Introduction
Background and Objectives
Gastroesophageal reflux disease (GERD) is a prevalent chronic condition characterized by regurgitating stomach contents into the esophagus, leading to symptoms such as heartburn and acid reflux. GERD is a significant public health issue worldwide, with an increasing prevalence over the past decades. It has been linked to complications such as esophagitis, Barrett’s esophagus, and an elevated risk of esophageal adenocarcinoma. The importance of advancing research in GERD cannot be understated, as understanding its pathophysiology, management strategies, and potential complications remains critical for improving patient outcomes [
1].
Globally, GERD affects millions of people, with varying prevalence rates across different regions. The condition is prevalent in Western countries, where lifestyle factors such as diet, obesity, and sedentary behavior contribute to its high incidence. In contrast, the prevalence in Asian countries has been historically lower, but recent studies indicate a rising trend, possibly due to changes in dietary habits and increasing rates of obesity [
2].
In Europe and North America, GERD is one of the most common gastrointestinal disorders. The high prevalence in these regions has led to significant healthcare costs and a substantial burden on healthcare systems. Research in these areas has focused on understanding the impact of lifestyle factors, such as diet and physical activity, on GERD symptoms and developing effective management strategies [
3]. Additionally, there is a growing interest in the role of genetic predisposition and the microbiome in the development of GERD.
In Asia, the prevalence of GERD has been increasing over the past few decades. This rise is attributed to rapid urbanization, changes in dietary patterns, and an increase in obesity rates. Asian countries have also seen a growing interest in GERD research, with studies exploring the unique clinical characteristics and risk factors in this population. There is a particular focus on the role of Helicobacter pylori infection, which is more common in Asia and may have a protective effect against GERD [
4].
Despite the growing volume of research on GERD, the field faces several challenges, including variability in diagnostic criteria, management strategies, and a need for more robust clinical guidelines. Moreover, while significant progress has been made in understanding GERD’s underlying mechanisms and treatment options, collaboration among researchers remains crucial for further advancements. Co-authorship network analysis offers valuable insights into the collaborative landscape of GERD research by elucidating the relationships between researchers and identifying key contributors and influential research groups. This study aims to analyze the structure and evolution of co-authorship networks in GERD research from 2000 to 2023, providing a comprehensive overview of collaboration patterns and identifying critical nodes within the network.
Scope of the Study
This study examines publications related to GERD research indexed in the Web of Science (WoS) Core Collection database between 2000 and 2023. A total of 19,463 articles were selected for analysis, providing a comprehensive overview of the collaborative landscape within this specialized field over the past two decades. The dataset ensures the inclusion of the most recent publications (as of September 2024). The analysis will focus on constructing and evaluating co-authorship networks using macro-level indicators such as network density (the ratio of actual to possible connections), clustering coefficient (the degree to which nodes tend to cluster together), number of components (distinct connected subgroups within the network), and average path length (the average distance between nodes). At the micro-level, I will assess degree centrality (the number of direct connections each node has), closeness centrality (how close a node is to all other nodes), and betweenness centrality (the extent to which a node lies on the shortest path between other nodes). These metrics will help illuminate the structure and dynamics of researcher collaborations in this field.
Significance of the Study
This study contributes significantly to the field of GERD research by providing a detailed analysis of the collaborative network structures and their evolution over time. By identifying major researchers and research institutions, the study highlights the key players who drive GERD research. Moreover, understanding the progression of international collaborative research and its impact on the field can guide future efforts to foster global partnerships. This analysis not only delineates the structural characteristics of the co-authorship networks but also sheds light on the current state and future directions of GERD research.
The findings of this study have the potential to inform strategic decisions by researchers, funding bodies, and policymakers by identifying areas where collaboration can be strengthened. Furthermore, by mapping the network changes over the years, this study provides insights into the dynamics of research collaboration, including the emergence of new research clusters and shifts in research focus. Ultimately, this study underscores the importance of international collaboration in advancing GERD research and highlights the evolving landscape of scientific cooperation within this field.
Material and Methods
The present study investigates the co-authorship patterns in GERD research papers. I utilized the WoS Core Collection database, conducting a "Topic Search" with the keyword " gastroesophageal reflux disease" to analyze a total of 19,463 articles published between 2000 and 2023 (as of September 2024). In this analysis, I examined who collaborated with whom in co-authoring these papers. I conducted network analysis using the Python programming language (version 3.10.5) within the integrated development environment (IDE) PyCharm (software version 2022.1.3). This study employed methodology-established principles of social network analysis [
5]. I carried out the analysis in two main parts:
Macro-level Metrics:
Network Density: Calculated as the ratio of the number of edges to the maximum possible edges Between all nodes.
Clustering Coefficient: Measured the extent to which nodes form clusters by considering the number of edges among neighboring nodes and calculating the average.
Components: Identified and counted the number of subgraphs (components) where nodes are mutually connected.
Average Path Length: Evaluated the average "distance" between nodes by calculating the overall average path length in the network [
6].
Micro-level Metrics:
Degree Centrality: Measured the importance of each node by counting the number of edges it has in the network.
Closeness Centrality: Defined as the inverse of the sum of the shortest path lengths from a node to all other nodes, measuring how close each node is to others in the network.
Betweenness Centrality: Assessed the extent to which a node lies on the shortest paths between other nodes, indicating its importance in information transmission within the network [
6,
7].
The significance of these macro-level metrics in understanding the structure of scientific collaboration networks and these micro-level centrality measures in scientific collaboration networks has been well documented and used [
6,
7]. Through these analyses, I can identify collaborative relationships and influential researchers in GERD research. This information may be useful for understanding research trends and planning future collaborative studies.
Results
The study analyzed the co-authorship network of researchers in the field of GERD research, focusing on the periods from 2000 to 2023. The analysis was conducted using data from the WoS Core Collection and utilized both macro and micro-level network metrics to understand the evolution of collaborative networks in this field.
2000-2009: Network Analysis
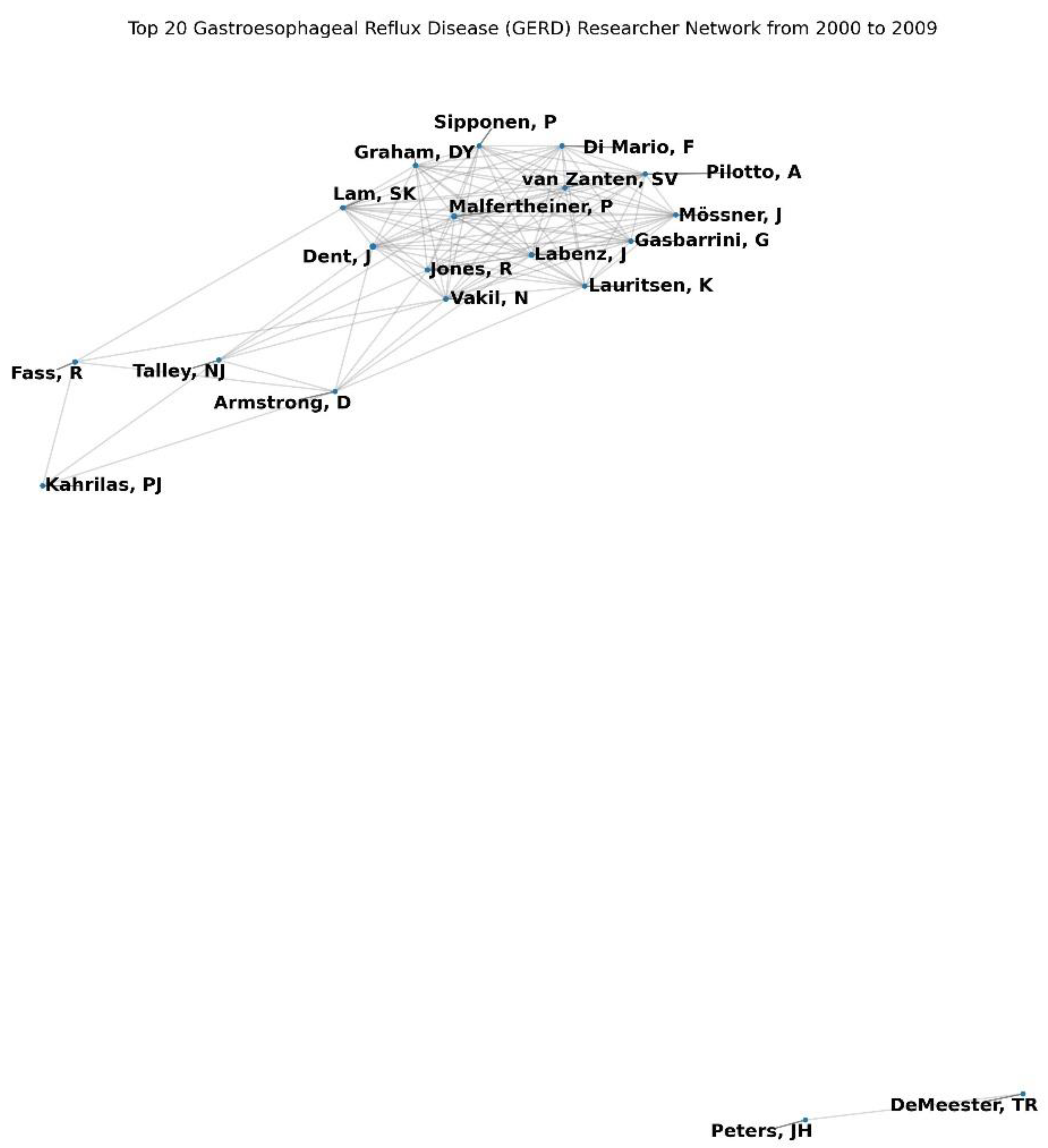
During the 2000–2009 period, the GERD co-authorship network consisted of numerous nodes and links, representing authors and their collaborative relationships (
Figure 1). The network density was extremely low at 0.000485 (
Table 1), indicating that only 0.0485% of the potential collaborations were realized. The average clustering coefficient was 0.879 (
Table 1), suggesting that authors tended to cluster together in collaborative groups (
Figure 1). The network was fragmented into 1,468 components (
Table 1), with the average distance between nodes being infinite, reflecting the disconnection between many parts of the network [
8].
In terms of micro-level metrics, the top authors by degree centrality, indicating the number of direct collaborations, included Dent, J (0.0114), Malfertheiner, P (0.0097), and Labenz, J (0.008). These authors were highly influential in terms of their collaborative reach within the network (
Table 2). Closeness centrality, which measures how quickly an author can connect with others, highlighted Dent, J (0.1004), Kahrilas, PJ (0.0988), and Talley, NJ (0.0970) as central figures (
Table 3). Betweenness centrality, reflecting the extent to which an author serves as a bridge in the network, identified Dent, J (0.0203), Fass, R (0.0149), and Kahrilas, PJ (0.0147) as key intermediaries facilitating connections between otherwise disconnected authors (
Table 4).
2010-2019: Network Analysis
In the subsequent decade (2010–2019), the GERD co-authorship network expanded, yet it remained sparse with a network density of 0.000514 (
Table 1), slightly higher than the previous period but still indicating limited collaboration among potential connections (
Figure 2). The average clustering coefficient increased to 0.898 (
Table 1), demonstrating a stronger tendency for authors to form tight-knit clusters (
Figure 2). The number of components rose to 2,492 (
Table 1), reflecting the network's continued fragmentation, and the average distance remained infinite, indicating persistent structural gaps [
8].
The degree centrality analysis for this period revealed that Shaheen, Nicholas J. (0.0185), De Giorgio, Roberto (0.0135), and Di Sabatino, Antonio (0.0131) were among the most collaborative authors, engaging with numerous peers (
Table 2). Closeness centrality scores highlighted Gyawali, C. Prakash (0.1341), Bredenoord, Albert J. (0.1334), and Vaezi, Michael F. (0.1332) as being strategically positioned to reach others efficiently (
Table 3). Betweenness centrality underscored the importance of authors such as Verheij, J (0.026), Bredenoord, Albert J. (0.0240), and Kahrilas, Peter J. (0.0188), who played significant roles in connecting disparate parts of the network (
Table 4).
2020-2023: Network Analysis
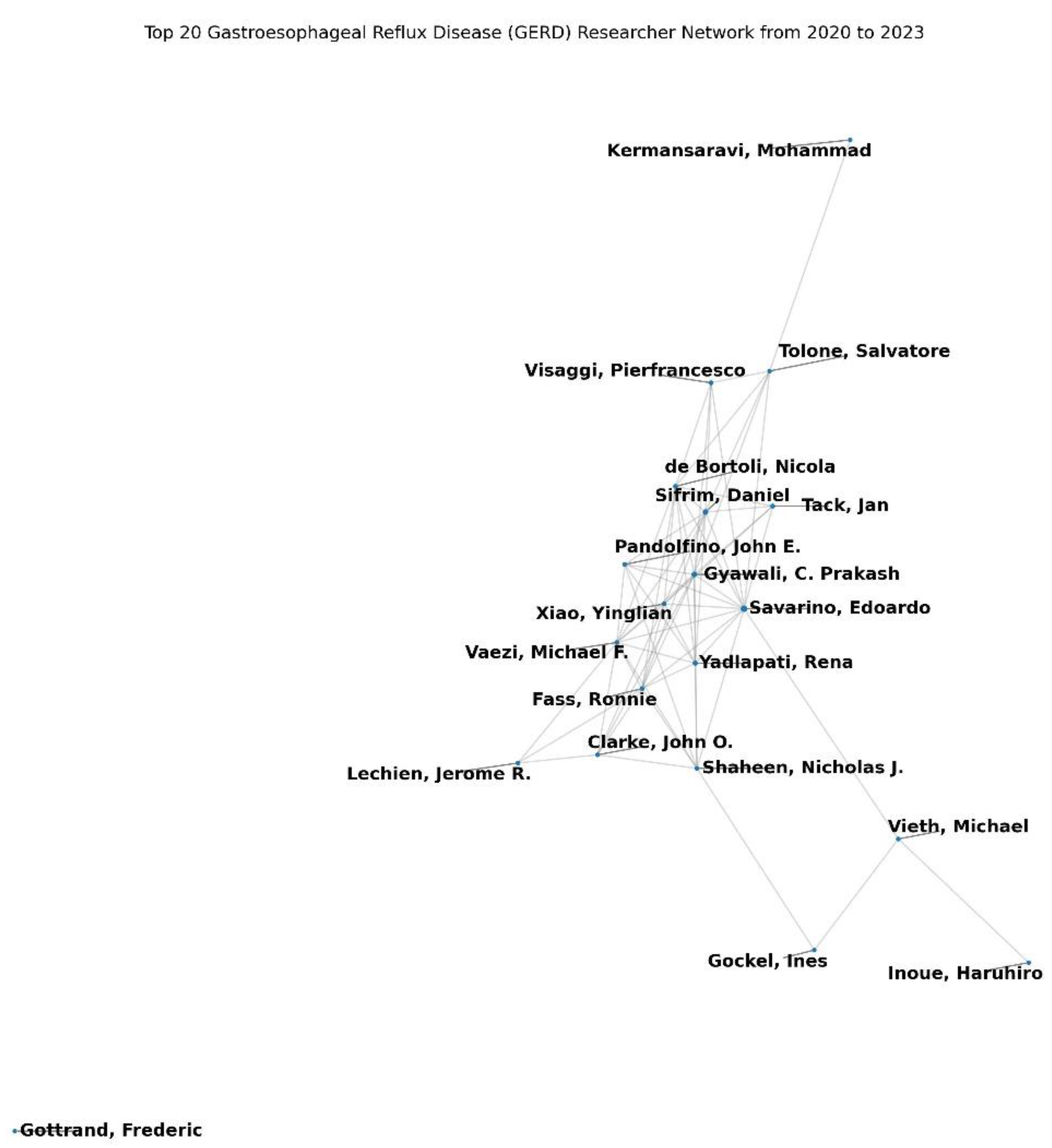
The GERD co-authorship network analysis from 2000 to 2023 revealed a complex structure of collaborative relationships. The network density was extremely low at 0.000414 (
Table 1), indicating that only 0.0414% of potential collaborations were realized. This suggests a sparse network with considerable untapped collaborative potential (
Figure 3). The average clustering coefficient was high at 0.921 (
Table 1), demonstrating that authors tended to form tight-knit collaborative groups (
Figure 3). The network was highly fragmented, consisting of 2,172 components (
Table 1), with the average distance between nodes being infinite, reflecting significant disconnection between many parts of the network [
8].
At the micro level, the top authors by degree centrality, indicating the number of direct collaborations, included Savarino, Edoardo (0.0119), Gyawali, C. Prakash (0.0081), and Sifrim, Daniel (0.0081). These authors were highly influential in terms of their collaborative reach within the network (
Table 2). Closeness centrality, which measures how quickly an author can connect with others, highlighted Savarino, Edoardo (0.1271), Gyawali, C. Prakash (0.1262), and Yadlapati, Rena (0.1226) as central figures (
Table 3). Betweenness centrality, reflecting the extent to which an author serves as a bridge in the network, identified Savarino, Edoardo (0.0198), Gyawali, C. Prakash (0.0177), and Lin, Lin (0.0129) as key intermediaries facilitating connections between otherwise disconnected authors (
Table 4).
Discussion
This study examined the structure of co-authorship networks in GERD research from 2000 to 2023, using data from the WoS Core Collection. The analysis revealed significant insights into the collaborative landscape of GERD research, highlighting both the strengths and weaknesses of the existing network structures over time.
From 2000 to 2009, the GERD research co-authorship network displayed low connectivity, characterized by a sparse network with a density of only 0.000485. This suggests collaborative efforts were relatively limited, with most potential co-authorships remaining unrealized. Despite this, the clustering coefficient was relatively high at 0.879, indicating that when collaborations did occur, they were often within closely-knit groups of researchers. This high clustering tendency may suggest that collaborations were largely driven by small, focused research teams rather than extensive interdisciplinary or international collaborations. In the subsequent decade (2010-2019), the network density was 0.000514, indicating a relatively sparse network with limited collaborative efforts in GERD research. The average clustering coefficient was notably high at 0.898, suggesting that while collaborations existed, they were primarily within tightly knit research groups rather than across the broader network. The large number of components (2,492) and the infinite average path length indicate significant fragmentation within the network, with many researchers working in isolated subgroups without connections to the broader research community. During the most recent period (2020-2023), the co-authorship network showed a slight decline in density to 0.000414, reflecting a continued sparsity in collaborative connections. However, the average clustering coefficient increased to 0.921, indicating a trend towards even more closely connected clusters within the network, albeit without a substantial increase in overall interconnectedness. The number of components decreased to 2,172, reflecting a modest improvement in network integration, but the average distance remained infinite, highlighting persistent gaps between research groups.
Analyzing micro-level metrics revealed that researchers with a high degree centrality played pivotal roles in facilitating collaborations across different periods in GERD research. For instance, during 2000-2009, Dent, J (0.0114) and Malfertheiner, P (0.0097) were among the highest in degree centrality, emphasizing their influence in connecting researchers within the network. Similarly, from 2010-2019, Shaheen, Nicholas J. (0.0185) and De Giorgio, Roberto (0.0135) were key figures, and from 2020-2023, Savarino, Edoardo (0.0119) and Gyawali, C. Prakash (0.0081) emerged as central nodes. Their consistent presence underscores the central role these individuals played in fostering research connections across the years.
In terms of closeness centrality, Dent, J (0.1004) and Kahrilas, PJ (0.0988) were prominent during 2000-2009, suggesting their well-positioned roles within the network, which allowed them to effectively disseminate information and ideas. In 2010-2019, Gyawali, C. Prakash (0.1341) and Bredenoord, Albert J. (0.1334) stood out, while in 2020-2023, Savarino, Edoardo (0.1271) and Gyawali, C. Prakash (0.1262) maintained strong positions, indicating their ability to connect rapidly with other researchers. Betweenness centrality analysis identified several researchers as critical bridges within the network. For 2000-2009, Dent, J (0.0203) and Fass, R (0.0149) were key intermediaries, facilitating the flow of knowledge across otherwise disconnected groups. From 2010-2019, Verheij, J. (0.026) and Bredenoord, Albert J. (0.0240) played similar roles, and in 2020-2023, Savarino, Edoardo (0.0198) and Gyawali, C. Prakash (0.0177) acted as essential connectors, enhancing the overall connectivity of the GERD research community. The analysis indicates that while GERD research networks have become increasingly connected over time, evidenced by high clustering coefficients across all periods (0.879, 0.898, 0.921), there is still room for growth, particularly in fostering more global and interdisciplinary collaborations. The relatively low network densities (0.000485, 0.000514, 0.000414) and high number of components suggest that while the networks are interconnected, the overall percentage of realized collaborations remains limited, highlighting the potential for expanding research partnerships. The findings underscore the importance of key researchers who act as central nodes within the network. These individuals often drive the direction of research and significantly influence the development of collaborative projects. Efforts to encourage broader participation and integrate emerging researchers into the network could further enhance the robustness and reach of GERD research.
In conclusion, this study provides a comprehensive overview of the evolution of co-authorship networks in GERD research from 2000 to 2023. The findings highlight both progress and ongoing challenges in the field, emphasizing the need for continued efforts to foster collaboration and integration within the GERD research community. By identifying key contributors and understanding the structural dynamics of research networks, this analysis offers valuable insights that can guide future collaborative initiatives and research strategies in GERD.
Conclusion
The co-authorship network analysis of GERD research from 2000 to 2023 reveals a sparse and fragmented collaborative landscape, with significant room for improvement in researcher cooperation and connectivity. Despite the global prevalence of GERD and its substantial impact on public health, the co-authorship network exhibited a consistently low density across all periods studied, indicating that only a small fraction of potential collaborations between researchers have been realized. Specifically, the network density remained below 0.001 in each period, demonstrating a vast number of unexploited collaborative opportunities.
The analysis highlighted a persistent trend of high clustering within smaller groups, as evidenced by average clustering coefficients exceeding 0.87 across all decades. This suggests that while authors tend to form tight-knit collaborative clusters, these clusters are largely isolated from one another, contributing to the overall disconnection of the network. The number of components consistently increased over time, reaching 2,172 in the 2020-2023 period, and the average path length between nodes remained infinite, indicating significant structural gaps and limited pathways for information flow across the broader network.
At the micro level, key researchers with high degree, closeness, and betweenness centrality scores were identified in each period, underscoring their pivotal roles in facilitating collaboration and information dissemination within their respective clusters. For example, in the most recent period (2020-2023), Savarino, Edoardo (Italy), Gyawali, C. Prakash (USA), and Sifrim, Daniel (UK) emerged as influential figures with extensive collaborative reach, as reflected by their high centrality metrics. However, the reliance on a limited number of central nodes for connectivity underscores the network's vulnerability to disruptions, as these key individuals serve as critical bridges in otherwise disconnected segments of the network.
The findings from this study underscore the need for enhanced collaborative efforts and broader integration among GERD researchers worldwide. Increasing inter-cluster collaborations could significantly improve the cohesion of the network, facilitating more efficient knowledge sharing and accelerating scientific advancements in GERD research. As the prevalence of GERD continues to rise globally, particularly in Asia, fostering robust international collaborations and expanding the network's connectivity will be crucial for addressing the complex challenges associated with this condition. Ultimately, this study highlights the evolving landscape of GERD research collaboration and provides a roadmap for strategic efforts to strengthen the global research network in this field.
Institutional Review Board Statement
not applicable for this article.
Conflicts of Interest
none.
Abbreviations
GERD, Gastroesophageal Reflux Disease; WoS, Web of Science; IDE, Integrated Development Environment
References
- Boulton, Katie H. A.;Dettmar, Peter W. A narrative review of the prevalence of gastroesophageal reflux disease (GERD). Ann. Esophagus 2022, 5, 7. [CrossRef]
- Dent J, El-Serag HB, Wallander MA, Johansson S. Epidemiology of gastro-oesophageal reflux disease: a systematic review. Gut 2005, 54, 710–717. [CrossRef] [PubMed]
- El-Serag HB, Sweet S, Winchester CC, Dent J. Update on the epidemiology of gastro-oesophageal reflux disease: a systematic review. Gut 2014, 63, 871–880. [CrossRef] [PubMed]
- Jung, HK. Epidemiology of gastroesophageal reflux disease in Asia: a systematic review. J Neurogastroenterol Motil. 2011, 17, 14–27. [Google Scholar] [CrossRef] [PubMed]
- Wasserman, S., & Faust, K. (1994) Social network analysis: Methods and applications. Cambridge University Press. [CrossRef]
- Newman, M. 'Scientific collaboration networks. II. Shortest paths, weighted networks, and centrality'. Phys. Rev. E 2001, 64, 016132. [Google Scholar] [CrossRef]
- Newman, ME. The structure of scientific collaboration networks. Proc Natl Acad Sci USA 2001, 98, 404–409. [Google Scholar] [CrossRef] [PubMed]
- Barabasi AL, Albert R. Emergence of scaling in random networks. Science 1999, 286, 509–512. [CrossRef] [PubMed]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).