, Debeaujon; J-M, Routaboul; A, Alboresi; B, Weisshaar; L, Lepiniec. MYBL2 is a new regulator of flavonoid biosynthesis in Arabidopsis thaliana. The Plant Journal: For Cell and Molecular Biology 2008, 55, 940–953. [Google Scholar] [CrossRef]
- Eichenberger M, Schwander T; S, Hüppi; J, Kreuzer; PRE, Mittl; F, Peccati; G, Jiménez-Osés; M, Naesby; RM, Buller. The catalytic role of glutathione transferases in heterologous anthocyanin biosynthesis. Nature Catalysis 2023, 6, 927–938. [Google Scholar] [CrossRef]
- Fattorini R, Khojayori FN; G, Mellers; E, Moyroud; E, Herrero; RT, Kellenberger; R, Walker; Q, Wang; L, Hill; BJ, Glover. Complex petal spot formation in the Beetle Daisy (Gorteria diffusa) relies on spot-specific accumulation of malonylated anthocyanin regulated by paralogous GdMYBSG6 transcription factors. New Phytologist n/a, 2024. [Google Scholar] [CrossRef]
- Feild TS, Lee DW; NM, Holbrook. Why Leaves Turn Red in Autumn. The Role of Anthocyanins in Senescing Leaves of Red-Osier Dogwood. Plant Physiology 2001, 127, 566–574. [Google Scholar] [CrossRef]
- Feng K, Xu Z-S; J-X, Liu; J-W, Li; F, Wang; A-S, Xiong. Isolation, purification, and characterization of AgUCGalT1, a galactosyltransferase involved in anthocyanin galactosylation in purple celery (Apium graveolens L.). Planta 2018, 247, 1363–1375. [Google Scholar] [CrossRef]
- Flachowsky H, Szankowski I, Fischer TC, Richter K, Peil A, Höfer M, Dörschel C, Schmoock S, Gau AE, Halbwirth H, et al.2010. Transgenic apple plants overexpressing the Lc gene of maize show an altered growth habit and increased resistance to apple scab and fire blight. Planta 231: 623–635. [CrossRef]
- Ford CM, Boss PK; PB, Høj. Cloning and Characterization of Vitis viniferaUDP-Glucose:Flavonoid 3-O-Glucosyltransferase, a Homologue of the Enzyme Encoded by the Maize Bronze-1Locus That May Primarily Serve to Glucosylate Anthocyanidins in Vivo *. Journal of Biological Chemistry 1998, 273, 9224–9233. [Google Scholar] [CrossRef]
- Francisco RM, Regalado A, Ageorges A, Burla BJ, Bassin B, Eisenach C, Zarrouk O, Vialet S, Marlin T, Chaves MM, et al.2013. ABCC1, an ATP binding cassette protein from grape berry, transports anthocyanidin 3-O-Glucosides. The Plant Cell 25: 1840–1854. [CrossRef]
- Franco-Zorrilla JM, Valli A; M, Todesco; I, Mateos; MI, Puga; I, Rubio-Somoza; A, Leyva; D, Weigel; JA, García; J, Paz-Ares. Target mimicry provides a new mechanism for regulation of microRNA activity. Nature Genetics 2007, 39, 1033–1037. [Google Scholar] [CrossRef]
- Fraser CM, Thompson MG; AM, Shirley; J, Ralph; JA, Schoenherr; T, Sinlapadech; MC, Hall; C, Chapple. Related Arabidopsis Serine Carboxypeptidase-Like Sinapoylglucose Acyltransferases Display Distinct But Overlapping Substrate Specificities. Plant Physiology 2007, 144, 1986–1999. [Google Scholar] [CrossRef]
- Frommann J-F, Pucker B; LM, Sielmann; C, Müller; B, Weisshaar; R, Stracke; R, Schweiger. Metabolic fingerprinting reveals roles of Arabidopsis thaliana BGLU1, BGLU3 and BGLU4 in glycosylation of various flavonoids. : 2024.01.30.577901. [CrossRef]
- Frydman A, Liberman R; DV, Huhman; M, Carmeli-Weissberg; M, Sapir-Mir; R, Ophir; L, W. Sumner; Y, Eyal. The molecular and enzymatic basis of bitter/non-bitter flavor of citrus fruit: evolution of branch-forming rhamnosyltransferases under domestication. The Plant Journal 2013, 73, 166–178. [Google Scholar] [CrossRef]
- Frydman A, Weisshaus O; M, Bar-Peled; DV, Huhman; LW, Sumner; FR, Marin; E, Lewinsohn; R, Fluhr; J, Gressel; Y, Eyal. Citrus fruit bitter flavors: isolation and functional characterization of the gene Cm1,2RhaT encoding a 1,2 rhamnosyltransferase, a key enzyme in the biosynthesis of the bitter flavonoids of citrus. The Plant Journal: For Cell and Molecular Biology 2004, 40, 88–100. [Google Scholar] [CrossRef]
- Garcia JE, Hannah L; M, Shrestha; M, Burd; AG, Dyer. Fly pollination drives convergence of flower coloration. New Phytologist 2022, 233, 52–61. [Google Scholar] [CrossRef]
- Gebhardt YH, Witte S; H, Steuber; U, Matern; S, Martens. Evolution of Flavone Synthase I from Parsley Flavanone 3β-Hydroxylase by Site-Directed Mutagenesis. Plant Physiology 2007, 144, 1442–1454. [Google Scholar] [CrossRef]
- Gerchman Y, Dodek I; R, Petichov; Y, Yerushalmi; A, Lerner; T, Keasar. Beyond pollinator attraction: extra-floral displays deter herbivores in a Mediterranean annual plant. Evolutionary Ecology 2012, 26, 499–512. [Google Scholar] [CrossRef]
- Gervasi DDL, Schiestl FP. Real-time divergent evolution in plants driven by pollinators. Nature Communications 2017, 8, 14691. [Google Scholar] [CrossRef]
- Gierer A, Meinhardt H. A theory of biological pattern formation. Kybernetik 1972, 12, 30–39. [Google Scholar] [CrossRef]
- Givnish TJ. 1990. Leaf Mottling: Relation to Growth Form and Leaf Phenology and Possible Role as Camouflage. Functional Ecology 4: 463–474. [CrossRef]
- Glover BJ, Walker RH; E, Moyroud; SF, Brockington. How to spot a flower. New Phytologist 2013, 197, 687–689. [Google Scholar] [CrossRef]
- Gollop R, Farhi S; A, Perl. Regulation of the leucoanthocyanidin dioxygenase gene expression in Vitis vinifera. Plant Science 2001, 161, 579–588. [Google Scholar] [CrossRef]
- Gong W-C, Liu Y-H; C-M, Wang; Y-Q, Chen; K, Martin; L-Z, Meng. Why Are There so Many Plant Species That Transiently Flush Young Leaves Red in the Tropics? Frontiers in Plant Science 11, 2020. [Google Scholar] [CrossRef]
- Gonzalez A, Brown M, Hatlestad G, Akhavan N, Smith T, Hembd A, Moore J, Montes D, Mosley T, Resendez J, et al.2016. TTG2 controls the developmental regulation of seed coat tannins in Arabidopsis by regulating vacuolar transport steps in the proanthocyanidin pathway. Developmental Biology 419: 54–63. [CrossRef]
- Gonzalez A, Zhao M; JM, Leavitt; AM, Lloyd. Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. The Plant Journal 2008, 53, 814–827. [Google Scholar] [CrossRef]
- Goodman CD, Casati P; V, Walbot. A Multidrug Resistance–Associated Protein Involved in Anthocyanin Transport in Zea mays. The Plant Cell 2004, 16, 1812–1826. [Google Scholar] [CrossRef]
- Gou J-Y, Felippes FF; C-J, Liu; D, Weigel; J-W, Wang. Negative Regulation of Anthocyanin Biosynthesis in Arabidopsis by a miR156-Targeted SPL Transcription Factor. The Plant Cell 2011, 23, 1512–1522. [Google Scholar] [CrossRef]
- Gould KS. 1993. Leaf Heteroblasty in Pseudopanax crassifolius: Functional Significance of Leaf Morphology and Anatomy. Annals of Botany 71: 61–70. [CrossRef]
- Gould KS. 2004. Nature’s Swiss Army Knife: The Diverse Protective Roles of Anthocyanins in Leaves. Journal of Biomedicine and Biotechnology 2004: 314–320. [CrossRef]
- Gould KS, Dudle DA; HS, Neufeld. Why some stems are red: cauline anthocyanins shield photosystem II against high light stress. Journal of Experimental Botany 2010, 61, 2707–2717. [Google Scholar] [CrossRef]
- Gould KS, Jay-Allemand C; BA, Logan; Y, Baissac; LPR, Bidel. When are foliar anthocyanins useful to plants? Re-evaluation of the photoprotection hypothesis using Arabidopsis thaliana mutants that differ in anthocyanin accumulation. Environmental and Experimental Botany 2018, 154, 11–22. [Google Scholar] [CrossRef]
- Gould KS, Kuhn DN; DW, Lee; SF, Oberbauer. Why leaves are sometimes red. Nature 1995, 378, 241–242. [Google Scholar] [CrossRef]
- Gould KS, McKelvie J; KR, Markham. Do anthocyanins function as antioxidants in leaves? Imaging of H2O2 in red and green leaves after mechanical injury. Plant, Cell & Environment 2002a, 25, 1261–1269. [Google Scholar] [CrossRef]
- Gould KS, Neill SO; TC, Vogelmann. A unified explanation for anthocyanins in leaves? In: Advances in Botanical Research. Academic Press, 167–192; 2002b. [Google Scholar] [CrossRef]
- Gronquist M, Bezzerides A; A, Attygalle; J, Meinwald; M, Eisner; T, Eisner. Attractive and defensive functions of the ultraviolet pigments of a flower (Hypericum calycinum). Proceedings of the National Academy of Sciences of the United States of America 2001, 98, 13745–13750. [Google Scholar] [CrossRef]
- Grotewold E. 2006. The genetics and biochemistry of floral pigments. Annual Review of Plant Biology 57: 761–780. [CrossRef]
- Gu K-D, Wang C-K; D-G, Hu; Y-J, Hao. How do anthocyanins paint our horticultural products? Scientia Horticulturae 2019, 249, 257–262. [Google Scholar] [CrossRef]
- Hajiboland R, Farhanghi F. Remobilization of boron, photosynthesis, phenolic metabolism and anti-oxidant defense capacity in boron-deficient turnip (Brassica rapa L.) plants. Soil Science and Plant Nutrition 2010, 56, 427–437. [Google Scholar] [CrossRef]
- Harborne JB. 1962. Anthocyanins and their sugar components. Fortschritte Der Chemie Organischer Naturstoffe = Progress in the Chemistry of Organic Natural Products. Progres Dans La Chimie Des Substances Organiques Naturelles 20: 165–199. [CrossRef]
- Harborne JB. 1982. Introduction to Ecological Biochemistry.
- Harmer SL, Hogenesch JB; M, Straume; HS, Chang; B, Han; T, Zhu; X, Wang; JA, Kreps; SA, Kay. Orchestrated transcription of key pathways in Arabidopsis by the circadian clock. Science (New York, N.Y.) 2000, 290, 2110–2113. [Google Scholar] [CrossRef]
- Hellens RP, Moreau C, Lin-Wang K, Schwinn KE, Thomson SJ, Fiers MWEJ, Frew TJ, Murray SR, Hofer JMI, Jacobs JME, et al.2010. Identification of Mendel’s White Flower Character. PLOS ONE 5: e13230. [CrossRef]
- Henry A, Chopra S; DG, Clark; JP, Lynch. Responses to low phosphorus in high and low foliar anthocyanin coleus (Solenostemon scutellarioides) and maize (Zea mays). Functional Plant Biology 2012, 39, 255–265. [Google Scholar] [CrossRef]
- Hichri I, Barrieu F; J, Bogs; C, Kappel; S, Delrot; V, Lauvergeat. Recent advances in the transcriptional regulation of the flavonoid biosynthetic pathway. Journal of Experimental Botany 2011, 62, 2465–2483. [Google Scholar] [CrossRef]
- Ho WW, Smith SD. Molecular evolution of anthocyanin pigmentation genes following losses of flower color. BMC Evolutionary Biology 2016, 16, 98. [Google Scholar] [CrossRef]
- Hoballah ME, Gübitz T; J, Stuurman; L, Broger; M, Barone; T, Mandel; A, Dell’Olivo; M, Arnold; C, Kuhlemeier. Single Gene–Mediated Shift in Pollinator Attraction in Petunia. The Plant Cell 2007, 19, 779–790. [Google Scholar] [CrossRef] [PubMed]
- Hoch WA, Zeldin EL; BH, McCown. Physiological significance of anthocyanins during autumnal leaf senescence. Tree Physiology 2001, 21, 1–8. [Google Scholar] [CrossRef]
- Hsu Y-H, Tagami T; K, Matsunaga; M, Okuyama; T, Suzuki; N, Noda; M, Suzuki; H, Shimura. Functional characterization of UDP-rhamnose-dependent rhamnosyltransferase involved in anthocyanin modification, a key enzyme determining blue coloration in Lobelia erinus. The Plant Journal: For Cell and Molecular Biology 2017, 89, 325–337. [Google Scholar] [CrossRef]
- Hughes NM, Connors MK; MH, Grace; MA, Lila; BN, Willans; AJ, Wommack. The same anthocyanins served four different ways: Insights into anthocyanin structure-function relationships from the wintergreen orchid, Tipularia discolor. Plant Science 2021, 303, 110793. [Google Scholar] [CrossRef]
- Hughes NM, Neufeld HS; KO, Burkey. Functional role of anthocyanins in high-light winter leaves of the evergreen herb Galax urceolata. New Phytologist 2005, 168, 575–587. [Google Scholar] [CrossRef]
- Hughes NM, Vogelmann TC; WK, Smith. Optical effects of abaxial anthocyanin on absorption of red wavelengths by understorey species: revisiting the back-scatter hypothesis. Journal of Experimental Botany 2008, 59, 3435–3442. [Google Scholar] [CrossRef]
- Ichiishi S, Nagamitsu T; Y, Kondo; T, Iwashina; K, Kondo; N, Tagashira. Effects of Macro-components and Sucrose in the Medium on in vitro Red-color Pigmentation in Dionaea muscipula Ellis and Drosera spathulata Laill. Plant Biotechnology 1999, 16, 235–238. [Google Scholar] [CrossRef]
- Ichino T, Fuji K, Ueda H, Takahashi H, Koumoto Y, Takagi J, Tamura K, Sasaki R, Aoki K, Shimada T, et al.2014. GFS9/TT9 contributes to intracellular membrane trafficking and flavonoid accumulation in Arabidopsis thaliana. The Plant Journal: For Cell and Molecular Biology 80: 410–423. [CrossRef]
- Ide J-Y. 2022. Why do red/purple young leaves suffer less insect herbivory: tests of the warning signal hypothesis and the undermining of insect camouflage hypothesis. Arthropod-Plant Interactions 16: 567–581. [CrossRef]
- Irani NG, Grotewold E. Light-induced morphological alteration in anthocyanin-accumulating vacuoles of maize cells. BMC Plant Biology 2005, 5, 7. [Google Scholar] [CrossRef] [PubMed]
- Ishihara H, Tohge T; P, Viehöver; AR, Fernie; B, Weisshaar; R, Stracke. Natural variation in flavonol accumulation in Arabidopsis is determined by the flavonol glucosyltransferase BGLU6. Journal of Experimental Botany 2016, 67, 1505–1517. [Google Scholar] [CrossRef]
- Jaakola L. 2013. New insights into the regulation of anthocyanin biosynthesis in fruits. Trends in Plant Science 18: 477–483. [CrossRef]
- Jaakola L, Poole M, Jones MO, Kämäräinen-Karppinen T, Koskimäki JJ, Hohtola A, Häggman H, Fraser PD, Manning K, King GJ, et al.2010. A SQUAMOSA MADS Box Gene Involved in the Regulation of Anthocyanin Accumulation in Bilberry Fruits. Plant Physiology 153: 1619–1629. [CrossRef]
- Jeong S-W, Das PK, Jeoung SC, Song J-Y, Lee HK, Kim Y-K, Kim WJ, Park YI, Yoo S-D, Choi S-B, et al.2010. Ethylene Suppression of Sugar-Induced Anthocyanin Pigmentation in Arabidopsis1[C][W][OA]. Plant Physiology 154: 1514–1531. [CrossRef]
- Jezek M, Allan AC; JJ, Jones; C-M, Geilfus. Why do plants blush when they are hungry? New Phytologist 2023, 239, 494–505. [Google Scholar] [CrossRef]
- Jezek M, Zörb C; N, Merkt; C-M, Geilfus. Anthocyanin Management in Fruits by Fertilization. Journal of Agricultural and Food Chemistry 2018, 66, 753–764. [Google Scholar] [CrossRef] [PubMed]
- Jiang X, Huang K, Zheng G, Hou H, Wang P, Jiang H, Zhao X, Li M, Zhang S, Liu Y, et al.2018. CsMYB5a and CsMYB5e from Camellia sinensis differentially regulate anthocyanin and proanthocyanidin biosynthesis. Plant Science 270: 209–220. [CrossRef]
- Jiang L, Yue M, Liu Y, Zhang N, Lin Y, Zhang Y, Wang Y, Li M, Luo Y, Zhang Y, et al.2023. A novel R2R3-MYB transcription factor FaMYB5 positively regulates anthocyanin and proanthocyanidin biosynthesis in cultivated strawberries (Fragaria × ananassa). Plant Biotechnology Journal 21: 1140–1158. [CrossRef]
- Jiu S, Guan L, Leng X, Zhang K, Haider MS, Yu X, Zhu X, Zheng T, Ge M, Wang C, et al.2021. The role of VvMYBA2r and VvMYBA2w alleles of the MYBA2 locus in the regulation of anthocyanin biosynthesis for molecular breeding of grape (Vitis spp.) skin coloration. Plant Biotechnology Journal 19: 1216–1239. [CrossRef]
- Jokioja J, Yang B; KM, Linderborg. Acylated anthocyanins: A review on their bioavailability and effects on postprandial carbohydrate metabolism and inflammation. Comprehensive Reviews in Food Science and Food Safety 2021, 20, 5570–5615. [Google Scholar] [CrossRef] [PubMed]
- Jonsson LMV, Aarsman MEG; J, van Diepen; P, de Vlaming; N, Smit; AW, Schram. Properties and genetic control of anthocyanin 5-O-glucosyltransferase in flowers of Petunia hybrida. Planta 1984, 160, 341–347. [Google Scholar] [CrossRef]
- Ju Z, Sun W; X, Meng; L, Liang; Y, Li; T, Zhou; H, Shen; X, Gao; L, Wang. Isolation and functional characterization of two 5-O-glucosyltransferases related to anthocyanin biosynthesis from Freesia hybrida. Plant Cell, Tissue and Organ Culture (PCTOC) 2018, 135, 99–110. [Google Scholar] [CrossRef]
- Juniper B. 1993. Flamboyant flushes: a reinterpretation of non-green flush colours in leaves. International dendrology society yearbook: 49–57.
- Kalaji HM, Bąba W, Gediga K, Goltsev V, Samborska IA, Cetner MD, Dimitrova S, Piszcz U, Bielecki K, Karmowska K, et al.2018. Chlorophyll fluorescence as a tool for nutrient status identification in rapeseed plants. Photosynthesis Research 136: 329–343. [CrossRef]
- Kamsteeg J, Brederode J van; van, Nigtevecht G. Identification, Properties and Genetic Control of UDP-ʟ-Rhamnose: Anthocyanidin 3-O-Glucoside, 6″-O-Rhamnosyltransferase Isolated from Retals of the Red Campion (Silene dioica). Zeitschrift für Naturforschung C 1980, 35, 249–257. [Google Scholar] [CrossRef]
- Kang X, Mikami R; Y, Akita. Characterization of 5-O-glucosyltransferase involved in anthocyanin biosynthesis in Cyclamen purpurascens. Plant Biotechnology 2021, 38, 263–268. [Google Scholar] [CrossRef]
- Karageorgou P, Buschmann C; Y, Manetas. Red leaf color as a warning signal against insect herbivory: Honest or mimetic? Flora - Morphology, Distribution, Functional Ecology of Plants 2008, 203, 648–652. [Google Scholar] [CrossRef]
- Karageorgou P, Manetas Y. The importance of being red when young: anthocyanins and the protection of young leaves of Quercus coccifera from insect herbivory and excess light. Tree Physiology 2006, 26, 613–621. [Google Scholar] [CrossRef]
- Karban R, Baldwin IT. Induced Responses to Herbivory. University of Chicago Press. 1997. [Google Scholar]
- Karppinen K, Lafferty DJ; NW, Albert; N, Mikkola; T, McGhie; AC, Allan; BM, Afzal; H, Häggman; RV, Espley; L, Jaakola. MYBA and MYBPA transcription factors co-regulate anthocyanin biosynthesis in blue-coloured berries. New Phytologist 2021, 232, 1350–1367. [Google Scholar] [CrossRef]
- Kaur S, Kumari A; N, Sharma; AK, Pandey; M, Garg. Physiological and molecular response of colored wheat seedlings against phosphate deficiency is linked to accumulation of distinct anthocyanins. Plant physiology and biochemistry: PPB 2022, 170, 338–349. [Google Scholar] [CrossRef]
- Kawai Y, Ono E; M, Mizutani. Evolution and diversity of the 2–oxoglutarate-dependent dioxygenase superfamily in plants. The Plant Journal 2014, 78, 328–343. [Google Scholar] [CrossRef]
- Kim S, Chen J, Cheng T, Gindulyte A, He J, He S, Li Q, Shoemaker BA, Thiessen PA, Yu B, et al.2023. PubChem 2023 update. Nucleic Acids Research 51: D1373–D1380. [CrossRef]
- Kim S, Hwang G; S, Lee; J-Y, Zhu; I, Paik; TT, Nguyen; J, Kim; E, Oh. High Ambient Temperature Represses Anthocyanin Biosynthesis through Degradation of HY5. Frontiers in Plant Science 2017, 8, 1787. [Google Scholar] [CrossRef]
- Kim D-H, Park S; J-Y, Lee; S-H, Ha; J-G, Lee; S-H, Lim. A Rice B-Box Protein, OsBBX14, Finely Regulates Anthocyanin Biosynthesis in Rice. International Journal of Molecular Sciences 2018, 19, 2190. [Google Scholar] [CrossRef]
- Kitamura S, Shikazono N; A, Tanaka. TRANSPARENT TESTA 19 is involved in the accumulation of both anthocyanins and proanthocyanidins in Arabidopsis. The Plant Journal: For Cell and Molecular Biology 2004, 37, 104–114. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi NI, Saito T; N, Iwata; Y, Ohmae; R, Iwata; K, Tanoi; TM, Nakanishi. Leaf senescence in rice due to magnesium deficiency mediated defect in transpiration rate before sugar accumulation and chlorosis. Physiologia Plantarum 2013, 148, 490–501. [Google Scholar] [CrossRef]
- Koeslin-Findeklee F, Rizi VS; MA, Becker; S, Parra-Londono; M, Arif; S, Balazadeh; B, Mueller-Roeber; R, Kunze; WJ, Horst. Transcriptomic analysis of nitrogen starvation- and cultivar-specific leaf senescence in winter oilseed rape (Brassica napus L.). Plant Science: An International Journal of Experimental Plant Biology 2015, 233, 174–185. [Google Scholar] [CrossRef]
- Koseki M, Goto K; C, Masuta; A, Kanazawa. The Star-type Color Pattern in Petunia hybrida ‘Red Star’ Flowers is Induced by Sequence-Specific Degradation of Chalcone Synthase RNA. Plant and Cell Physiology 2005, 46, 1879–1883. [Google Scholar] [CrossRef] [PubMed]
- Kovinich N, Kayanja G; A, Chanoca; K, Riedl; MS, Otegui; E, Grotewold. Not all anthocyanins are born equal: distinct patterns induced by stress in Arabidopsis. Planta 2014, 240, 931–940. [Google Scholar] [CrossRef]
- Kovinich N, Saleem A; JT, Arnason; B, Miki. Functional characterization of a UDP-glucose:flavonoid 3-O-glucosyltransferase from the seed coat of black soybean (Glycine max (L.) Merr.). Phytochemistry 2010, 71, 1253–1263. [Google Scholar] [CrossRef]
- Kranz HD, Denekamp M, Greco R, Jin H, Leyva A, Meissner RC, Petroni K, Urzainqui A, Bevan M, Martin C, et al.1998. Towards functional characterisation of the members of the R2R3-MYB gene family from Arabidopsis thaliana. The Plant Journal: For Cell and Molecular Biology 16: 263–276. [CrossRef]
- Krause GH, Virgo A; K, Winter. High susceptibility to photoinhibition of young leaves of tropical forest trees. Planta 1995, 197, 583–591. [Google Scholar] [CrossRef]
- Kroon J, Souer E; A, De Graaff; Y, Xue; J, Mol; R, Koes. Cloning and structural analysis of the anthocyanin pigmentation locus Rt of Petunia hybrida: characterization of insertion sequences in two mutant alleles. The Plant Journal 1994, 5, 69–80. [Google Scholar] [CrossRef]
- Kumar V, Sharma SS. Nutrient Deficiency-dependent Anthocyanin Development in Spirodela Polyrhiza L. Schleid. Biologia Plantarum 1999, 42, 621–624. [Google Scholar] [CrossRef]
- Kytridis V-P, Manetas Y. Mesophyll versus epidermal anthocyanins as potential in vivo antioxidants: evidence linking the putative antioxidant role to the proximity of oxy-radical source. Journal of Experimental Botany 2006, 57, 2203–2210. [Google Scholar] [CrossRef] [PubMed]
- LaFountain AM, Yuan Y-W. Repressors of anthocyanin biosynthesis. New Phytologist 2021, 231, 933–949. [Google Scholar] [CrossRef]
- Landi M, Tattini M; KS, Gould. Multiple functional roles of anthocyanins in plant-environment interactions. Environmental and Experimental Botany 2015, 119, 4–17. [Google Scholar] [CrossRef]
- Lasin P, Weise A; A, Reinders; JM, Ward. Arabidopsis Sucrose Transporter AtSuc1 introns act as strong enhancers of expression. Plant & Cell Physiology 2020, 61, 1054–1063. [Google Scholar] [CrossRef]
- Lawanson AO, Akindele BB; PB, Fasalojo; BL, Akpe. Time-course of anthocyanin formation during deficiencies of nitrogen, phosphorus and potassium in seedlings of zea mays Linn. var. E.S. 1. Zeitschrift für Pflanzenphysiologie 1972, 66, 251–253. [Google Scholar] [CrossRef]
- Lea US, Slimestad R; P, Smedvig; C, Lillo. Nitrogen deficiency enhances expression of specific MYB and bHLH transcription factors and accumulation of end products in the flavonoid pathway. Planta 2007, 225, 1245–1253. [Google Scholar] [CrossRef] [PubMed]
- Lee DW. 2002. Anthocyanins in autumn leaf senescence. In: Advances in Botanical Research. Academic Press, 147–165. [CrossRef]
- Lee DW, Brammeier S; AP, Smith. The Selective Advantages of Anthocyanins in Developing Leaves of Mango and Cacao. Biotropica 1987, 19, 40–49. [Google Scholar] [CrossRef]
- Lee DW, Collins TM. Phylogenetic and Ontogenetic Influences on the Distribution of Anthocyanins and Betacyanins in Leaves of Tropical Plants. International Journal of Plant Sciences 2001, 162, 1141–1153. [Google Scholar] [CrossRef]
- Lee DW, O’Keefe J; NM, Holbrook; TS, Feild. Pigment dynamics and autumn leaf senescence in a New England deciduous forest, eastern USA. Ecological Research 2003, 18, 677–694. [Google Scholar] [CrossRef]
- Lei KJ, Zhang L; XY, Du; Y, An; GH, Chang; GY, An. A chalcone synthase controls the verticillium disease resistance response in both Arabidopsis thaliana and cotton. European Journal of Plant Pathology 2018, 152, 769–781. [Google Scholar] [CrossRef]
- Lei KJ, Zhou H; DL, Gu; GY, An. The involvement of abscisic acid-insensitive mutants in low phosphate stress responses during rhizosphere acidification, anthocyanin accumulation and Pi homeostasis in Arabidopsis. Plant Science: An International Journal of Experimental Plant Biology 2022, 322, 111358. [Google Scholar] [CrossRef]
- Lev-Yadun S. 2001. Aposematic (warning) Coloration Associated with Thorns in Higher Plants. Journal of Theoretical Biology 210: 385–388. [CrossRef]
- Lev-Yadun S. 2003. Why do some thorny plants resemble green zebras? Journal of Theoretical Biology 224: 483–489. [CrossRef]
- Lev-Yadun S. 2016. Biochemical Evidence of Convergent Evolution of Aposematic Coloration in Thorny, Spiny and Prickly Plants. In: Lev-Yadun S, ed. Defensive (anti-herbivory) Coloration in Land Plants. Cham: Springer International Publishing, 183–183. [CrossRef]
- Lev-Yadun S. 2021. Avoiding rather than resisting herbivore attacks is often the first line of plant defence. Biological Journal of the Linnean Society 134: 775–802. [CrossRef]
- Lev-Yadun S. 2024. Visual-, Olfactory-, and Nectar-Taste-Based Flower Aposematism. Plants 13: 391. [CrossRef]
- Lev-Yadun S, Dafni A; MA, Flaishman; M, Inbar; I, Izhaki; G, Katzir; G, Ne’eman. Plant coloration undermines herbivorous insect camouflage. BioEssays: News and Reviews in Molecular, Cellular and Developmental Biology 2004, 26, 1126–1130. [Google Scholar] [CrossRef] [PubMed]
- Lev-Yadun S, Gould KS. Role of Anthocyanins in Plant Defence. In: Winefield C, Davies K, Gould K, eds. Anthocyanins: Biosynthesis, Functions, and Applications. New York, NY: Springer, 22–28. [CrossRef]
- Lev-Yadun S, Inbar M. Defensive ant, aphid and caterpillar mimicry in plants? Biological Journal of the Linnean Society 2002, 77, 393–398. [Google Scholar] [CrossRef]
- Lev-Yadun S, Ne’eman G; I, Izhaki. Unripe red fruits may be aposematic. Plant Signaling & Behavior 2009, 4, 836–841. [Google Scholar] [CrossRef]
- Lev-Yadun S, Silva JA. Defensive coloration in plants: a review of current ideas about anti-herbivore coloration strategies. 2006. [Google Scholar]
- Li S, Dong Y; D, Li; S, Shi; N, Zhao; J, Liao; Y, Liu; H, Chen. Eggplant transcription factor SmMYB5 integrates jasmonate and light signaling during anthocyanin biosynthesis. Plant Physiology 2024, 194, 1139–1165. [Google Scholar] [CrossRef] [PubMed]
- Li B, Fan R, Guo S, Wang P, Zhu X, Fan Y, Chen Y, He K, Kumar A, Shi J, et al.2019a. The Arabidopsis MYB transcription factor, MYB111 modulates salt responses by regulating flavonoid biosynthesis. Environmental and Experimental Botany 166: 103807. [CrossRef]
- Li Y, Kong F; Z, Liu; L, Peng; Q, Shu. PhUGT78A22, a novel glycosyltransferase in Paeonia ‘He Xie’, can catalyze the transfer of glucose to glucosylated anthocyanins during petal blotch formation. BMC Plant Biology 2022, 22, 405. [Google Scholar] [CrossRef]
- Li P, Li Y-J; F-J, Zhang; G-Z, Zhang; X-Y, Jiang; H-M, Yu; B-K, Hou. The Arabidopsis UDP-glycosyltransferases UGT79B2 and UGT79B3, contribute to cold, salt and drought stress tolerance via modulating anthocyanin accumulation. The Plant Journal 2017, 89, 85–103. [Google Scholar] [CrossRef] [PubMed]
- Li D-D, Ni R; P-P, Wang; X-S, Zhang; P-Y, Wang; T-T, Zhu; C-J, Sun; C-J, Liu; H-X, Lou; A-X, Cheng. Molecular Basis for Chemical Evolution of Flavones to Flavonols and Anthocyanins in Land Plants. Plant Physiology 2020a, 184, 1731–1743. [Google Scholar] [CrossRef]
- Li X, Ouyang X, Zhang Z, He L, Wang Y, Li Y, Zhao J, Chen Z, Wang C, Ding L, et al.2019b. Over-expression of the red plant gene R1 enhances anthocyanin production and resistance to bollworm and spider mite in cotton. Molecular Genetics and Genomics 294: 469–478. [CrossRef]
- Li Y, Shan X; L, Zhou; R, Gao; S, Yang; S, Wang; L, Wang; X, Gao. The R2R3-MYB Factor FhMYB5 From Freesia hybrida Contributes to the Regulation of Anthocyanin and Proanthocyanidin Biosynthesis. Frontiers in Plant Science 9, 2019c. [Google Scholar]
- Li Y, Van den Ende W; F, Rolland. Sucrose induction of anthocyanin biosynthesis is mediated by DELLA. Molecular Plant 2014, 7, 570–572. [Google Scholar] [CrossRef]
- Li C, Wu J; K-D, Hu; S-W, Wei; H-Y, Sun; L-Y, Hu; Z, Han; G-F, Yao; H, Zhang. PyWRKY26 and PybHLH3 cotargeted the PyMYB114 promoter to regulate anthocyanin biosynthesis and transport in red-skinned pears. Horticulture Research 2020b, 7, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Liang J, He J. Protective role of anthocyanins in plants under low nitrogen stress. Biochemical and Biophysical Research Communications 2018, 498, 946–953. [Google Scholar] [CrossRef]
- Liu Y, Liu J; Y, Qi; A, Zhang; Z, Liu; X, Ren. Identification and characterization of AcUFGT6b, a xylosyltransferase involved in anthocyanin modification in red-fleshed kiwifruit (Actinidia chinensis). Plant Cell, Tissue and Organ Culture (PCTOC) 2019a, 138, 257–271. [Google Scholar] [CrossRef]
- Liu H, Shu Q; K, Lin-Wang; AC, Allan; RV, Espley; J, Su; M, Pei; J, Wu. The PyPIF5-PymiR156a-PySPL9-PyMYB114/MYB10 module regulates light-induced anthocyanin biosynthesis in red pear. Molecular Horticulture 2021, 1, 14. [Google Scholar] [CrossRef]
- Liu H, Su J; Y, Zhu; G, Yao; AC, Allan; C, Ampomah-Dwamena; Q, Shu; K, Lin-Wang; S, Zhang; J, Wu. The involvement of PybZIPa in light-induced anthocyanin accumulation via the activation of PyUFGT through binding to tandem G-boxes in its promoter. Horticulture Research 2019b, 6, 134. [Google Scholar] [CrossRef] [PubMed]
- Liu W, Wang Y, Yu L, Jiang H, Guo Z, Xu H, Jiang S, Fang H, Zhang J, Su M, et al.2019c. MdWRKY11 Participates in Anthocyanin Accumulation in Red-Fleshed Apples by Affecting MYB Transcription Factors and the Photoresponse Factor MdHY5. Journal of Agricultural and Food Chemistry 67: 8783–8793. [CrossRef]
- Lloyd A, Brockman A; L, Aguirre; A, Campbell; A, Bean; A, Cantero; A, Gonzalez. Advances in the MYB–bHLH–WD Repeat (MBW) Pigment Regulatory Model: Addition of a WRKY Factor and Co-option of an Anthocyanin MYB for Betalain Regulation. Plant and Cell Physiology 2017, 58, 1431–1441. [Google Scholar] [CrossRef]
- Lloyd AM, Walbot V; RW, Davis. Arabidopsis and Nicotiana Anthocyanin Production Activated by Maize Regulators R and C1. Science 1992, 258, 1773–1775. [Google Scholar] [CrossRef] [PubMed]
- Lloyd JC, Zakhleniuk OV. Responses of primary and secondary metabolism to sugar accumulation revealed by microarray expression analysis of the Arabidopsis mutant, pho3. Journal of Experimental Botany 2004, 55, 1221–1230. [Google Scholar] [CrossRef]
- Lo Piccolo E, Landi M, Pellegrini E, Agati G, Giordano C, Giordani T, Lorenzini G, Malorgio F, Massai R, Nali C, et al.2018. Multiple Consequences Induced by Epidermally-Located Anthocyanins in Young, Mature and Senescent Leaves of Prunus. Frontiers in Plant Science 9. [CrossRef]
- Logan BA, Stafstrom WC; MJL, Walsh; JS, Reblin; KS, Gould. Examining the photoprotection hypothesis for adaxial foliar anthocyanin accumulation by revisiting comparisons of green- and red-leafed varieties of coleus (Solenostemon scutellarioides). Photosynthesis Research 2015, 124, 267–274. [Google Scholar] [CrossRef]
- Loreti E, Povero G; G, Novi; C, Solfanelli; A, Alpi; P, Perata. Gibberellins, jasmonate and abscisic acid modulate the sucrose-induced expression of anthocyanin biosynthetic genes in Arabidopsis. New Phytologist 2008, 179, 1004–1016. [Google Scholar] [CrossRef] [PubMed]
- Lotkowska ME, Tohge T; AR, Fernie; G-P, Xue; S, Balazadeh; B, Mueller-Roeber. The Arabidopsis Transcription Factor MYB112 Promotes Anthocyanin Formation during Salinity and under High Light Stress. Plant Physiology 2015, 169, 1862–1880. [Google Scholar] [CrossRef]
- Lozoya-Gloria E, Cuéllar-González F; N, Ochoa-Alejo. Anthocyanin metabolic engineering of Euphorbia pulcherrima: advances and perspectives. Frontiers in Plant Science 14, 2023. [Google Scholar]
- Lunau K, Maier EJ. Innate colour preferences of flower visitors. Journal of Comparative Physiology A 1995, 177, 1–19. [Google Scholar] [CrossRef]
- Luo J, Nishiyama Y, Fuell C, Taguchi G, Elliott K, Hill L, Tanaka Y, Kitayama M, Yamazaki M, Bailey P, et al.2007. Convergent evolution in the BAHD family of acyl transferases: identification and characterization of anthocyanin acyl transferases from Arabidopsis thaliana. The Plant Journal 50: 678–695. [CrossRef]
- Ma H, Yang T; Y, Li; J, Zhang; T, Wu; T, Song; Y, Yao; J, Tian. The long noncoding RNA MdLNC499 bridges MdWRKY1 and MdERF109 function to regulate early-stage light-induced anthocyanin accumulation in apple fruit. The Plant Cell 2021, 33, 3309–3330. [Google Scholar] [CrossRef] [PubMed]
- Mabry TJ, Turner BL. Chemical Investigations of the Batidaceae. TAXON 1964, 13, 197–200. [Google Scholar] [CrossRef]
- Manetas Y. 2006. Why some leaves are anthocyanic and why most anthocyanic leaves are red? Flora 3: 163–177. [CrossRef]
- Manetas Y, Petropoulou Y; GK, Psaras; A, Drinia. Exposed red (anthocyanic) leaves of Quercus coccifera display shade characteristics. Functional plant biology: FPB 2003, 30, 265–270. [Google Scholar] [CrossRef] [PubMed]
- Marinova K, Pourcel L; B, Weder; M, Schwarz; D, Barron; J-M, Routaboul; I, Debeaujon; M, Klein. The Arabidopsis MATE transporter TT12 acts as a vacuolar flavonoid/H+ -antiporter active in proanthocyanidin-accumulating cells of the seed coat. The Plant Cell 2007, 19, 2023–2038. [Google Scholar] [CrossRef]
- Marin-Recinos MF, Pucker B. Genetic factors explaining anthocyanin pigmentation differences. BMC Plant Biology 2024, 24, 627. [Google Scholar] [CrossRef]
- Mark Hodges D, Nozzolillo C. Anthocyanin and Anthocyanoplast Content of Cruciferous Seedlings Subjected to Mineral Nutrient Deficiencies. Journal of Plant Physiology 1996, 147, 749–754. [Google Scholar] [CrossRef]
- Marquardt C. 1835. Die Farben der Blüthen. Habicht.
- Martens S, Forkmann G; L, Britsch; F, Wellmann; U, Matern; R, Lukačin. Divergent evolution of flavonoid 2-oxoglutarate-dependent dioxygenases in parsley 1. FEBS Letters 2003, 544, 93–98. [Google Scholar] [CrossRef]
- Matile P. 2000. Biochemistry of Indian summer: physiology of autumnal leaf coloration. Experimental Gerontology 35: 145–158. [CrossRef]
- Matile Ph, Flach BM-P; BM, Eller. Autumn Leaves of Ginkgo biloba L.: Optical Properties, Pigments and Optical Brighteners. Botanica Acta 1992, 105, 13–17. [Google Scholar] [CrossRef]
- Mato M, Ozeki Y; Y, Itoh; D, Higeta; K, Yoshitama; S, Teramoto; R, Aida; N, Ishikura; M, Shibata. Isolation and Characterization of a cDNA Clone of UDP-Galactose: Flavonoid 3-0-Galactosyltransferase (UF3GaT) Expressed in Vigna mungo Seedlings. Plant and Cell Physiology 1998, 39, 1145–1155. [Google Scholar] [CrossRef] [PubMed]
- Matsuba Y, Sasaki N, Tera M, Okamura M, Abe Y, Okamoto E, Nakamura H, Funabashi H, Takatsu M, Saito M, et al.2010. A Novel Glucosylation Reaction on Anthocyanins Catalyzed by Acyl-Glucose–Dependent Glucosyltransferase in the Petals of Carnation and Delphinium[C][W]. The Plant Cell 22: 3374–3389. [CrossRef]
- McClintock B. 1950. The Origin and Behavior of Mutable Loci in Maize. Proceedings of the National Academy of Sciences of the United States of America 36: 344–355.
- Mendel G. 1865. Versuche uber pflanzen-hybriden. Vorgelegt in den Sitzungen.
- Meng J, Wang H; R, Chi; Y, Qiao; J, Wei; Y, Zhang; M, Han; Y, Wang; H, Li. The eTM–miR858–MYB62-like module regulates anthocyanin biosynthesis under low-nitrogen conditions in Malus spectabilis. New Phytologist 2023, 238, 2524–2544. [Google Scholar] [CrossRef] [PubMed]
- Meng L-S, Xu M-K, Wan W, Yu F, Li C, Wang J-Y, Wei Z-Q, Lv M-J, Cao X-Y, Li Z-Y, et al.2018. Sucrose Signaling Regulates Anthocyanin Biosynthesis Through a MAPK Cascade in Arabidopsis thaliana. Genetics 210: 607–619. [CrossRef]
- Merzlyak MN, Chivkunova OB. Light-stress-induced pigment changes and evidence for anthocyanin photoprotection in apples. Journal of Photochemistry and Photobiology. B, Biology 2000, 55, 155–163. [Google Scholar] [CrossRef]
- Miller R, Owens SJ; B, Rørslett. Plants and colour: Flowers and pollination. Optics & Laser Technology 2011, 43, 282–294. [Google Scholar] [CrossRef]
- Miyahara T, Sakiyama R; Y, Ozeki; N, Sasaki. Acyl-glucose-dependent glucosyltransferase catalyzes the final step of anthocyanin formation in Arabidopsis. Journal of Plant Physiology 2013, 170, 619–624. [Google Scholar] [CrossRef]
- Miyahara T, Takahashi M; Y, Ozeki; N, Sasaki. Isolation of an acyl-glucose-dependent anthocyanin 7-O-glucosyltransferase from the monocot Agapanthus africanus. Journal of Plant Physiology 2012, 169, 1321–1326. [Google Scholar] [CrossRef]
- Modolo LV, Blount JW; L, Achnine; MA, Naoumkina; X, Wang; RA, Dixon. A functional genomics approach to (iso)flavonoid glycosylation in the model legume Medicago truncatula. Plant Molecular Biology 2007, 64, 499–518. [Google Scholar] [CrossRef] [PubMed]
- Modolo LV, Li L; H, Pan; JW, Blount; RA, Dixon; X, Wang. Crystal Structures of Glycosyltransferase UGT78G1 Reveal the Molecular Basis for Glycosylation and Deglycosylation of (Iso)flavonoids. Journal of Molecular Biology 2009, 392, 1292–1302. [Google Scholar] [CrossRef]
- Moghe GD, Last RL. Something Old, Something New: Conserved Enzymes and the Evolution of Novelty in Plant Specialized Metabolism. Plant Physiology 2015, 169, 1512–1523. [Google Scholar] [CrossRef]
- Mollier A, Pellerin S. Maize root system growth and development as influenced by phosphorus deficiency. Journal of Experimental Botany 1999, 50, 487–497. [Google Scholar] [CrossRef]
- Montefiori M, Comeskey DJ; M, Wohlers; TK, McGhie. Characterization and Quantification of Anthocyanins in Red Kiwifruit (Actinidia spp.). Journal of Agricultural and Food Chemistry 2009, 57, 6856–6861. [Google Scholar] [CrossRef]
- Moreau C, Ambrose MJ; L, Turner; L, Hill; THN, Ellis; JMI, Hofer. The B gene of pea encodes a defective flavonoid 3’,5’-hydroxylase, and confers pink flower color. Plant Physiology 2012, 159, 759–768. [Google Scholar] [CrossRef] [PubMed]
- Morishita T, Kojima Y; T, Maruta; A, Nishizawa-Yokoi; Y, Yabuta; S, Shigeoka. Arabidopsis NAC transcription factor, ANAC078, regulates flavonoid biosynthesis under high-light. Plant & Cell Physiology 2009, 50, 2210–2222. [Google Scholar] [CrossRef]
- Morita Y, Takagi K, Fukuchi-Mizutani M, Ishiguro K, Tanaka Y, Nitasaka E, Nakayama M, Saito N, Kagami T, Hoshino A, et al.2014. A chalcone isomerase-like protein enhances flavonoid production and flower pigmentation. The Plant Journal 78: 294–304. [CrossRef]
- Mueller LA, Goodman CD; RA, Silady; V, Walbot. AN9, a Petunia Glutathione S-Transferase Required for Anthocyanin Sequestration, Is a Flavonoid-Binding Protein. Plant Physiology 2000, 123, 1561–1570. [Google Scholar] [CrossRef] [PubMed]
- Mur LA. 1995. Characterization of Members of the myb Gene Family of Transcription Factors from Petunia hybrida.
- Murai Y, Kokubugata G; M, Yokota; J, Kitajima; T, Iwashina. Flavonoids and anthocyanins from six Cassytha taxa (Lauraceae) as taxonomic markers. Biochemical Systematics and Ecology 2008, 36, 745–748. [Google Scholar] [CrossRef]
- Nagata T, Todoriki S; T, Masumizu; I, Suda; S, Furuta; Z, Du; S, Kikuchi. Levels of active oxygen species are controlled by ascorbic acid and anthocyanin in Arabidopsis. Journal of Agricultural and Food Chemistry 2003, 51, 2992–2999. [Google Scholar] [CrossRef]
- Naing AH, Ai TN; KB, Lim; IJ, Lee; CK, Kim. Overexpression of Rosea1 From Snapdragon Enhances Anthocyanin Accumulation and Abiotic Stress Tolerance in Transgenic Tobacco. Frontiers in Plant Science 9, 2018. [Google Scholar] [CrossRef]
- Nakano Y, Asada K. Spinach chloroplasts scavenge hydrogen peroxide on illumination. Plant and Cell Physiology 1980, 21, 1295–1307. [Google Scholar] [CrossRef]
- Nakatsuka T, Sato K; H, Takahashi; S, Yamamura; M, Nishihara. Cloning and characterization of the UDP-glucose:anthocyanin 5-O-glucosyltransferase gene from blue-flowered gentian. Journal of Experimental Botany 2008, 59, 1241–1252. [Google Scholar] [CrossRef]
- Naya L, Paul S; O, Valdés-López; AB, Mendoza-Soto; B, Nova-Franco; G, Sosa-Valencia; JL, Reyes; G, Hernández. Regulation of copper homeostasis and biotic interactions by microRNA 398b in common bean. PloS One 2014, 9, e84416. [Google Scholar] [CrossRef]
- Neill S, Gould KS. Optical properties of leaves in relation to anthocyanin concentration and distribution. Canadian Journal of Botany 2000, 77, 1777–1782. [Google Scholar] [CrossRef]
- Neill SO, Gould KS; PA, Kilmartin; KA, Mitchell; KR, Markham. Antioxidant activities of red versus green leaves in Elatostema rugosum. Plant, Cell & Environment 2002, 25, 539–547. [Google Scholar] [CrossRef]
- Ni R, Zhu T-T; X-S, Zhang; P-Y, Wang; C-J, Sun; Y-N, Qiao; H-X, Lou; A-X, Cheng. Identification and evolutionary analysis of chalcone isomerase-fold proteins in ferns. Journal of Experimental Botany 2020, 71, 290–304. [Google Scholar] [CrossRef] [PubMed]
- Niu J, Zhang G; W, Zhang; V, Goltsev; S, Sun; J, Wang; P, Li; F, Ma. Anthocyanin concentration depends on the counterbalance between its synthesis and degradation in plum fruit at high temperature. Scientific Reports 2017, 7, 7684. [Google Scholar] [CrossRef]
- Nowak MS, Harder B; SN, Meckoni; R, Friedhoff; K, Wolff; B, Pucker. Genome sequence and RNA-seq analysis reveal genetic basis of flower coloration in the giant water lily Victoria cruziana. : 2024.06.15.599162. [CrossRef]
- Nozue M, Yamada K; T, Nakamura; H, Kubo; M, Kondo; M, Nishimura. Expression of a Vacuolar Protein (VP24) in Anthocyanin-Producing Cells of Sweet Potato in Suspension Culture. Plant Physiology 1997, 115, 1065–1072. [Google Scholar] [CrossRef]
- Nvsvrot T, Yang X; Y, Zhang; L, Huang; G, Cai; Y, Ding; W, Ren; N, Wang. The PdeWRKY65-UGT75L28 gene module negatively regulates lignin biosynthesis in poplar petioles. Industrial Crops and Products 2023, 191, 115937. [Google Scholar] [CrossRef]
- Oberbaueri SF, Starr G. The role of anthocyanins for photosynthesis of Alaskan arctic evergreens during snowmelt. In: Advances in Botanical Research. Academic Press, 129–145. [CrossRef]
- Oberrath R, Böhning-Gaese K. Floral color change and the attraction of insect pollinators in lungwort (Pulmonaria collina). Oecologia 1999, 121, 383–391. [Google Scholar] [CrossRef]
- Ogata J u. n., Sakamoto T; M, Yamaguchi; S, Kawanobu; K, Yoshitama. Isolation and characterization of anthocyanin 5-O-glucosyltransferase from flowers of Dahlia variabilis. Journal of Plant Physiology 2001, 158, 709–714. [Google Scholar] [CrossRef]
- Owen CR, Bradshaw HD. Induced mutations affecting pollinator choice in Mimulus lewisii (Phrymaceae). Arthropod-Plant Interactions 2011, 5, 235–244. [Google Scholar] [CrossRef]
- Padmavati M, Sakthivel N; KV, Thara; AR, Reddy. Differential sensitivity of rice pathogens to growth inhibition by flavonoids. Phytochemistry 1997, 46, 499–502. [Google Scholar] [CrossRef]
- Paz-Ares J, Ghosal D; U, Wienand; PA, Peterson; H, Saedler. The regulatory c1 locus of Zea mays encodes a protein with homology to myb proto-oncogene products and with structural similarities to transcriptional activators. The EMBO Journal 1987, 6, 3553–3558. [Google Scholar] [CrossRef] [PubMed]
- Peng M, Bi Y-M; T, Zhu; SJ, Rothstein. Genome-wide analysis of Arabidopsis responsive transcriptome to nitrogen limitation and its regulation by the ubiquitin ligase gene NLA. Plant Molecular Biology 2007, 65, 775–797. [Google Scholar] [CrossRef] [PubMed]
- Pesch M, Dartan B; R, Birkenbihl; IE, Somssich; M, Hülskamp. Arabidopsis TTG2 Regulates TRY Expression through Enhancement of Activator Complex-Triggered Activation. The Plant Cell 2014, 26, 4067–4083. [Google Scholar] [CrossRef]
- Pringsheim N. 1881. Untersuchungen über Lichtwirkung und Chlorophyllfunction in der Pflanze. DMW - Deutsche Medizinische Wochenschrift 7: 245–248. [CrossRef]
- Pucker B, Iorizzo M. Apiaceae FNS I originated from F3H through tandem gene duplication. PLOS ONE 2023, 18, e0280155. [Google Scholar] [CrossRef]
- Pucker B, Reiher F; HM, Schilbert. Automatic Identification of Players in the Flavonoid Biosynthesis with Application on the Biomedicinal Plant Croton tiglium. Plants 2020, 9, 1103. [Google Scholar] [CrossRef]
- Pucker B, Selmar D. Biochemistry and Molecular Basis of Intracellular Flavonoid Transport in Plants. Plants (Basel, Switzerland) 2022, 11, 963. [Google Scholar] [CrossRef] [PubMed]
- Pucker B, Walker-Hale N; J, Dzurlic; WC, Yim; JC, Cushman; A, Crum; Y, Yang; SF, Brockington. Multiple mechanisms explain loss of anthocyanins from betalain-pigmented Caryophyllales, including repeated wholesale loss of a key anthocyanidin synthesis enzyme. New Phytologist 2024, 241, 471–489. [Google Scholar] [CrossRef]
- Quattrocchio F, Verweij W; A, Kroon; C, Spelt; J, Mol; R, Koes. PH4 of Petunia is an R2R3 MYB protein that activates vacuolar acidification through interactions with basic-helix-loop-helix transcription factors of the anthocyanin pathway. The Plant Cell 2006, 18, 1274–1291. [Google Scholar] [CrossRef]
- Ramsay NA, Glover BJ. MYB-bHLH-WD40 protein complex and the evolution of cellular diversity. Trends in Plant Science 2005, 10, 63–70. [Google Scholar] [CrossRef]
- Rausher MD. 2008. Evolutionary Transitions in Floral Color. International Journal of Plant Sciences 169: 7–21. [CrossRef]
- Rempel A, Choudhary N; B, Pucker. KIPEs3: Automatic annotation of biosynthesis pathways. PLOS ONE 2023, 18, e0294342. [Google Scholar] [CrossRef]
- Renner SS, Zohner CM. The occurrence of red and yellow autumn leaves explained by regional differences in insolation and temperature. New Phytologist 2019, 224, 1464–1471. [Google Scholar] [CrossRef]
- Richter R, Dietz A; J, Foster; J, Spaethe; A, Stöckl. Flower patterns improve foraging efficiency in bumblebees by guiding approach flight and landing. Functional Ecology 2023, 37, 763–777. [Google Scholar] [CrossRef]
- Ridley HN. 1930. The dispersal of plants throughout the world.
- Ruxton GD, Schaefer HM. Floral colour change as a potential signal to pollinators. Current Opinion in Plant Biology 2016, 32, 96–100. [Google Scholar] [CrossRef] [PubMed]
- Saad KR, Kumar G; SN, Mudliar; P, Giridhar; NP, Shetty. Salt Stress-Induced Anthocyanin Biosynthesis Genes and MATE Transporter Involved in Anthocyanin Accumulation in Daucus carota Cell Culture. ACS Omega 2021, 6, 24502–24514. [Google Scholar] [CrossRef] [PubMed]
- Schaefer HM, Rolshausen G. Plants on red alert: do insects pay attention? BioEssays: News and Reviews in Molecular, Cellular and Developmental Biology 2006, 28, 65–71. [Google Scholar] [CrossRef] [PubMed]
- Schilbert HM, Schöne M; T, Baier; M, Busche; P, Viehöver; B, Weisshaar; D, Holtgräwe. Characterization of the Brassica napus Flavonol Synthase Gene Family Reveals Bifunctional Flavonol Synthases. Frontiers in Plant Science 12, 2021. [Google Scholar]
- Seitz C, Ameres S; G, Forkmann. Identification of the molecular basis for the functional difference between flavonoid 3’-hydroxylase and flavonoid 3’,5’-hydroxylase. FEBS letters 2007, 581, 3429–3434. [Google Scholar] [CrossRef]
- Shang Y, Venail J; S, Mackay; PC, Bailey; KE, Schwinn; PE, Jameson; CR, Martin; KM, Davies. The molecular basis for venation patterning of pigmentation and its effect on pollinator attraction in flowers of Antirrhinum. New Phytologist 2011, 189, 602–615. [Google Scholar] [CrossRef]
- Sheehan H, Feng T, Walker-Hale N, Lopez-Nieves S, Pucker B, Guo R, Yim WC, Badgami R, Timoneda A, Zhao L, et al.2020. Evolution of l-DOPA 4,5-dioxygenase activity allows for recurrent specialisation to betalain pigmentation in Caryophyllales. New Phytologist 227: 914–929. [CrossRef]
- Sheehan H, Moser M; U, Klahre; K, Esfeld; A, Dell’Olivo; T, Mandel; S, Metzger; M, Vandenbussche; L, Freitas; C, Kuhlemeier. MYB-FL controls gain and loss of floral UV absorbance, a key trait affecting pollinator preference and reproductive isolation. Nature Genetics 2016, 48, 159–166. [Google Scholar] [CrossRef]
- Shi M-Z, Xie D-Y. Features of anthocyanin biosynthesis in pap1-D and wild-type Arabidopsis thaliana plants grown in different light intensity and culture media conditions. Planta 2010, 231, 1385–1400. [Google Scholar] [CrossRef]
- Shin DH, Choi M; K, Kim; G, Bang; M, Cho; S-B, Choi; G, Choi; Y-I, Park. HY5 regulates anthocyanin biosynthesis by inducing the transcriptional activation of the MYB75/PAP1 transcription factor in Arabidopsis. FEBS Letters 2013, 587, 1543–1547. [Google Scholar] [CrossRef]
- Shin J, Park E; G, Choi. PIF3 regulates anthocyanin biosynthesis in an HY5-dependent manner with both factors directly binding anthocyanin biosynthetic gene promoters in Arabidopsis. The Plant Journal 2007, 49, 981–994. [Google Scholar] [CrossRef] [PubMed]
- Sinkkonen A. 2008. Red Reveals Branch Die-back in Norway Maple Acer platanoides. Annals of Botany 102: 361–366. [CrossRef]
- Sivankalyani V, Feygenberg O; S, Diskin; B, Wright; N, Alkan. Increased anthocyanin and flavonoids in mango fruit peel are associated with cold and pathogen resistance. Postharvest Biology and Technology 2016, 111, 132–139. [Google Scholar] [CrossRef]
- Sivitz AB, Reinders A; JM, Ward. Arabidopsis Sucrose Transporter AtSUC1 Is Important for Pollen Germination and Sucrose-Induced Anthocyanin Accumulation. Plant Physiology 2008, 147, 92–100. [Google Scholar] [CrossRef] [PubMed]
- Smillie RM, Hetherington SE. Photoabatement by Anthocyanin Shields Photosynthetic Systems from Light Stress. Photosynthetica 1999, 36, 451–463. [Google Scholar] [CrossRef]
- Smith SD. 2010. Using phylogenetics to detect pollinator-mediated floral evolution. New Phytologist 188: 354–363. [CrossRef]
- Solfanelli C, Poggi A; E, Loreti; A, Alpi; P, Perata. Sucrose-specific induction of the anthocyanin biosynthetic pathway in Arabidopsis. Plant Physiology 2006, 140, 637–646. [Google Scholar] [CrossRef]
- Stiles EW. 1982. Fruit Flags: Two Hypotheses. The American Naturalist 120: 500–509.
- Stone BC. 1979. Protective Coloration of Young Leaves in Certain Malaysian Palms. Biotropica. [CrossRef]
- Stracke R, Werber M; B, Weisshaar. The R2R3-MYB gene family in Arabidopsis thaliana. Current Opinion in Plant Biology 2001, 4, 447–456. [Google Scholar] [CrossRef]
- Su M, Zuo W; Y, Wang; W, Liu; Z, Zhang; N, Wang; X, Chen. The WKRY transcription factor MdWRKY75 regulates anthocyanins accumulation in apples (Malus domestica). Functional plant biology: FPB 2022, 49, 799–809. [Google Scholar] [CrossRef]
- Sun Y, Ji K, Liang B, Du Y, Jiang L, Wang J, Kai W, Zhang Y, Zhai X, Chen P, et al.2017. Suppressing ABA uridine diphosphate glucosyltransferase (SlUGT75C1) alters fruit ripening and the stress response in tomato. The Plant Journal 91: 574–589. [CrossRef]
- Sun Y, Li H; J-R, Huang. Arabidopsis TT19 Functions as a Carrier to Transport Anthocyanin from the Cytosol to Tonoplasts. Molecular Plant 2012, 5, 387–400. [Google Scholar] [CrossRef]
- Sun SG, Liao K; J, Xia; YH, Guo. Floral colour change in Pedicularis monbeigiana (Orobanchaceae). Plant Systematics and Evolution 2005, 255, 77–85. [Google Scholar] [CrossRef]
- Tan H, Luo X; J, Lu; L, Wu; Y, Li; Y, Jin; X, Peng; X, Xu; J, Li; W, Zhang. The long noncoding RNA LINC15957 regulates anthocyanin accumulation in radish. Frontiers in Plant Science 14, 2023. [Google Scholar]
- Tao R, Bai S; J, Ni; Q, Yang; Y, Zhao; Y, Teng. The blue light signal transduction pathway is involved in anthocyanin accumulation in ‘Red Zaosu’ pear. Planta 2018, 248, 37–48. [Google Scholar] [CrossRef]
- Tatsuzawa F, Saito N; N, Murata; K, Shinoda; A, Shigihara; T, Honda. 6-Hydroxypelargonidin glycosides in the orange–red flowers of Alstroemeria. Phytochemistry 2003, 62, 1239–1242. [Google Scholar] [CrossRef] [PubMed]
- Tavares S, Vesentini D; JC, Fernandes; RB, Ferreira; O, Laureano; JM, Ricardo-Da-Silva; S, Amâncio. Vitis vinifera secondary metabolism as affected by sulfate depletion: diagnosis through phenylpropanoid pathway genes and metabolites. Plant physiology and biochemistry: PPB 2013, 66, 118–126. [Google Scholar] [CrossRef]
- Teng S, Keurentjes J; L, Bentsink; M, Koornneef; S, Smeekens. Sucrose-Specific Induction of Anthocyanin Biosynthesis in Arabidopsis Requires the MYB75/PAP1 Gene. Plant Physiology 2005, 139, 1840–1852. [Google Scholar] [CrossRef] [PubMed]
- Thimm O, Bläsing O; Y, Gibon; A, Nagel; S, Meyer; P, Krüger; J, Selbig; LA, Müller; SY, Rhee; M, Stitt. MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. The Plant Journal: For Cell and Molecular Biology 2004, 37, 914–939. [Google Scholar] [CrossRef]
- Timoneda A, Feng T; H, Sheehan; N, Walker-Hale; B, Pucker; S, Lopez-Nieves; R, Guo; S, Brockington. The evolution of betalain biosynthesis in Caryophyllales. New Phytologist 2019, 224, 71–85. [Google Scholar] [CrossRef] [PubMed]
- Tohge T, Nishiyama Y, Hirai MY, Yano M, Nakajima J, Awazuhara M, Inoue E, Takahashi H, Goodenowe DB, Kitayama M, et al.2005. Functional genomics by integrated analysis of metabolome and transcriptome of Arabidopsis plants over-expressing an MYB transcription factor. The Plant Journal: For Cell and Molecular Biology 42: 218–235. [CrossRef]
- Trunschke J, Lunau K; GH, Pyke; Z-X, Ren; H, Wang. Flower Color Evolution and the Evidence of Pollinator-Mediated Selection. Frontiers in Plant Science 2021, 12, 617851. [Google Scholar] [CrossRef]
- Turcek F. 1963. Color preference in fruit- and seed-eating birds. Proc Int Ornithol Congr 13: 285–292.
- Turing AM. 1952. The chemical basis of morphogenesis. Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences 237: 37–72. [CrossRef]
- Van der Niet T, Peakall R; SD, Johnson. Pollinator-driven ecological speciation in plants: new evidence and future perspectives. Annals of Botany 2014, 113, 199–212. [Google Scholar] [CrossRef]
- Verweij W, Spelt CE; M, Bliek; M, de Vries; N, Wit; M, Faraco; R, Koes; FM, Quattrocchio. Functionally Similar WRKY Proteins Regulate Vacuolar Acidification in Petunia and Hair Development in Arabidopsis. The Plant Cell 2016, 28, 786–803. [Google Scholar] [CrossRef]
- Verweij W, Spelt C; G-P, Di Sansebastiano; J, Vermeer; L, Reale; F, Ferranti; R, Koes; F, Quattrocchio. An H+ P-ATPase on the tonoplast determines vacuolar pH and flower colour. Nature Cell Biology 2008, 10, 1456–1462. [Google Scholar] [CrossRef]
- Waki T, Mameda R, Nakano T, Yamada S, Terashita M, Ito K, Tenma N, Li Y, Fujino N, Uno K, et al.2020. A conserved strategy of chalcone isomerase-like protein to rectify promiscuous chalcone synthase specificity. Nature Communications 11: 870. [CrossRef]
- Walker AR, Lee E; SP, Robinson. Two new grape cultivars, bud sports of Cabernet Sauvignon bearing pale-coloured berries, are the result of deletion of two regulatory genes of the berry colour locus. Plant Molecular Biology 2006, 62, 623–635. [Google Scholar] [CrossRef]
- Wang H, Fan W; H, Li; J, Yang; J, Huang; P, Zhang. Functional characterization of Dihydroflavonol-4-reductase in anthocyanin biosynthesis of purple sweet potato underlies the direct evidence of anthocyanins function against abiotic stresses. PloS One 2013, 8, e78484. [Google Scholar] [CrossRef] [PubMed]
- Wang T-J, Huang S; A, Zhang; P, Guo; Y, Liu; C, Xu; W, Cong; B, Liu; Z-Y, Xu. JMJ17-WRKY40 and HY5-ABI5 modules regulate the expression of ABA-responsive genes in Arabidopsis. The New Phytologist 2021, 230, 567–584. [Google Scholar] [CrossRef] [PubMed]
- Waterman PG, Ross JA; DB, McKey. Factors affecting levels of some phenolic compounds, digestibility, and nitrogen content of the mature leaves ofBarteria fistulosa (Passifloraceae). Journal of Chemical Ecology 1984, 10, 387–401. [Google Scholar] [CrossRef]
- Wegener CB, Jansen G. Soft-rot Resistance of Coloured Potato Cultivars (Solanum tuberosum L.): The Role of Anthocyanins. Potato Research 2007, 50, 31–44. [Google Scholar] [CrossRef]
- Weiss MR. 1991. Floral colour changes as cues for pollinators. Nature 354: 227–229. [CrossRef]
- Weiss D. 2000. Regulation of flower pigmentation and growth: Multiple signaling pathways control anthocyanin synthesis in expanding petals. Physiologia Plantarum 110: 152–157. [CrossRef]
- Weng J-K, Noel JP. The Remarkable Pliability and Promiscuity of Specialized Metabolism. Cold Spring Harbor Symposia on Quantitative Biology 2012, 77, 309–320. [Google Scholar] [CrossRef] [PubMed]
- Werlein H-D, Kütemeyer C; G, Schatton; EM, Hubbermann; K, Schwarz. Influence of elderberry and blackcurrant concentrates on the growth of microorganisms. Food Control 2005, 16, 729–733. [Google Scholar] [CrossRef]
- Wheldale M. 1916. The anthocyanin pigments of plants.
- Williamson GB. 1982. Plant mimicry: evolutionary constraints. Biological Journal of the Linnean Society 18: 49–58. [CrossRef]
- Willmer P, Stanley DA; K, Steijven; IM, Matthews; CV, Nuttman. Bidirectional flower color and shape changes allow a second opportunity for pollination. Current biology: CB 2009, 19, 919–923. [Google Scholar] [CrossRef]
- Winkel-Shirley B. 2001. Flavonoid Biosynthesis. A Colorful Model for Genetics, Biochemistry, Cell Biology, and Biotechnology. Plant Physiology 126: 485–493. [CrossRef]
- Wolff K, Friedhoff R; JM, Horz; B, Pucker. Genome sequence of the medicinal and ornamental plant Digitalis purpurea reveals the molecular basis of flower color variation. : 2024.02.14.580303. [CrossRef]
- Wolff K, Pucker B. Dark Side of Anthocyanin Pigmentation. 2024. [Google Scholar] [CrossRef]
- Wolf-Saxon ER, Moorman CC; A, Castro; A, Ruiz-Rivera; JP, Mallari; JR, Burke. Regulatory ligand binding in plant chalcone isomerase-like (CHIL) proteins. The Journal of biological chemistry 2023, 299, 104804. [Google Scholar] [CrossRef]
- Wong DCJ, Wang Z; J, Perkins; X, Jin; GE, Marsh; EG, John; R, Peakall. Molecular Ecology; e17334: n/a, 2024. [Google Scholar] [CrossRef]
- Xu Z-S, Ma J; F, Wang; H-Y, Ma; Q-X, Wang; A-S, Xiong. Identification and characterization of DcUCGalT1, a galactosyltransferase responsible for anthocyanin galactosylation in purple carrot (Daucus carota L.) taproots. Scientific Reports 2016, 6, 27356. [Google Scholar] [CrossRef]
- Xu F, Ning Y; W, Zhang; Y, Liao; L, Li; H, Cheng; S, Cheng. An R2R3-MYB transcription factor as a negative regulator of the flavonoid biosynthesis pathway in Ginkgo biloba. Functional & Integrative Genomics 2014, 14, 177–189. [Google Scholar] [CrossRef]
- Xu Z, Rothstein SJ. ROS-Induced anthocyanin production provides feedback protection by scavenging ROS and maintaining photosynthetic capacity in Arabidopsis. Plant Signaling & Behavior 2018, 13, e1451708. [Google Scholar] [CrossRef]
- Yabuta Y, Motoki T; K, Yoshimura; T, Takeda; T, Ishikawa; S, Shigeoka. Thylakoid membrane-bound ascorbate peroxidase is a limiting factor of antioxidative systems under photo-oxidative stress. The Plant Journal 2002, 32, 915–925. [Google Scholar] [CrossRef] [PubMed]
- Yamasaki H, Sakihama Y; N, Ikehara. Flavonoid-Peroxidase Reaction as a Detoxification Mechanism of Plant Cells against H2O2. Plant Physiology 1997, 115, 1405–1412. [Google Scholar] [CrossRef]
- Yamazaki M, Yamagishi E; Z, Gong; M, Fukuchi-Mizutani; Y, Fukui; Y, Tanaka; T, Kusumi; M, Yamaguchi; K, Saito. Two flavonoid glucosyltransferases from Petunia hybrida: molecular cloning, biochemical properties and developmentally regulated expression. Plant Molecular Biology 2002, 48, 401–411. [Google Scholar] [CrossRef]
- Yao G, Ming M, Allan AC, Gu C, Li L, Wu X, Wang R, Chang Y, Qi K, Zhang S, et al.2017. Map-based cloning of the pear gene MYB114 identifies an interaction with other transcription factors to coordinately regulate fruit anthocyanin biosynthesis. The Plant Journal 92: 437–451. [CrossRef]
- Yonekura-Sakakibara K, Fukushima A, Nakabayashi R, Hanada K, Matsuda F, Sugawara S, Inoue E, Kuromori T, Ito T, Shinozaki K, et al.2012. Two glycosyltransferases involved in anthocyanin modification delineated by transcriptome independent component analysis in Arabidopsis thaliana. The Plant Journal 69: 154–167. [CrossRef]
- Yonekura-Sakakibara K, Tohge T; F, Matsuda; R, Nakabayashi; H, Takayama; R, Niida; A, Watanabe-Takahashi; E, Inoue; K, Saito. Comprehensive Flavonol Profiling and Transcriptome Coexpression Analysis Leading to Decoding Gene–Metabolite Correlations in Arabidopsis. The Plant Cell 2008, 20, 2160–2176. [Google Scholar] [CrossRef] [PubMed]
- Yoo J, Shin DH; M-H, Cho; T-L, Kim; SH, Bhoo; T-R, Hahn. An ankyrin repeat protein is involved in anthocyanin biosynthesis in Arabidopsis. Physiologia Plantarum 2011, 142, 314–325. [Google Scholar] [CrossRef]
- Yu J, Qiu K; W, Sun; T, Yang; T, Wu; T, Song; J, Zhang; Y, Yao; J, Tian. A long noncoding RNA functions in high-light-induced anthocyanin accumulation in apple by activating ethylene synthesis. Plant Physiology 2022, 189, 66–83. [Google Scholar] [CrossRef]
- Zhang Y, Butelli E, De Stefano R, Schoonbeek H, Magusin A, Pagliarani C, Wellner N, Hill L, Orzaez D, Granell A, et al.2013. Anthocyanins Double the Shelf Life of Tomatoes by Delaying Overripening and Reducing Susceptibility to Gray Mold. Current Biology 23: 1094–1100. [CrossRef]
- Zhang S, Chen J; T, Jiang; X, Cai; H, Wang; C, Liu; L, Tang; X, Li; X, Zhang; J, Zhang. Genetic mapping, transcriptomic sequencing and metabolic profiling indicated a glutathione S-transferase is responsible for the red-spot-petals in Gossypium arboreum. TAG. Theoretical and applied genetics. Theoretische und angewandte Genetik 2022a, 135, 3443–3454. [Google Scholar] [CrossRef]
- Zhang G, Chen D; T, Zhang; A, Duan; J, Zhang; C, He. Transcriptomic and functional analyses unveil the role of long non-coding RNAs in anthocyanin biosynthesis during sea buckthorn fruit ripening. DNA Research 2018, 25, 465–476. [Google Scholar] [CrossRef]
- Zhang M, Zhang X, Wang H, Ye M, Liu Y, Song Z, Du T, Cao H, Song L, Xiao X, et al.2022b. Identification and Analysis of Long Non-Coding RNAs Related to UV-B-Induced Anthocyanin Biosynthesis During Blood-Fleshed Peach (Prunus persica) Ripening. Frontiers in Genetics 13.
- Zhao J, Dixon RA. MATE Transporters Facilitate Vacuolar Uptake of Epicatechin 3′-O-Glucoside for Proanthocyanidin Biosynthesis in Medicago truncatula and Arabidopsis. The Plant Cell 2009, 21, 2323–2340. [Google Scholar] [CrossRef]
- Zheng X, Om K, Stanton KA, Thomas D, Cheng PA, Eggert A, Simmons E, Yuan Y-W, Conradi Smith GD, Puzey JR, et al.2021. The regulatory network for petal anthocyanin pigmentation is shaped by the MYB5a/NEGAN transcription factor in Mimulus. Genetics 217. [CrossRef]
- Zhou H, He J; Y, Zhang; H, Zhao; X, Sun; X, Chen; X, Liu; Y, Zheng; H, Lin. RHA2b-mediated MYB30 degradation facilitates MYB75-regulated, sucrose-induced anthocyanin biosynthesis in Arabidopsis seedlings. Plant Communications 2024, 5, 100744. [Google Scholar] [CrossRef] [PubMed]
- Zhou H, Lin-Wang K; H, Wang; C, Gu; AP, Dare; RV, Espley; H, He; AC, Allan; Y, Han. Molecular genetics of blood-fleshed peach reveals activation of anthocyanin biosynthesis by NAC transcription factors. The Plant Journal 2015, 82, 105–121. [Google Scholar] [CrossRef] [PubMed]
- Zhou L-L, Shi M-Z; D-Y, Xie. Regulation of anthocyanin biosynthesis by nitrogen in TTG1-GL3/TT8-PAP1-programmed red cells of Arabidopsis thaliana. Planta 2012, 236, 825–837. [Google Scholar] [CrossRef]
- Zhu H-F, Fitzsimmons K; A, Khandelwal; RG, Kranz. CPC, a single-repeat R3 MYB, is a negative regulator of anthocyanin biosynthesis in Arabidopsis. Molecular Plant 2009, 2, 790–802. [Google Scholar] [CrossRef] [PubMed]
- Zhu L, Li X; X, Hu; X, Wu; Y, Liu; Y, Yang; Y, Zang; H, Tang; C, Wang; J, Xu. Quality Characteristics and Anthocyanin Profiles of Different Vitis amurensis Grape Cultivars and Hybrids from Chinese Germplasm. Molecules 2021, 26, 6696. [Google Scholar] [CrossRef]
- Zirngibl M-E, Araguirang GE; A, Kitashova; K, Jahnke; T, Rolka; C, Kühn; T, Nägele; AS, Richter. Triose phosphate export from chloroplasts and cellular sugar content regulate anthocyanin biosynthesis during high light acclimation. Plant Communications 2023, 4, 100423. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).




