1. Introduction
Multiple factors contribute to cancer development, resistance, and recurrence [
1]; however, the accumulation of genetic aberrations in crucial genes plays a central role in this process leading to uncontrolled intracellular signaling networks [
2]. Among them [
1,
3], the oncogene ZNF217 has recently emerged as a potential therapeutic target. ZNF217 has been reported to foster tumorigenesis by several mechanisms, including sustaining cell proliferation, enabling replicative immortality, and resisting cell death [
4]. In cancer later stages, it also induces epithelial-mesenchymal transition (EMT) and chemotherapy resistance [
5]. The dysregulation of different pathways is involved in ZNF217 deleterious effects, among which PI3K/AKT, mitogen-activated protein kinase (MAPK), Janus kinase/signal transducers and activators of transcription 3 (JAK/STAT3), and transforming growth factor beta (TGF-β) ones [
5]. ZNF217 aberrant expression has been highlighted in several tumors, such as breast, colon, liver, and thyroid ones, and correlated to therapeutic resistance and patients’ poor outcomes [
5,
6]. Therefore, targeting ZNF217 could be a promising strategy to fight cancer and its related chemoresistance. So far, only one drug, namely triciribine, has shown to counteract the ZNF217-driven deleterious effects [
7]. Therefore, it is still necessary to discover new compounds able to target ZNF217, so improving the prognosis of cancer patients.
Plants represent an important source of novel bioactive compounds which can help in the prevention and treatment of cancer, thus acting as chemopreventive agents [
8]. They can be exploited in healthy people to prevent cancer occurrence, in high-risk subjects to avoid the progression of premalignant lesions, and as adjuvant treatments in oncologic or post-treated patients [
9]. Among compounds of potential interest for both chemopreventive effects and eco-friendly recover, cucurbitacins have recently emerged. These are highly oxidized tetracyclic triterpenoids distributed in several plant families (e.g., Cucurbitaceae and Elaeocarpaceae) where they act as defense compounds [
10]. They have attracted a great attention due to their broad range of pharmacological effects, which include anticancer and chemosensitizer ones [
11]. So far, several cucurbitacins have been isolated [
12]. Cucurbitacin B, E, and I represent the most investigated compounds; however, also CucD deserves attention [
13]. Indeed, it has shown antiproliferative activity in variety of cancers, including human adult T-cell leukemia, breast, cervical, prostate, gastric, lung, and liver ones, by modulating several pathways involved in cancer cell proliferation, such as PI3K/AKT/mTOR, MAPKs, JAK2/STAT3, ROS/p38, NF-kappa B, and EGFR ones, which are linked to ZNF217 expression [
14,
15,
16,
17,
18]. Despite that, the modulatory effect of Cucurbitacin D on ZNF217 expression has not been investigated yet.
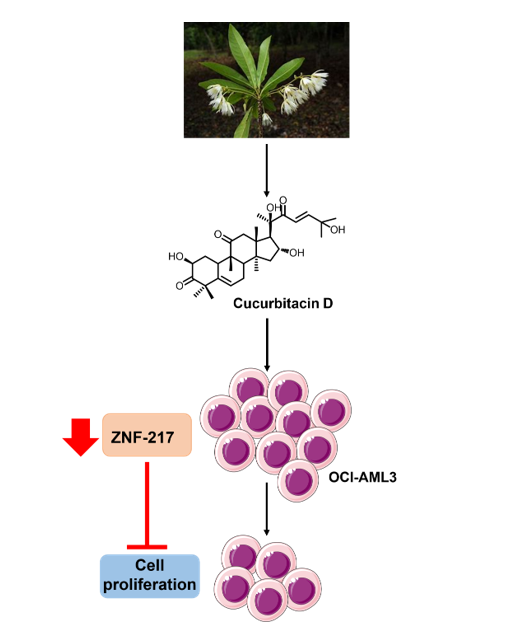
Therefore, the objectives of the present study is to evaluated the existing relationship between the chemopreventive properties of Cucurbitacins and the modulation of ZNF217 in acute myeloid leukemia and other hematology cancer cell lines that have been shown to express ZNF217. To this aim, CucD (1), isocucD (2), and CucI (3) were isolated from the aerial parts of the Vietnamese plant
Elaeocarpus hainanensis Oliv. (
E. hainanensis), an ornamental plant highly used in Asian urban landscape and garden [
19,
20]. The use of
Elaeocarpus has been known in oriental medicine [
19] and the traditional medicine use of
E. hainanensis has been reported in Institutum Botanicum Academiae Sinicae, in Iconographia Cormophytorum Sinicorum and in Flora of China. Indeed, the recovery of cucurbitacins from the pruning process could represent an interesting recycling strategy which allow to obtain them for therapeutic purposes without endangering the ecosystem biodiversity. This approach acquires greater importance also considering the difficulties to obtain CuD by synthesis, owing to its complex chemical structure [
21]. Afterwards, purified compounds were tested on nucleophosmin (NPM)-mutated acute myeloid leukemia (AML) and other hematology cancer cell lines to study their effects on viability, apoptosis, and cell cycle progression. At last, the influence of cucurbitacins on the expression of ZNF217 was evaluated too. The significance of the study lies in the identification of physiologically active compounds from terrestrial plants and in the pharmacology of compounds of natural origin.
2. Results
2.1. Isolation of Cucurbitacins from E. hainanensis
Compounds 1, 2, and mixture of 1 + 3 were isolated from the methanolic extract of Elaeocarpus hainanensis Oliv. twigs and leaves (E. hainanensis) by using combined column chromatographic separations with appropriate mobile phases. Their structures were identified as cucurbitacin D (CucD, 1) [
22,
23], 3-epi-isocucurbitacin D (isocucD, 2)[
23], and mixture of cucurbitacin D and cucurbitacin I (CucD + CucI, 1 + 3) [
22,
23] by NMR spectroscopic analysis and comparison with those of reported data [
22,
23,
24] (see details of NMR data in
Table S1 in Supporting Information). The ratio of compounds 1 and 3 in the mixture (1:3 =1:1) was determined by the proton integral intensity (H-6).
2.2. Effect of Cucurbitacin D (CucD) on the Number and Survival of OCI-AML-3 Cells
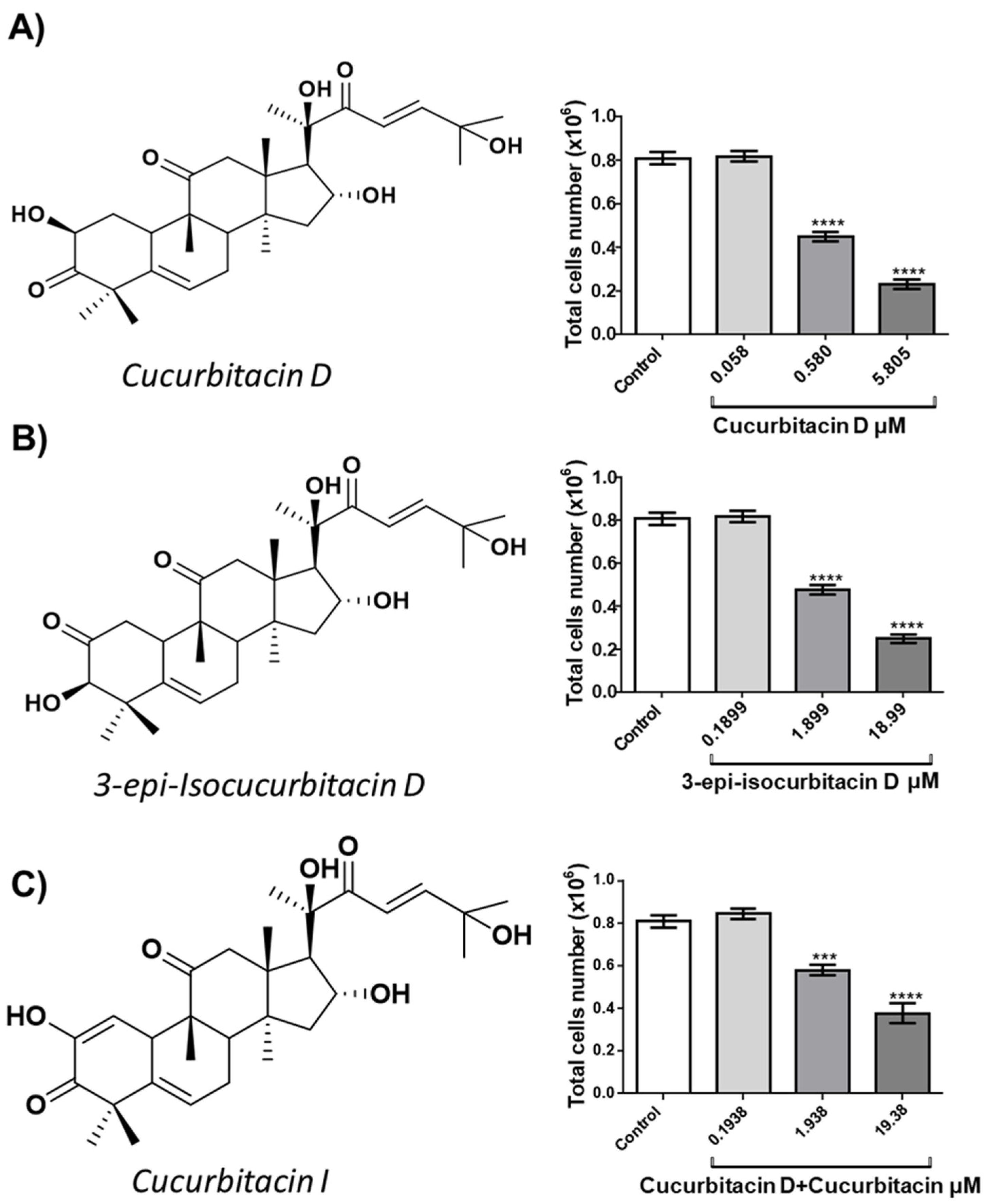
OCI-AML3 (OCI) acute myeloid leukemia cells were cultured with different concentrations of either CucD (0.058, 0.58 and 5,805 μM, IC50: 0.58 μM), isocucD (0.1899, 1.899 or 18.99 μM, IC50 = 1.899 μM) or the mixture CucD + CucI, (0,1938, 1.938 and 19.38 μM, IC50 = 1.938 μM). After 24 hours, the cells were collected and counted.
Figure 1 shows the chemical structures (on the left side) and the number of the harvested cells (right side). All three cucurbitacins resulted in a significant reduction of the OCI cell number although CucD was the most powerful since the minimum effective concentration was 0.3 μg/mL. Since at 24 hours, the effects were already evident, we focused on this time without carrying out time-course experiments. Since our compounds resulted in a positive effect, we only added negative control to avoid false positives. While we did not add positive controls, which were necessary if the results were negative (to avoid false negatives). Furthermore, being three, each compound acted as a positive control for the others.
2.3. Effect of CucD and IsocucD on Apoptosis of OCI-AML-3 Cells
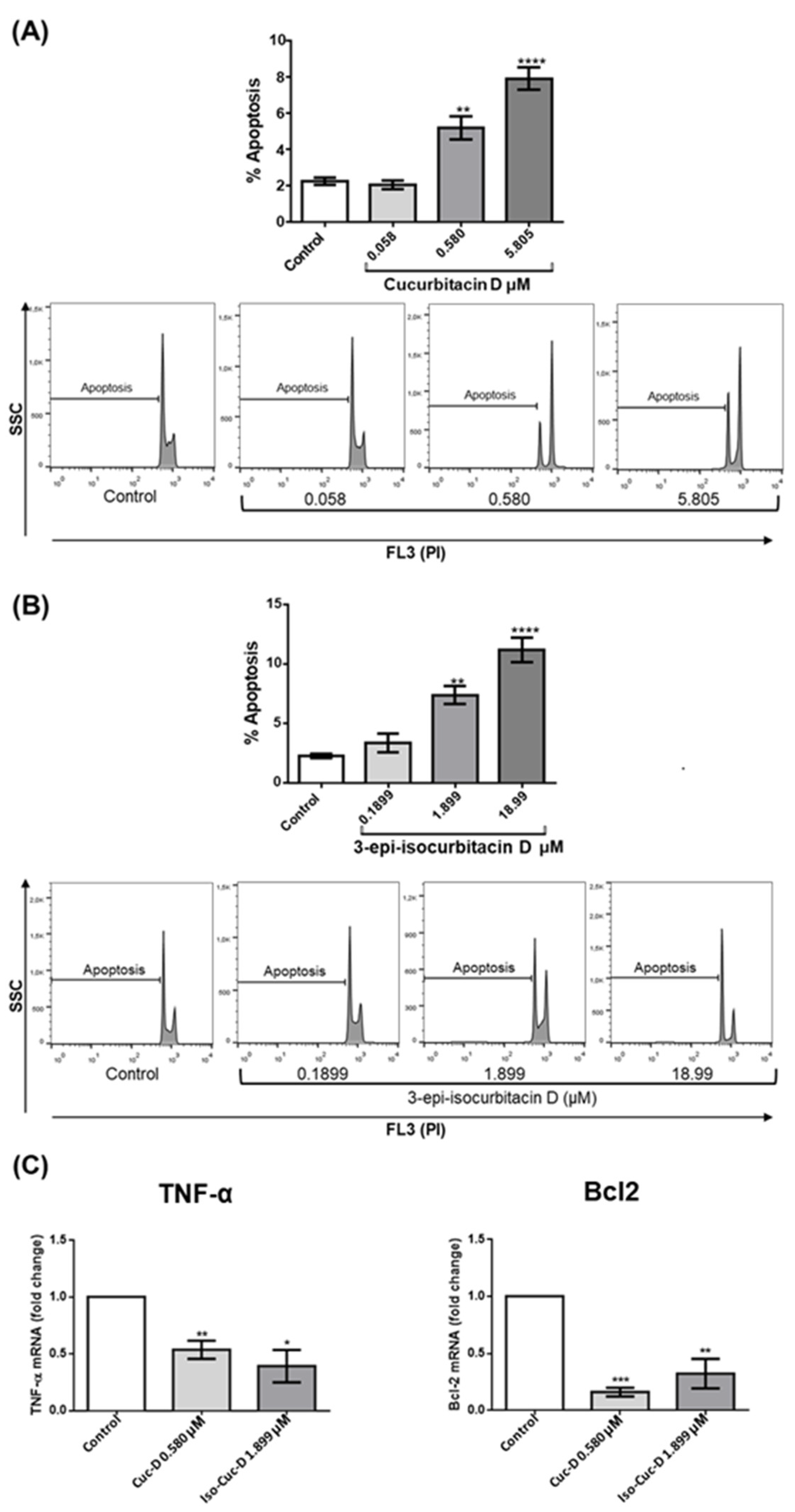
The reduction in cell number induced by the isolated compounds may be the result of increased cell death, decreased cell proliferation, or both. Therefore, we first investigated the possible induction of apoptotic cell death by the two most powerful compounds, namely CucD and IsocucD. To this aim, we stained cell nuclei with propidium iodide (PI) and performed a cytofluorimeter analysis, after exclusion of necrotic cells by size (FSC) and elimination of doublets. Results showed that the same concentrations that resulted in reduction of cell number (0.58 and 5.805 μM for CucD; 1.899 and 18.99 μM for IsocucD) also promoted a significant increase in apoptosis (
Figure 2A and 2B). Due to the low level of apoptosis, its increase can only be detected by measurement and is not detectable by eyes. So, the tested cucurbitacins promoted the reduction in the number of OCI cells, at least in part, through an increase of apoptosis.
The increased CucD- and IsoCucD-mediated apoptosis of OCI-AML3 cells prompted us to investigate the molecules involved in apoptotic cell death. Apoptosis is activated by at least two different pathways. The mitochondrial (i.e., intrinsic) pathway leads to down-regulation of anti-apoptotic molecules, such as Bcl2, with the following sequential release of cytochrome c from mitochondria and activation of caspase-9, which directly cleaves and activates caspase-3. The second (i.e., extrinsic) pathway involves activation of caspase-8 that is triggered by the stimulation of death receptors such as TNFR by its ligand TNF-α [
25]. We cultured OCI-AML3 cells for 24 h with the vehicle DMSO (control), CucD or IsoCucD before extracting RNA for real-time analyses. The significant differences were calculated on five independent experiments.
Figure 2 shows that the pro-apoptotic molecule TNF-α was significantly down-regulated, whereas the intrinsic anti-apoptotic molecule Bcl2 was significantly down-regulated. These results indicate that the TNF-α-dependent extrinsic apoptotic pathway was inhibited by CucD and IsoCucD treatment, whereas the Bcl2-dependent intrinsic apoptotic pathway was induced by treatment with CucD and IsoCucD.
2.4. Effect of CucD and IsocucD on the Cell Cycle of OCI-AML-3 Cells
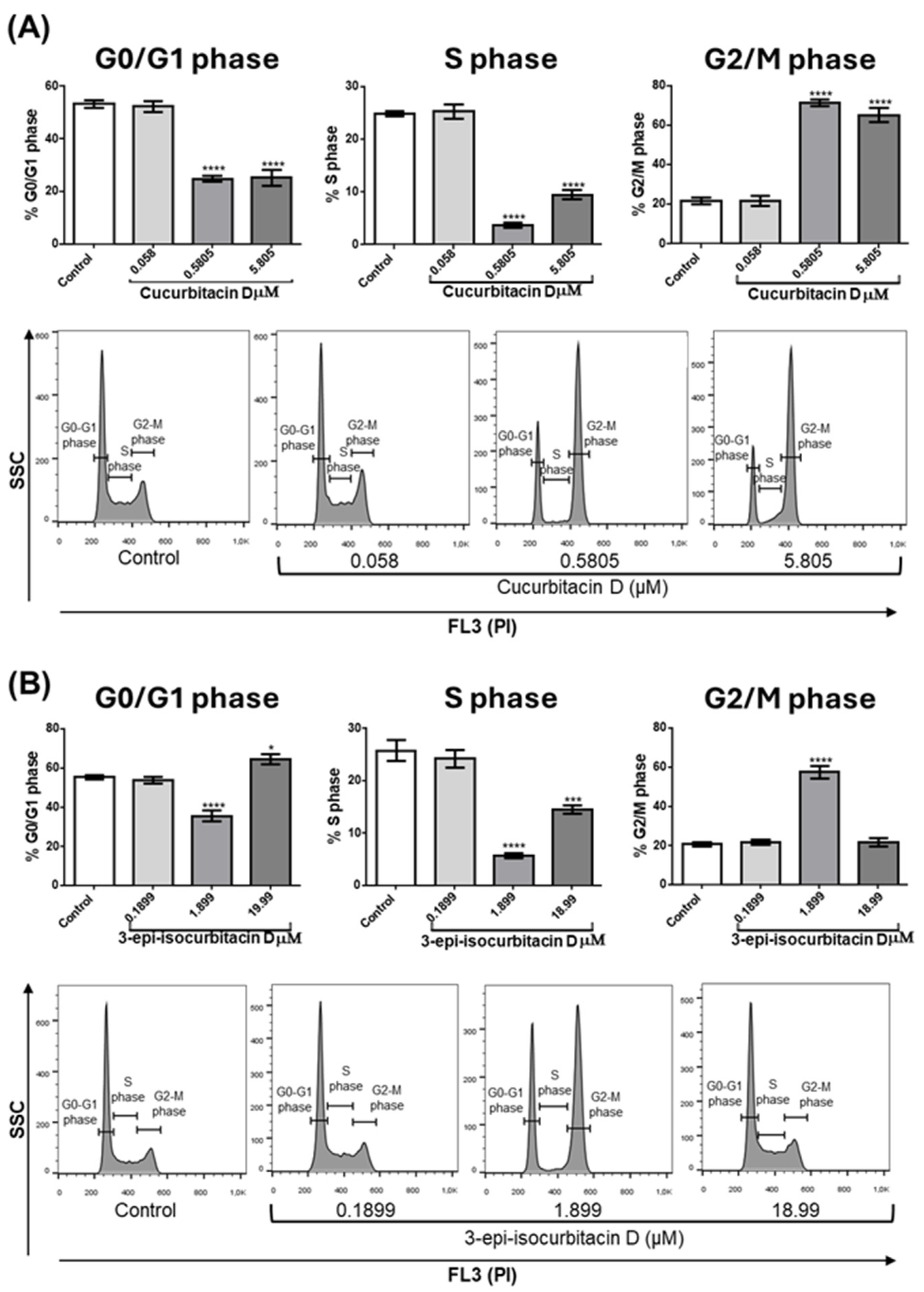
Another possible cause of cell number reduction is that cucurbitacins can interfere with cell cycle progression and consequently with cell proliferation. To analyze this, OCI cells were treated with tested compounds for 24 hours; then, cells were collected, stained with PI and the cell cycle was analyzed by flow cytometry.
Figure 3A (quantitative analysis) and 3B (representative experiment) show that at concentrations of 0.58 and 5.805 μM CucD significantly reduced the percentage of cells in the G0/G1 phase (left Bar panel) and in the S phase (middle bar panel), whereas significantly increased the percentage of cells in the G2/M phase (right panel). A slight difference was seen with IsocucD, since this compound significantly decreased cells in G0/G1 phase at 1.899 μM but determined a small, although significant, increase at 18.99 μM (C, left panel). IsocucD, as CucD, significantly decreased the number of cells in S phase at both effective concentrations (C, middle panel), whereas increased the number of cells in G2/M phase at 1.899 but not at 18.99 μM (C, right panel). So, the tested compounds not only caused an increase in apoptosis but also a blockage of the entry into the cell cycle (phase G0/G1) and of DNA synthesis (S phase) with consequent accumulation of cells in the G2/M phase (mitosis). These results suggest that tested compounds decreased cell number by acting on both the survival and proliferation of leukemia cells. Given the previous experiments, we chose the minimum effective concentration for all subsequent experiments.
2.5. Effects of CucD and isoCucD on Cell Proliferation Pathways
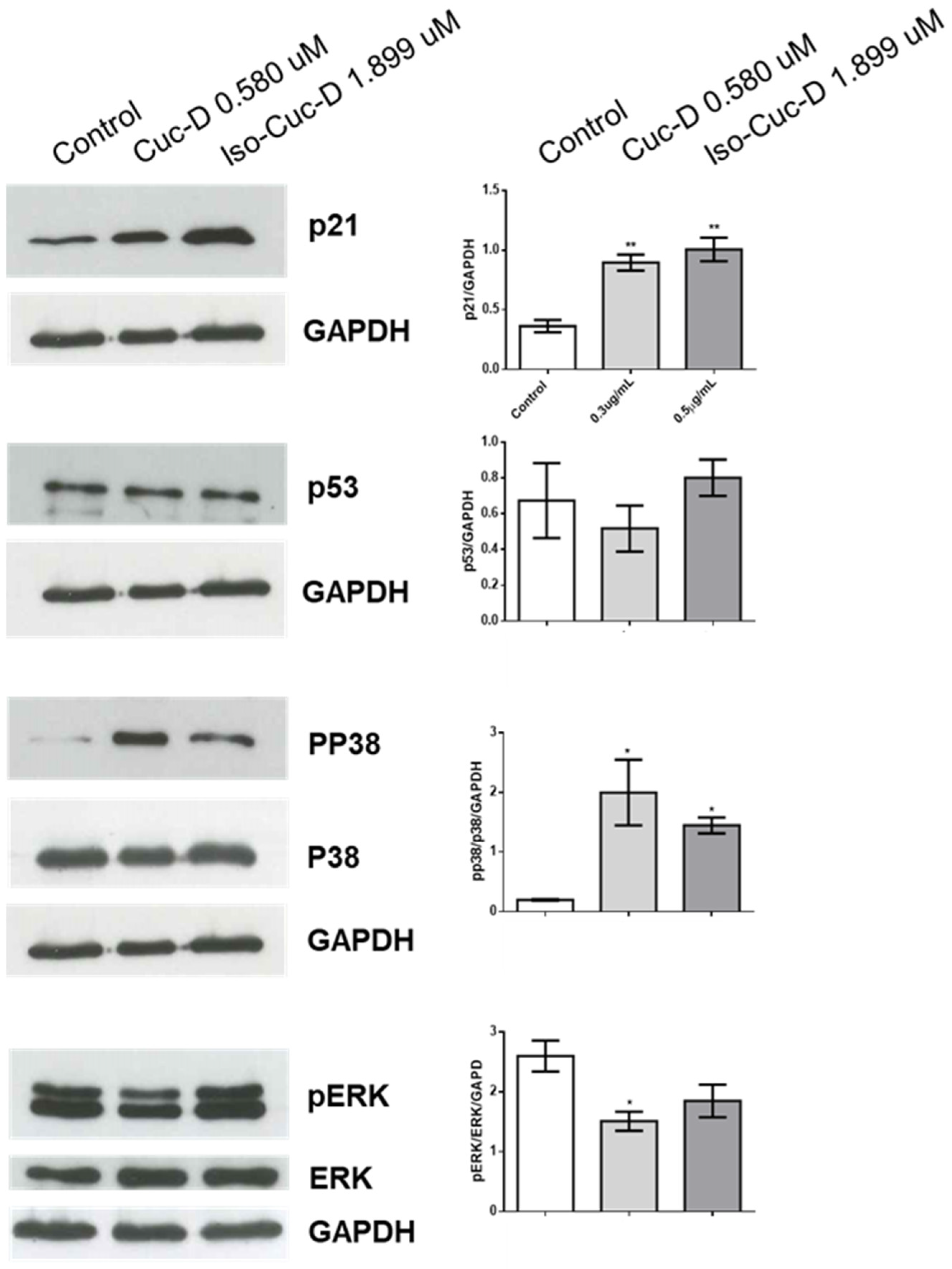
We next analyzed potential mechanisms by which CucD and IsoCucD affected the cell cycle using Western blotting to measure expression of p21 in CucD- or isoCudD-treated OCI-AML3 cells. Protein bands from western blotting of 5 independent experiments were quantitated and, as shown in Figure 6, both CucD and IsoCucD significantly upregulated p21 at the tested concentrations. Because p21 is regulated by p53 [
18,
19], we also investigated whether CucD or iso CucD treatment induces p53 expression in OCI-AML3 cells. We found that both compounds did not affect the expression of p53, suggesting that p21-dependent, p53-independent pathway is at least partially involved for the effect of CucD and IsoCucD concentrations on cell cycle arrest. We also investigated the potential roles of ERK and p38 in CucD- and IsoCucD-induced p21 expression using Western blotting.
Figure 4 shows that levels of phosphorylated ERK significantly decreased after CucD but the decrease with IsoCucD treatment did not reach the significancy when compared to DMSO treatment (control). The presence of two bands in phosphorylated ERK is the result of different levels of phosphorylation. Conversely, levels of phosphorylated p38 significantly increased after Both CucD and IsoCucD treatment. Thus, CucD- and IsoCucD-dependent changes in p21 expression in OCI-AML3 cells are possibly related, at least in part, to changes in MAPK pathway signaling.
2.6. Effect of CucD and IsocucD on the Expression of ZNF-217 Gene
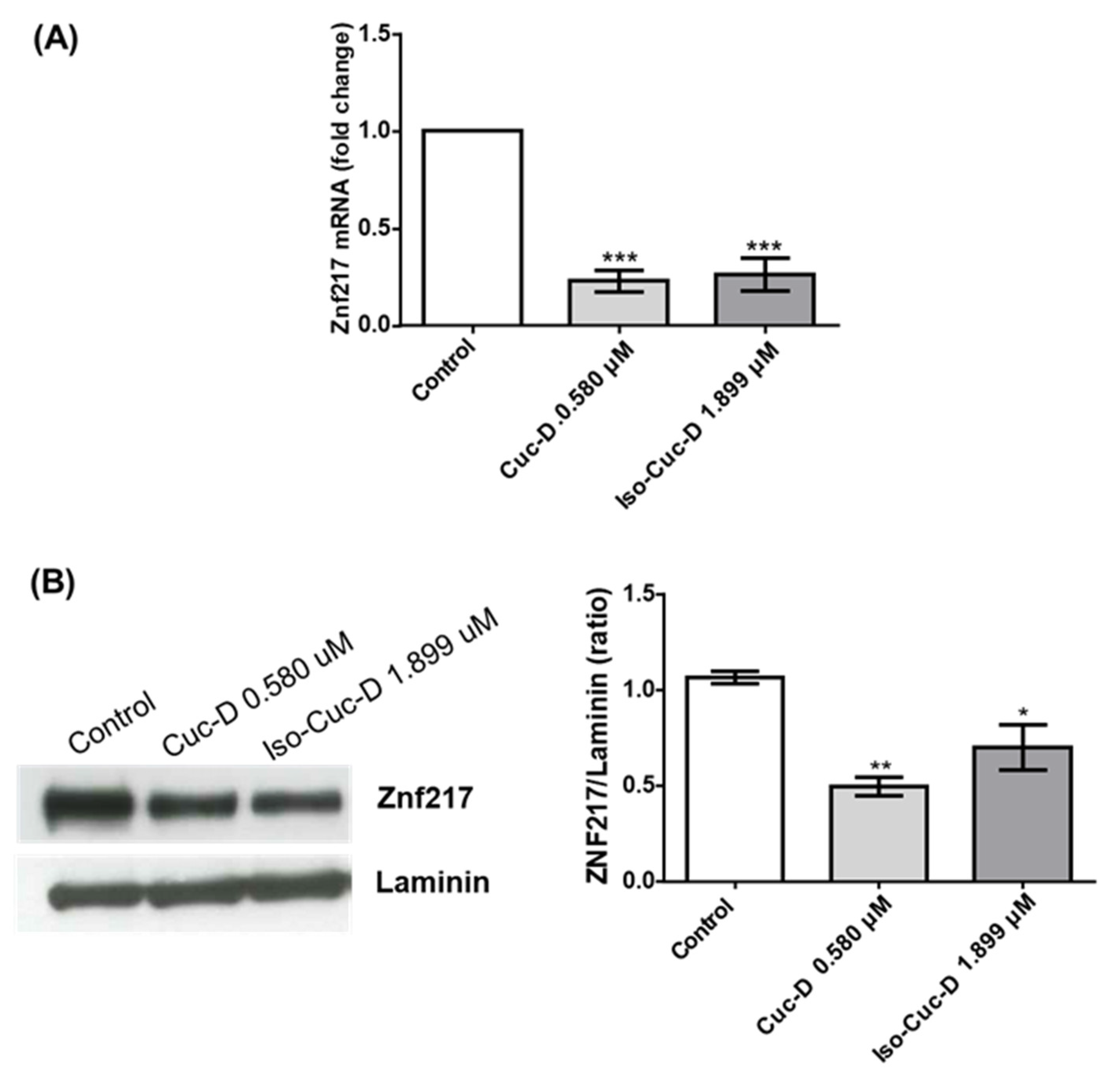
We tested the effects of cucurbitacins on ZNF217 after a screening of this and other genes that are currently under study in our lab. Therefore, in the subsequent experiments, we investigated its possible modulation on OCI cell line. To this aim, cells were cultured with CucD or IsocucD for 24 hours in four independent experiments. Then, cells were collected and RNA and proteins were extracted. ZNF217 expression was assessed by both real-time PCR and western blotting. As can be seen from
Figure 5A, ZNF217 mRNA was expressed by acute myeloid leukemia OCI cells and both CucD and IsocucD significantly downregulated its expression at their minimum effective concentration (0.3 and 0.5 μg/mL for CucD and IsocucD, respectively). These results were confirmed by western blotting experiments (
Figure 4B). Indeed, ZNF217 protein was expressed by OCI cells and both compounds were able to significantly decrease its expression, being CucD more effective with respect to IsocucD.
2.7. Effect of CucD and IsocucD on Other Cancer Cell Lines
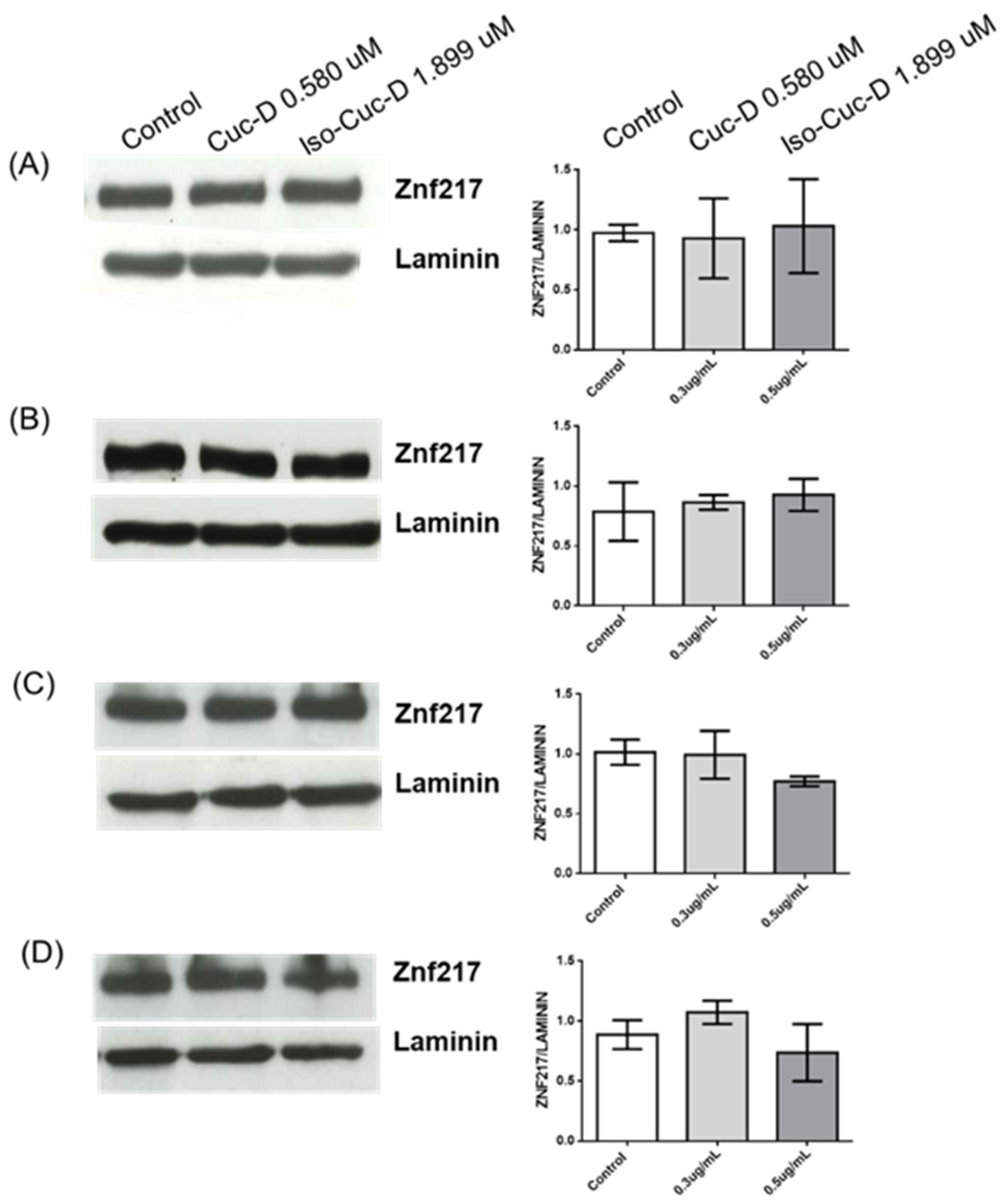
To understand if the expression of ZNF217 and its modulation by the tested compounds were limited to OCI cells, 3 independent experiments have been performed on 4 additional cell lines, namely PGA1 and MEC1, chronic lymphatic leukemias, U937, another acute myeloid leukemia cell line, and Jurkat, a T-cell lymphoma. The cells were cultured in the absence and presence of CucD or IsocucD. After 24 hours, they were collected and the protein extracted to perform western blotting analysis. As showed in
Figure 6, all the cell lines expressed ZNF217 but, contrary to OCI, both compounds did not decrease ZNF217 expression. Thus, while ZNF217 is expressed by cancer cells of various nature, the tested cucurbitacins selectively decreased ZNF217 only in acute myeloid leukemia OCI-AML3 cells.
2.8. Effect of ZNF217 Silencing on OCI-AML3 Cells (Contribution of ZNF217 Down-Modulation to the Effects of Isolated Compounds on OCI-AML3 Cells)
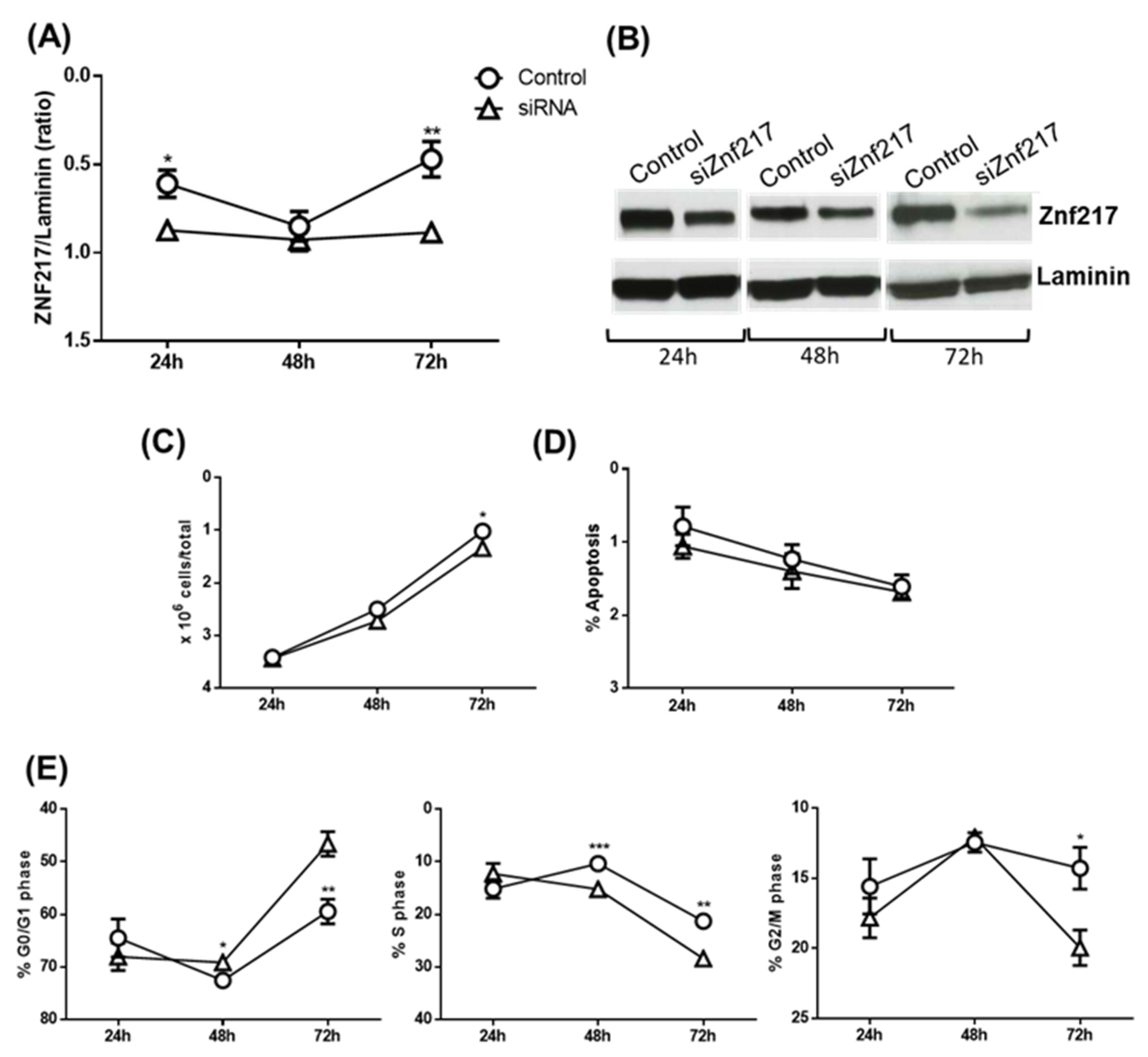
Considering the above, in the subsequent experiments we tried to understand if the downregulation of ZNF217 by the tested substances was responsible for the pro-apoptotic and/or anti-proliferative effects. To do this, the ZNF217 gene was significantly silenced after 24 and 72 hours of siRNA treatment (
Figure 7A and 7B). The ZNF-217 silencing was associated with a significant, although slight, decrease of cell number after 72 hours (
Figure 7C), which was not due to an increase of apoptosis (
Figure 7D) but rather to a significant increase of cells in G0/G1 phase (
Figure 6E, left panel, 48 and 72 hours of treatment) and a decrease in S and G2/M phases (Middle and left panel, respectively). Thus, these experiments suggest that the downregulation of ZNF217 is responsible, at least in part, only for cell cycle blocking but not for the pro-apoptotic action of CucD and IsocucD.
3. Discussion
Although the activity of cucurbitacin derivatives has been extensively studied in acute myeloid leukemia, in this work we highlight significant new knowledge. The first is that the oncogene ZNF217 is expressed by AML cells. Until now, this gene, involved in the development of tumors and their metastatic spread, had never been documented in these cancer cells. In addition, we document that cucurbitacins down-regulates it significantly, and so far only one other molecule with this effect has been documented. Furthermore, the effect we document occurs only in AML cells mutated for NPM, whose mutation supports the development of 30% of AML cases.
The ZNF217 protein belongs to the Kruppel-like family of zinc finger transcription factors. It confers several functions on cancer cells such as promotion of proliferation, evasion of growth suppressors, replicative immortality capacity, resistance to apoptosis, enrichment of cancer stem cells, drug resistance and activation of invasion and metastasis. As far as is known, it exerts its oncogenic functions thanks to an excessive increase in its expression. Indeed, its aberrant expression has been highlighted in several tumors, such as breast, colon, liver, and thyroid ones [
5]. Moreover, patients with ZNF217-positive tumors present poor outcome with a worse relapse-free survival and overall survival than ZNF217-negative ones [
6]. Therefore, targeting ZNF217 oncogene could be a promising strategy not only to fight cancer and its related chemoresistance but also as a potential biomarker for its early diagnosis.
Considering the above, in the present study we have analyzed the effect of cucurbitacins isolated from E. hainanensis on the expression of the ZNF217 oncogene in acute myeloid leukemia and other leukemia cells. Indeed, preliminary experiments allowed us to identify for the first time the expression of ZNF217 oncogene in acute myeloid leukemia cells. This intrigued us to evaluate its modulation as potential mechanism of cucurbitacin antiproliferative properties.
At first, cucurbitacin D, 3-epi-isocucurbitacin D or a mixture of cucurbitacin D and cucurbitacin I (see also supplementary files) was tested in AML cell model, showing a significant reduction of OCI-AML3 cell number after 24 hours of treatment. Among them, CucD was the most potent exerting its effect at the lowest concentration of 0.3 μg/mL. Subsequent experiments were carried out only on CucD and IsocucD to highlight the contribution of an increased apoptosis and/or an inhibition of proliferation on the observed cell reduction phenomenon. We documented that both factors were significantly operational. In fact, CucD increased apoptosis by about a third and isocucD by about 50%. Apoptosis augmented possibly because of the triggering of the intrinsic pathway since the expression of the antiapoptotic molecule Bcl2, belonging to this pathway, was decresead. This was further confirmed by the significant decrease of the TNF-α expression. TNF-, in fact, binds to the TNF receptor (TNFR), a type I transmembrane protein that belongs to the tumor necrosis factor/nerve growth factor receptor (TNF/NGFR) family [
26], and the triggering of TNFR activates the extrinsic apoptosis pathway. Cell cycle analysis highlighted the ability of both substances to block the cell cycle progression as they significantly decreased cells in the G0/G1 and S phases while the same cells accumulated in the G2/M phase. Thus, the reduction in the number of OCI cells was due to both an increase in cell death and a cell cycle blockage. The analysis of cell cycle pathways has shown that CucD and Iso CucD increased the expression of p21 but not p53, and the phosphorylation of members of the MAPK pathway such as p38 that increase p21 stability [
25] and decrease pERK that decreases p21 stability by promoting its degradation [
26]. Therefore, cucurbitacins, mainly CucD, were able to decrease the number of acute myeloid leukemia cells by acting on both apoptosis and proliferation.
Previous studies have investigated the potential usefulness of CucD and IsocucD in the management of cancer, although the latter in a less extent. Particularly, CucD and IsocucD were highlighted to exert antiproliferative activity in MCF7 breast cancer cells by partly disrupting Hsp90 client protein maturation [
27]. CucD has been shown to block cell cycle and to induce antiproliferative effects in endometrial and ovarian cancer cells by modulating pro- and antiapoptotic factors [
28]. Similar properties were also found in cervical cancer cells and were associated to the inhibition of STAT3 activation [
29]. In pancreatic cancer cells, CucD induced cell cycle arrest and apoptosis by triggering ROS generation and activation of p38 MAPK pathway [
15]. A downregulation of gene and proteins belonging to PI3K/AKT/mTOR, MAPK, and JAK2/STAT3 cascades has been highlighted in liver cancer 14. Alongside the proapoptotic effects, a modulation of autophagy has been reported in human gastric and T cell leukemia cells too [
18,
30,
31]. CucD has also reported able to counteract chemoresistance; particularly, it suppressed the proliferation of gemcitabine resistant pancreatic cancer cells [
31]. Moreover, it synergized cisplatin cytotoxicity in human lung tumor cells by inhibiting p-AKT, p-Erk, p-JNK, and p-ErbB3 signaling and suppressing STAT3 and NF-κB activity [
32]; similar effects were observed in doxorubicin-resistant human breast carcinoma cells [
17]. Finally, CucD allowed to overcome gefitinib resistance by modulating the EGFR expression [
16]. Surprisingly, most of the pathways affected by CucD are somehow related to the expression of ZNF217 gene. Indeed, ErbB3 gene is a direct target for ZNF217, and its overexpression determines the activation of both PI3K/Akt and Ras/MAPK survival pathway [
5,
31,
33]. Furthermore, EGF seems implicated in ZNF217-induced immortality; conversely, STAT3 seems to play a regulatory role on ZNF217 expression [
33].
Considering the above, in the subsequent set of experiments we investigated the effect of CucD on ZNF217, highlighting that CucD was able to significantly decrease its expression in OCI-AML3 cells. The involvement of CucD in decreasing ZNF217 expression is only concerned with the inhibition of proliferation and not with the increase of apoptosis as it was demonstrated by the results of gene silencing in AML cells. Of extreme interest, it is that the expression of this tumor-promoting oncogene was decreased by CucD only in acute myeloid leukemia cells compared to other hematologic cancers and, more importantly, that CucD decreased ZNF217 expression in nuclephosmin (NPM)-mutated AML cells, but not in other AML cells with no NPM mutation. NPM is a mutation present in about 30% of AML forms, so this finding opens the possibility that oncogenic expression of ZNF217 is linked to the NPM mutation. Noteworthy, despite notable advances have been done in the treatment of this frequent AML subtype in recent years, approximately 50% of AML patients with NPM1 mutation treated with conventional regimens eventually died of disease progression. Therefore, targeting ZNF217 could be a further strategy to counteract NPM1-AML so increasing patient survival. Obviously, this interesting possibility will have to be investigated further to confirm its veracity, to understand the mechanisms underlying it, and to evaluate the therapeutic possibilities of CucD in these forms of AML.
4. Materials and Methods
4.1. Plant Material
Elaeocarpus hainanensis Oliv. materials were collected in Ha Tinh province, Vietnam in November 2021. Elaeocarpus hainanensis Oliv. species was identified by botanics Dr. Do Ngoc Dai, Faculty of Agriculture, Forestry and Fishery, Nghe An University of Economics. The voucher specimen (No. 04TN.ICH.EH21) has been deposited in the Institute of Chemistry, VAST.
4.2. Cell Line Culture and Characterization
Acute myeloid leukemia (OCI-AML3, U-937, and HL-60), acute T-cell lymphoblastic leukemia (Jurkat), and chronic B-cell lymphocytic leukemia (MEC1, PGA1) cell lines of human origin were obtained from ATCC (Manassas, United States) and exploited as experimental models. OCI-AML3, U-937, HL-60, and MEC1 were maintained in RPMI-16140 medium while PGA1 in IMDM medium. In both media, 10% fetal bovine serum (FBS), 100 U/mL penicillin, and 100 μg/mL streptomycin were used as cofactors. Cells were grown under standard conditions (37 °C and 5% CO2) and the media were changed twice per week, as recommended by the suppliers. When the cells reached approximately 80% confluency, they were subcultured. In the experiments, cells were seeded into 24-wells plates, kept at concentration of 2 × 105 cells/mL, and treated with different concentrations of dimethyl sulfoxide (DMSO) or the test compounds (0.03 – 10 μg/mL) for 24h. DMSO was used at a maximum 1% v/v nontoxic concentration in the medium.
4.3. Analysis of Cell Viability and Cell Cycle Progression
Cells were counted manually using a hemocytometer. Cell count exceeded the number of 200 in four chambers so that the margin of error is reduced to negligible levels. Cell viability and cell cycle progression were analyzed by flow cytometry to determine DNA content of cell nuclei stained with propidium iodide (PI) after the exclusion of necrotic cells by forward light scatter (FCS). Cells were collected by centrifugation and washed in phosphate-buffered saline (PBS); DNA was stained by incubating the cells in H2O containing 50 μg/mL PI and incubated for 30 min at 4ºC. This allows direct DNA staining in PI hypotonic solution without the requirement of RNase treatment as the RNA is removed by hypotonic shock. Fluorescence was measured by flow cytometry using Coulter Epics XL-MCL equipment (Beckman Coulter Inc., Brea, CA, USA) and analyzed by FlowJo software.
4.4. Western Blotting
Proteins were extracted by RIPA buffer, separated by SDS-PAGE, and analyzed by Western blotting. Primary antibodies included anti-p21, anti-phospho-p38, anti-p38, anti-phospho-ERK, anti-ERK (Cell Signaling, MA, U.S.A.), anti-p53 (Santa Cruz Biotechnology, Dallas, TX), and anti-ZNF217 (Abcam, Cambridge, UK). Anti-laminin antibodies (Sigma-Aldrich, St. Louis, MO) and anti-GAPDH (OriGene, Rockville, USA) were used as controls. Secondary antibodies were labeled with horseradish peroxidase (Pierce/Thermo-Fisher Scientific, Waltham, MA). Antigen–antibody complexes were revealed by enhanced chemiluminescence by following the manufacturer’s instructions (Millipore, Billerica, MA). Western blotting films were scanned and band signal intensities were determined using ImageJ software (National Institutes of Health, Bethesda, MD).
4.5. Real-Time (RT)-PCR
RNA was isolated using the Qiagen RNeasy Plus Micro kit, and conversion of total RNA to cDNA was performed with the QuantiTect Reverse Transcription kit (Qiagen). RT-PCR was performed with the ABI-7300 Real-Time Cycler (Applied Biosystems, Foster City, CA, United States) and amplification was achieved using the TaqMan Assay (Hs00998133m1 for TGFb1, Hs00919915_m1 for Znf217, hs00174128 m1 for TNF-α, hs00187848_m1 for Bcl2 (Thermo Fisher Scientific, Waltham, MA, United States). The ΔCt method was used to determine the expression levels of TNF-α, TGF-β, BCL-2, and ZNF217.
4.6. Silencing
Lipofectamine RNAiMax Reagent (Invitrogen, CA, USA) was introduced for cell transfection with siRNA according to the manufacturer’s instructions. The two previously validated small interfering RNAs (siRNAs) targeting ZNF217 and a scrambled control (Life Technologies, CA, USA) were transfected into cells. OCI-AML3 cells were transfected with either Znf217 or scrambled control siRNA in culture medium for 24h, 48 h, and 72h. Serum-free Opti-MEM (Gibco, CA, USA) was adopted to dilute target plasmids and Lipofectamine, respectively. The cells were then controlled for Znf217 expression by western blotting and stained with propidium iodide (PI) for flow cytometry analysis.
4.7. Statistical Significance
All the data are expressed as mean ± standard error (SE) of at least two biological replicates in which at least two technical replicates per each concentration were performed. The statistical analysis was carried out by GraphPad Prism™ software (Version 6.00, GraphPad Software, Inc., San Diego, CA, USA). Statistical significance was determined using the one-way ANOVA test as specified in the figure legends. Differences were considered statistically significant according to the following criteria: ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ****p < 0.0001.
5. Conclusion
In conclusion, cucurbitacin D and 3-epiisocucurbitacin D, isolated by Elaeocarpus hainanensis decrease the number of OCI-AML3 acute myeloid leukemia cells by promoting apoptosis and inhibiting cell cycle progression. OCI-AML3 cells express the oncogene ZNF217 which is down-regulated by the studied cucurbitacins. The downregulation of ZNF217 seems to contribute only to the blockage of cell cycle progression and happened in NPM-mutated OCI-AML3 but not in other AML or other hematologic cancers. Overall, the obtained results suggest CucD as a promising molecule to counteract NPM-mutated AML hematologic cancer, although further studies are needed to deeply investigate the specificity of its mechanism of action.
Supplementary Materials
The following supporting information can be downloaded at the website of this paper posted on Preprints.org, Figure S1: 1H NMR (500 MHz, CDCl3) spectrum of CucD (1); Figure S2: 13C NMR (125 MHz, CDCl3) spectrum of CucD (1); Figure S3: 1H NMR (500 MHz, CD3OD) spectrum of IsocucD (2); Figure S4: 13C NMR (125 MHz, CD3OD) spectrum of IsocucD (2); Figure S5: DEPT-13C NMR (125 MHz, CD3OD) spectrum of IsocucD (2); Figure S6: 1H NMR (500 MHz, CD3OD) spectrum of mixture CucD + CucI (ratio, 1:1); Figure S7: 13C NMR (125 MHz, CD3OD) spectrum of mixture CucD + CucI; Figure S8: DEPT-13C NMR (125 MHz, CD3OD) spectrum of mixture CucD + CucI; Figure S9: Effect of Cucurbitacin D + Cucurbitacin I on OCI-AML3 cell apoptosis. Table S1: 13C (125 MHz) and 1H (500 MHz) NMR data of compounds 1 - 3 (CD3OD).
Author Contributions
Conceptualization, D.V.D. and TTT; Methodology, S.A.; Software, S.A.; Validation, A.F., E.A.; Formal Analysis, S.A., B.T.C.; Investigation, S.A., B.T.C., V.D.H., N.T.T.L., L.T.H.N.; Resources, D.V.D., T.T.T.; Data Curation, S.A., B.T.C.; Writing – Original Draft Preparation, D.V.D.; Writing – Review & Editing, D.V.D., S.D.G., T.T.T; Visualization, A.F., E.A., S.D.G.; Supervision, D.V.D.; Funding Acquisition, D.V.D., T.T.T.
Funding
This work was funded by Vietnam National Foundation for Science and Technology Development (NAFOSTED) [grant number 04/2022/TN]; and FM-AA (Funghi Medicinali Anti-Aging) Programma di Sviluppo Rurale per l’Umbria 2014-2020 – Misura 16 – Sottomisura 16.2.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Research data are available at the Department of Medicine and Surgery, University of Perugia, Perugia, Italy and at Institute of Chemistry, Vietnam Academy of Science and Technology, Hanoi, Vietnam.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Gyamfi, J., J. Kim, and J. Choi, Cancer as a Metabolic Disorder. Int J Mol Sci, 2022. 23(3). [CrossRef]
- Ahmad, E., et al., Molecular approaches in cancer. Clin Chim Acta, 2022. 537: p. 60-73. [CrossRef]
- Marin, J.J.G., et al., Expression of Chemoresistance-Associated ABC Proteins in Hepatobiliary, Pancreatic and Gastrointestinal Cancers. Cancers (Basel), 2022. 14(14). [CrossRef]
- Lu, J., et al., Clinicopathological and molecular characteristics of the alpha-fetoprotein-producing gastric cancer: emphasis on two major subtypes. APMIS, 2022. 130(3): p. 169-180. [CrossRef]
- Cohen, P.A., et al., The dark side of ZNF217, a key regulator of tumorigenesis with powerful biomarker value. Oncotarget, 2015. 6(39): p. 41566-81. [CrossRef]
- Shida, A., et al., Prognostic significance of ZNF217 expression in gastric carcinoma. Anticancer Res, 2014. 34(9): p. 4813-7.
- Alzahrani, A.S., PI3K/Akt/mTOR inhibitors in cancer: At the bench and bedside. Semin Cancer Biol, 2019. 59: p. 125-132. [CrossRef]
- Mosca, L., et al., Taxanes in cancer treatment: Activity, chemoresistance and its overcoming. Drug Resist Updat, 2021. 54: p. 100742. [CrossRef]
- Di Sotto, A., et al., Chemopreventive Potential of Caryophyllane Sesquiterpenes: An Overview of Preliminary Evidence. Cancers (Basel), 2020. 12(10).
- Zhao, G., et al., Metabolome and Transcriptome Analyses of Cucurbitacin Biosynthesis in Luffa (Luffa acutangula). Front Plant Sci, 2022. 13: p. 886870. [CrossRef]
- Hussain, H., et al., Cucurbitacins as Anticancer Agents: A Patent Review. Recent Pat Anticancer Drug Discov, 2019. 14(2): p. 133-143. [CrossRef]
- Ge, W., et al., Synthesis of Cucurbitacin B Derivatives as Potential Anti-Hepatocellular Carcinoma Agents. Molecules, 2018. 23(12). [CrossRef]
- Kaushik, U., V. Aeri, and S.R. Mir, Cucurbitacins - An insight into medicinal leads from nature. Pharmacogn Rev, 2015. 9(17): p. 12-8. [CrossRef]
- Luo, H., et al., Naturally occurring anti-cancer compounds: shining from Chinese herbal medicine. Chin Med, 2019. 14: p. 48. [CrossRef]
- Mehdi Uremis, M., et al., Cucurbitacin D Inhibits the Proliferation of HepG2 Cells and Induces Apoptosis by Modulating JAK/STAT3, PI3K/Akt/mTOR and MAPK Signaling Pathways. Curr Cancer Drug Targets, 2022. 22(11): p. 931-944. [CrossRef]
- Kim, M.S., et al., Cucurbitacin D Induces G2/M Phase Arrest and Apoptosis via the ROS/p38 Pathway in Capan-1 Pancreatic Cancer Cell Line. Evid Based Complement Alternat Med, 2020. 2020: p. 6571674.
- Hong, S.H., et al., Cucurbitacin D Overcomes Gefitinib Resistance by Blocking EGF Binding to EGFR and Inducing Cell Death in NSCLCs. Front Oncol, 2020. 10: p. 62. [CrossRef]
- Ku, J.M., et al., Cucurbitacin D induces cell cycle arrest and apoptosis by inhibiting STAT3 and NF-kappaB signaling in doxorubicin-resistant human breast carcinoma (MCF7/ADR) cells. Mol Cell Biochem, 2015. 409(1-2): p. 33-43.
- Nakanishi, T., et al., Autophagy is associated with cucurbitacin D-induced apoptosis in human T cell leukemia cells. Med Oncol, 2016. 33(4): p. 30. [CrossRef]
- Aryal, B., et al., Antidiabetic, Antimicrobial, and Molecular Profiling of Selected Medicinal Plants. Evid Based Complement Alternat Med, 2021. 2021: p. 5510099. [CrossRef]
- Cham, B.T., et al., Noncytotoxic 16,23-epoxycucurbitacin-type triterpenoids from Elaeocarpus hainanensis. Nat Prod Res, 2022: p. 1-5.
- Meng, D., et al., Cytotoxic cucurbitane-type triterpenoids from Elaeocarpus hainanensis. Planta Med, 2008. 74(14): p. 1741-4. [CrossRef]
- Bucknam, A.R. and G.C. Micalizio, Asymmetric De Novo Synthesis of a Cucurbitane Triterpenoid: Total Synthesis of Octanorcucurbitacin B. J Am Chem Soc, 2022. 144(19): p. 8493-8497. [CrossRef]
- Seger, C., et al., 1H and 13C NMR signal assignment of cucurbitacin derivatives from Citrullus colocynthis (L.) Schrader and Ecballium elaterium L. (Cucurbitaceae). Magn Reson Chem, 2005. 43(6): p. 489-91.
- Delfino, D.V., et al., Glucocorticoid-induced activation of caspase-8 protects the glucocorticoid-induced protein Gilz from proteasomal degradation and induces its binding to SUMO-1 in murine thymocytes. Cell Death Differ, 2011. 18(1): p. 183-90. [CrossRef]
- Nocentini, G., et al., Gene structure and chromosomal assignment of mouse GITR, a member of the tumor necrosis factor/nerve growth factor receptor family. DNA Cell Biol, 2000. 19(4): p. 205-17. [CrossRef]
- Hall, J.A., et al., Cucurbitacin D Is a Disruptor of the HSP90 Chaperone Machinery. J Nat Prod, 2015. 78(4): p. 873-9. [CrossRef]
- Kim, G.Y., et al., The stress-activated protein kinases p38 alpha and JNK1 stabilize p21(Cip1) by phosphorylation. J Biol Chem, 2002. 277(33): p. 29792-802. [CrossRef]
- Hwang, C.Y., C. Lee, and K.S. Kwon, Extracellular signal-regulated kinase 2-dependent phosphorylation induces cytoplasmic localization and degradation of p21Cip1. Mol Cell Biol, 2009. 29(12): p. 3379-89. [CrossRef]
- Ishii, T., et al., Cucurbitacin D induces growth inhibition, cell cycle arrest, and apoptosis in human endometrial and ovarian cancer cells. Tumour Biol, 2013. 34(1): p. 285-91. [CrossRef]
- Sikander, M., et al., Cucurbitacin D exhibits potent anti-cancer activity in cervical cancer. Sci Rep, 2016. 6: p. 36594. [CrossRef]
- Jafargholizadeh, N., S.J. Zargar, and Y. Aftabi, The cucurbitacins D, E, and I from Ecballium elaterium (L.) upregulate the LC3 gene and induce cell-cycle arrest in human gastric cancer cell line AGS. Iran J Basic Med Sci, 2018. 21(3): p. 253-259. [CrossRef]
- Sikander, M., et al., Novel Mechanistic Insight into the Anticancer Activity of Cucurbitacin D against Pancreatic Cancer (Cuc D Attenuates Pancreatic Cancer). Cells, 2019. 9(1). [CrossRef]
Figure 1.
Effect of cucurbitacins on OCI-AML3 cell count. On the left side, the chemical structures of cucurbitacin D (A), 3-epi-isocucurbitacin D (B), and cucurbitacin D + cucurbitacin I (C). On the right side, bars represent the number of viable cells counted after 24h of treatment with control vehicle (Control) or cucurbitacin D (A), 3-epi-isocucurbitacin D (B) or cucurbitacin D + cucurbitacin I (C) at the concentrations reported on the x-axis. Data from five independent experiments are reporter as mean ± SEM. ***p < 0.001 and **** p < 0.0001, significant lowering of cell viability with respect to the control (one-way ANOVA).
Figure 1.
Effect of cucurbitacins on OCI-AML3 cell count. On the left side, the chemical structures of cucurbitacin D (A), 3-epi-isocucurbitacin D (B), and cucurbitacin D + cucurbitacin I (C). On the right side, bars represent the number of viable cells counted after 24h of treatment with control vehicle (Control) or cucurbitacin D (A), 3-epi-isocucurbitacin D (B) or cucurbitacin D + cucurbitacin I (C) at the concentrations reported on the x-axis. Data from five independent experiments are reporter as mean ± SEM. ***p < 0.001 and **** p < 0.0001, significant lowering of cell viability with respect to the control (one-way ANOVA).
Figure 2.
Effect of CucD (A) or IsocucD (B) on OCI-AML3 Apoptosis. Bars represent the percentage of Apoptosis after 24h of treatment, either with control vehicle (Control), CucD (A), or IsocucD (B), at the concentrations reported on the x-axis. Histograms are from representative experiments in which propidium iodide (PI) staining shown on the x-axes on the logarithmic scale (FL3) are reported below the Bar panels. (C), Bars represent the fold change of TNF-α (left panel), and Bcl2 (right panel) after 24h of treatment, either with control vehicle (Control), CucD, or IsocucD, at the concentrations reported on the x-axis. Data from five independent experiments are reporter as mean ± SEM. **p < 0.01, ***p < 0,001 and **** p < 0.0001 significant increase of apoptotic rate with respect to the control (one-way ANOVA).
Figure 2.
Effect of CucD (A) or IsocucD (B) on OCI-AML3 Apoptosis. Bars represent the percentage of Apoptosis after 24h of treatment, either with control vehicle (Control), CucD (A), or IsocucD (B), at the concentrations reported on the x-axis. Histograms are from representative experiments in which propidium iodide (PI) staining shown on the x-axes on the logarithmic scale (FL3) are reported below the Bar panels. (C), Bars represent the fold change of TNF-α (left panel), and Bcl2 (right panel) after 24h of treatment, either with control vehicle (Control), CucD, or IsocucD, at the concentrations reported on the x-axis. Data from five independent experiments are reporter as mean ± SEM. **p < 0.01, ***p < 0,001 and **** p < 0.0001 significant increase of apoptotic rate with respect to the control (one-way ANOVA).
Figure 3.
Effect of CucD and IsocucD on OCI-AML3 cell cycle progression. (A) and (C). Bars represent the percentage of cells in different phases of cell cycle (G0/G1 – left panels-, S – middle panels-, G2/M – right panels) after 24h of treatment with DMSO vehicle (Control) or the tested compounds at the concentrations reported on the x-axis. (B) and (D). Histograms from representative experiments in which PI staining is shown on the x-axes on a logarithmic scale (FL2). Data from five independent experiments are reporter as mean ± SEM. **p < 0.01, ***p < 0.001, ****p < 0.0001 significant different with respect to the control (one-way ANOVA).
Figure 3.
Effect of CucD and IsocucD on OCI-AML3 cell cycle progression. (A) and (C). Bars represent the percentage of cells in different phases of cell cycle (G0/G1 – left panels-, S – middle panels-, G2/M – right panels) after 24h of treatment with DMSO vehicle (Control) or the tested compounds at the concentrations reported on the x-axis. (B) and (D). Histograms from representative experiments in which PI staining is shown on the x-axes on a logarithmic scale (FL2). Data from five independent experiments are reporter as mean ± SEM. **p < 0.01, ***p < 0.001, ****p < 0.0001 significant different with respect to the control (one-way ANOVA).
Figure 4.
Effects of CucD and IsoCucD on expression of proteins involved in the cell cycle. Western blot analysis illustrating expression of p21 (first panel starting from upper side), p53 (second line panels), phosphorylated p38 (pp38), total p38 (p38) (third line panels), phosphorylated ERK (pERK), total ERK (ERK) (fourth line panels) from OCI-AML3 cells treated with vehicle (DMSO), CucD, or IsoCucD for 24 h. Western blots on the left side are representative of five independent experiments quantified by bar graphs in the right side. GADPH served as a loading control. Data are reported as mean ± SEM. * p < 0.05, ** p < 0.01.
Figure 4.
Effects of CucD and IsoCucD on expression of proteins involved in the cell cycle. Western blot analysis illustrating expression of p21 (first panel starting from upper side), p53 (second line panels), phosphorylated p38 (pp38), total p38 (p38) (third line panels), phosphorylated ERK (pERK), total ERK (ERK) (fourth line panels) from OCI-AML3 cells treated with vehicle (DMSO), CucD, or IsoCucD for 24 h. Western blots on the left side are representative of five independent experiments quantified by bar graphs in the right side. GADPH served as a loading control. Data are reported as mean ± SEM. * p < 0.05, ** p < 0.01.
Figure 5.
(A). Real time PCR of ZNF217 transcripts in OCI-AML3 treated with vehicle (DMSO, Control), CucD or IsocucD for 24h. Gene expression was normalized to the expression of 18S and reported as relative to the normalized expression (white bar, fold=1). Data from five independent experiments are reported as mean ± SEM. ***p < 0.001 significant different with respect to the control (one-way ANOVA). (B). Expression of protein level of ZNF217 in OCI-AML3 treated with vehicle (DMSO, Control), Cuc-D or IsocucD for 24h. Laminin expression was used as normalized sample. One representative western blotting of four is shown on the left and the quantitative analysis on the right panel. ZNF217/Laminin ratio is calculated by densitometric quantification of the specific bands detected in four independent experiments. Data from four independent experiments are reported as mean ± SEM. *p < 0.05 and **p < 0.01 significant different with respect to the control (one-way ANOVA).
Figure 5.
(A). Real time PCR of ZNF217 transcripts in OCI-AML3 treated with vehicle (DMSO, Control), CucD or IsocucD for 24h. Gene expression was normalized to the expression of 18S and reported as relative to the normalized expression (white bar, fold=1). Data from five independent experiments are reported as mean ± SEM. ***p < 0.001 significant different with respect to the control (one-way ANOVA). (B). Expression of protein level of ZNF217 in OCI-AML3 treated with vehicle (DMSO, Control), Cuc-D or IsocucD for 24h. Laminin expression was used as normalized sample. One representative western blotting of four is shown on the left and the quantitative analysis on the right panel. ZNF217/Laminin ratio is calculated by densitometric quantification of the specific bands detected in four independent experiments. Data from four independent experiments are reported as mean ± SEM. *p < 0.05 and **p < 0.01 significant different with respect to the control (one-way ANOVA).
Figure 6.
Expression of protein level of ZNF217 in different cell lines treated with vehicle (DMSO, Control), Cuc-D or Isocuc-D for 24h: PGA1 (A), MEC1 (B), U9370 (C), Jurkat (D). Laminin expression was used as normalized sample. One representative western blotting of three is shown on the left of each bar panel. ZNF217/Laminin ratio is calculated by densitometric quantification of the specific bands detected in three independent experiments. Data (mean ± SEM) are reporter as fold change of samples treated with vehicle (DMSO), CucD or IsocucD.
Figure 6.
Expression of protein level of ZNF217 in different cell lines treated with vehicle (DMSO, Control), Cuc-D or Isocuc-D for 24h: PGA1 (A), MEC1 (B), U9370 (C), Jurkat (D). Laminin expression was used as normalized sample. One representative western blotting of three is shown on the left of each bar panel. ZNF217/Laminin ratio is calculated by densitometric quantification of the specific bands detected in three independent experiments. Data (mean ± SEM) are reporter as fold change of samples treated with vehicle (DMSO), CucD or IsocucD.
Figure 7.
Silencing ZNF217. (A) Expression of protein level of ZNF217 in OCI-AML3 cells transfected with ZNF217 (siRNA) for 24h, 48h, 72h. ZNF217 expression was measured by Western blot. Laminin expression was used as sample normalized. (B) A representative western blotting of five is shown. ZNF217/Laminin ratio is calculated by densitometric quantification of the specific bands detected in five independent experiments. Data (mean ± SEM) are reporter as fold change of samples transfected with ZNF217 (siRNA) *p < 0.05 and **p < 0.01 significant different with respect to the control (one-way ANOVA). (C) After 24h, 48h, and 72h from the addition of siRNA, OCI-AML3 cells were counted by trypan blue exclusion and (D) cells were stained by PI for apoptosis evaluation or (E) cell cycle analysis. Data from five independent experiments are reporter as mean ± SEM. *p < 0.05, ** p < 0.001, and *** p < 0.0001, significant different with respect to the control (one-way ANOVA).
Figure 7.
Silencing ZNF217. (A) Expression of protein level of ZNF217 in OCI-AML3 cells transfected with ZNF217 (siRNA) for 24h, 48h, 72h. ZNF217 expression was measured by Western blot. Laminin expression was used as sample normalized. (B) A representative western blotting of five is shown. ZNF217/Laminin ratio is calculated by densitometric quantification of the specific bands detected in five independent experiments. Data (mean ± SEM) are reporter as fold change of samples transfected with ZNF217 (siRNA) *p < 0.05 and **p < 0.01 significant different with respect to the control (one-way ANOVA). (C) After 24h, 48h, and 72h from the addition of siRNA, OCI-AML3 cells were counted by trypan blue exclusion and (D) cells were stained by PI for apoptosis evaluation or (E) cell cycle analysis. Data from five independent experiments are reporter as mean ± SEM. *p < 0.05, ** p < 0.001, and *** p < 0.0001, significant different with respect to the control (one-way ANOVA).
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).