1. Introduction
This paper is grounded in the purpose and drive behind one of the Sustainable Development Goals (SDG) outlined by the United Nations—a comprehensive set of 17 objectives to be achieved by 2030 [
1]. These goals collectively embody humanity’s pursuit of a sustainable future for both the planet and its inhabitants. Serving as a global framework, the 17 SDGs guide international endeavours to address the challenges posed by climate change while harmonising human ambitions for prosperity and improved quality of life. Central to this vision is the assurance of food security [
2], particularly for a significant portion of the global population residing in environmentally vulnerable areas susceptible to the impacts of climate and weather fluctuations.
This study holds significant importance in academia, commerce, strategy, and ecology, focusing on two main elements: rice crops and Vietnam. Therefore, it is crucial to comprehend the global trade dynamics of rice crops. Rice deemed a “Gift of God” due to its nutritional value, plays a vital role in ensuring food security and public health. According to estimates from the Food and Agriculture Organization Corporate Statistical Database (FAOSTAT) maintained by the statistics division of the Food and Agriculture Organization (FAO), less than 8% of global rice production is traded, emphasising the localised nature of this commodity. The water-intensive nature of rice crop production contributes to its substantial ecological impact. Rice crops are sensitive to temperature extremes, requiring an optimal range of 25-35°C, and even brief exposure to high temperatures can render rice spike-lets sterile. Adverse effects also arise from unexpected rainfall during the reproductive stage. Given the extended crop durations, rice crops are highly susceptible to untimely weather patterns and climate changes.
Rice cultivation plays a pivotal role in Vietnam’s agricultural landscape, contributing significantly to the country’s economy and food security. Vietnam is one of the world’s leading rice exporters, and rice is a staple in the Vietnamese diet, forming the foundation of many traditional dishes. The importance of rice in Vietnam is underscored by its cultural and historical significance, as rice has been a central element of Vietnamese life for centuries. According to the Food and Agriculture Organisation (FAO) [
3], Vietnam is among the top global producers of rice, with diverse varieties cultivated across the country’s vast paddy fields. The rice sector provides livelihoods for millions of farmers, supporting rural communities and fostering economic development. Additionally, the government’s initiatives to enhance rice production and ensure sustainability align with the country’s commitment to food security.
Hence, to achieve precise crop yield estimates, this investigation seeks to advance the integration of diverse data sources (including synthetic aperture radar (SAR), multi-spectral imaging (MSI), meteorological, and rainfall data) alongside a novel Deep Ensemble machine learning model named RicEns-Net within a specified region of Vietnam. The adoption of multi-modal data analysis techniques, such as those utilised in recent remote sensing studies for tasks like air quality assessment [
4], semantic segmentation [
5], land cover mapping [
6], and image classification [
7], has garnered significant attention. The significance of this research is amplified by its pertinence to “Precision Farming,” where modern hyperspectral sensors mounted on drones offer heightened flexibility in scheduling flights.
The paper introduces several novel contributions:
The research leverages four remote sensing image and data sources, namely SAR, MSI, land and sea satellite measurements, and precipitation and hydrology data, in conjunction with field measurements. This comprehensive approach enables the proposal of robust and useful prediction features via data engineering.
A rigorous data engineering process is conducted for over 100 engineered features, employing various techniques such as Pairwise Feature Independence Check using the test, statistical significance tests based on p-values, outlier removal, and correlation and variance thresholding. This process results in the identification of 14 extracted features for model training and testing stages, encompassing rainfall, solar radiation, surface temperature, spectral indices, direct spectral measurements from MSI data, and vertical-vertical polarisation radar reflectance from SAR imagery.
The paper introduces a new Deep Ensemble model named RicEns-Net, which merges four top-performing machine learning approaches. These include two commonly used in ecological informatics i.e. convolutional neural networks (CNN) and multilayer perception (MLP) architectures, along with less commonly explored DenseNet and Autoencoder (AE) architectures, which have received limited attention or have not been explored at all in previous research on crop yield prediction.
The developed RicEns-Net coupled with the novel engineered features exhibit superior performance compared to global winners of EY Data contests as well as state-of-the-art machine learning models of random forests (RF), Gradient Boost, CNN and support vector regression (SVR). This is evidenced by robust performance metrics such as absolute and square error, as well as training and test modelling goodness of fit measures.
The remaining sections of the document are structured as follows:
Section 2 provides a literature review on crop yield prediction, whilst
Section 3 outlines specifics about the dataset employed in this study. Details about the proposed RicEns-Net architecture and additional methodology information are presented in
Section 4, and
Section 5 showcases the results of the experimental analysis. Finally,
Section 6 wraps up the paper with concluding remarks and a discussion on the limitations of the proposed work and potential future research directions.
2. Related Works
Advancements have influenced the evolution of crop yield monitoring in sensor technology and data analysis methodologies. While early studies highlighted rainfall as the primary determinant of crop yield, a key shift occurred in 1968 with the recognition of soil moisture as a more reliable predictor by Baier and Robertson [
8]. Their work leverages spectral data to estimate crop yield based on vegetation health indicators. Over time, numerous vegetation indices (VIs) have been developed to assess vegetative conditions and physiological characteristics of crops. These VIs, including the Normalized Difference Vegetation Index (NDVI), Leaf Area Index (LAI) [
9], and Transformed Soil Adjusted Vegetation Index (TSAVI) [
10], play a crucial role in crop prediction models. Advancements in hyperspectral imaging have enabled the capture of fine-grained spectral data, facilitating the development of biochemical indices for quantifying plant constituents.
In the early 2000s, research surged leveraging imaging and machine learning technologies for crop yield prediction. Studies introduced novel methodologies, such as artificial neural networks (ANN) and SVR, to analyse remote sensing data and historical yield records. These approaches demonstrated superior accuracy compared to traditional models and prepared for more precise and scalable methods for estimating crop yields. Uno et al. [
11] analyse hyperspectral images of corn plots in Canada using statistical and ANN approaches, demonstrating the potential of ANNs in predicting yield with superior accuracy compared to conventional models. Li et al. [
12] introduce a methodology employing ANN models to predict corn and soybean yields in the United States “corn belt” region, achieving high prediction accuracy through historical yield data and NDVI time series. Bala and Islam [
13] estimate potato yields in Bangladesh using TERRA MODIS reflectance data and Vegetation Indices (VIs), demonstrating the effectiveness of VIs derived from remote sensing for early yield estimation. Li et al. [
14] employ SVR and multi-temporal Landsat TM NDVIs to predict winter wheat yield in China, showcasing the precision and effectiveness of SVR models in yield estimation. Stojanova et al. [
15] integrate LiDAR and Landsat satellite data using machine learning techniques to model vegetation characteristics in Slovenia. Their approach combines the precision of LiDAR data with the broad coverage of satellite data, facilitating effective forest management and monitoring processes.
Furthermore, Mosleh et al. [
16] evaluated the efficacy of remote sensing imagery in mapping rice areas and forecasting production, highlighting challenges such as spatial resolution limitations and issues with radar imagery. Johnson et al. [
17] developed crop yield forecasting models for the Canadian Prairies, revealing the superiority of satellite-derived vegetation indices, particularly NDVI, in predicting yield potential. Pantazi et al. [
18] proposed a model for winter wheat yield prediction, integrating soil spectroscopy and remote sensing data to visually depict yield-influencing factors. Ramos et al. [
19] introduced an optimised Random Forest algorithm for maize-crop yield prediction, emphasising the importance of vegetation indices like NDVI, NDRE, and GNDVI. Li et al. [
20] utilised extreme gradient boosting machine learning to accurately predict vegetation growth in China, achieving high predictive accuracy and demonstrating effectiveness under diverse conditions. Zhang et al. [
21] employed field-surveyed data to predict smallholder maize yield, with novel insights into the performance of various vegetation indices and machine learning techniques.
Recent studies have demonstrated the efficacy of utilising Sentinel-2 satellite imagery and machine learning techniques for predicting crop yields and mapping crop types in various agricultural settings. Son et al. [
22] employed Sentinel-2 image composites and machine learning algorithms to forecast rice crop yields in Taiwan, finding that Support Vector Machines (SVM) outperformed RF and ANN at the field level, indicating their potential for accurate yield predictions approximately one month before harvesting. Perich et al. [
23] utilised Sentinel-2 imagery to map crop yields at the pixel level in small-scale agriculture, with machine learning models utilising spectral index and raw reflectance data proving effective, even in the presence of cloudy satellite time series. Khan et al. [
24] combined ground-based surveys with Sentinel-2 satellite images and deep learning techniques to map crop types, achieving high accuracy in identifying staple crops like rice, wheat, and sugarcane within the first four weeks of sowing.
The diverse literature outcomes examined previously, along with numerous others, have employed a variety of data types from different sources and machine learning models. This diversity presents challenges in generalising techniques across different datasets, yet it also enhances performance for specific datasets. The adoption of multi-modal data usage, multi-modal AI techniques, and Ensemble methods has emerged as the current practice in this research field. Shahhosseini et al. [
25] explore the predictive performance of two novel CNN-DNN machine learning ensemble models for forecasting county-level corn yields across the US Corn Belt. By combining management, environmental, and historical yield data from 1980 to 2019, the study compares the effectiveness of homogeneous and heterogeneous ensemble creation methods, finding that homogeneous ensembles provide the most accurate yield predictions, offering the potential for the development of a reliable tool to aid agronomic decision-making. Gavahi et al. [
26] introduce DeepYield, a novel approach for crop yield forecasting that combines Convolutional Long Short-Term Memory (ConvLSTM) and 3-Dimensional CNN (3DCNN). By integrating spatiotemporal features extracted from remote sensing data, including MODIS Land Surface Temperature (LST), Surface Reflectance (SR), and Land Cover (LC), DeepYield outperforms traditional methods and demonstrates superior forecasting accuracy for soybean yields across the Contiguous United States (CONUS). Zare et al. [
27] investigate the impact of data assimilation techniques on improving crop yield predictions by assimilating LAI data into three single crop models and their multimode ensemble using a particle filtering algorithm. Results from a case study in southwestern Germany reveal that data assimilation significantly enhances LAI simulation accuracy and grain yield prediction, particularly for certain crop models, highlighting the potential for further improvements in data assimilation applications through regional model calibration and input uncertainty analysis.
Moreover, Gopi and Karthikeyan [
28] introduce the Red Fox Optimization with Ensemble Recurrent Neural Network for Crop Recommendation and Yield Prediction (RFOERNN-CRYP) model, which leverages deep learning methods to provide automated crop recommendations and yield predictions. By employing ensemble learning with three different deep learning models (LSTM, bidirectional LSTM (BiLSTM), and gated recurrent unit (GRU)) and optimising hyperparameters using the RFO algorithm, the proposed model demonstrates improved performance compared to individual classifiers, offering valuable support for farmers in decision-making processes related to crop cultivation. Lastly, Boppudi et al. [
29] propose a deep ensemble model for accurately predicting crop yields in India, addressing the challenge posed by variations in weather and environmental factors. The model (Deep Max Out, Bi-GRU and CNN) incorporates improved preprocessing techniques, feature selection using the IBS-BOA algorithm, and prediction through a combination of Deep Ensemble Model and Ensemble classifiers, resulting in significantly reduced error rates compared to existing methods.
In the sequel, we introduce a novel framework for predicting crop yields, named as “RicEns-Net.” This framework incorporates advanced data engineering processes involving five unique data sources, namely Sentinel 1/2/3, NASA’s Goddard Earth Sciences (GES) Data and Information Services Centre (DISC), and field measurements. The novelty of RicEns-Net lies in its integration of these diverse and rich sources of multi-modal data, comprising 15 features selected from a pool of over 100, within an advanced Deep Ensemble model. This model encompasses widely-used CNN and MLP architectures, as well as less explored DenseNet and Autoencoder architectures, which have been infrequently utilised or entirely unexplored in existing literature.
3. Study Region and Data Details
As stated earlier, this study begins by employing the dataset offered by EY for the 2023 iteration of their Open Science Data Challenge [
30]. Within this section, we examine the provided data, outline our process for selecting features, and describe the methods used to create meaningful data features tailored to the objectives of this paper.
3.1. Study Area & Rice Crop Details
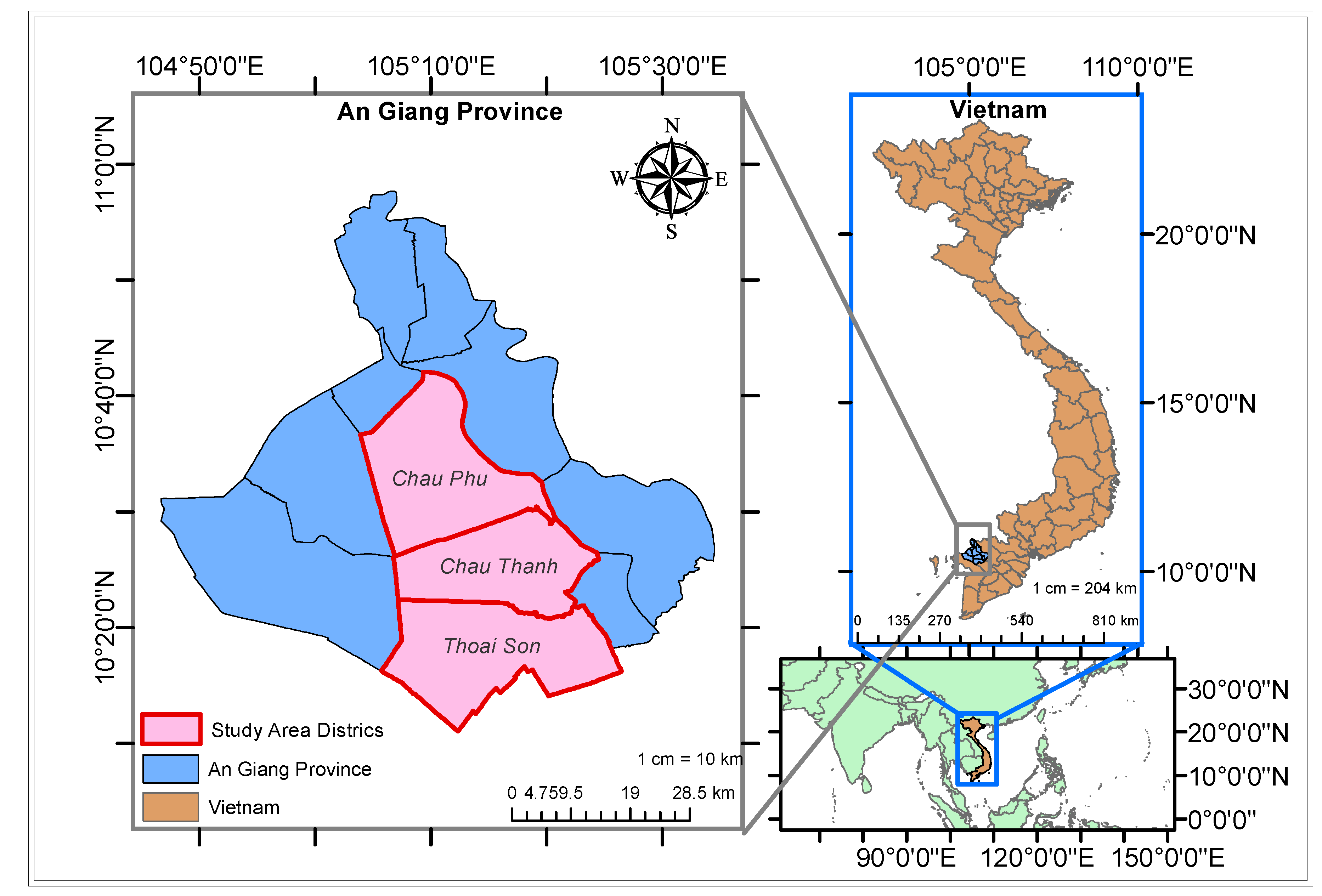
The dataset encompasses information from 557 farm sites situated in Chau Thanh, Thoai Son, and Chau Phu districts within the province of An Giang in Vietnam (see
Figure 1). The study province of An Giang relies significantly on agriculture as a cornerstone of its economy. Notably, An Giang province is situated in the Mekong River delta region, crucial for providing irrigation to support rice cultivation. The dataset, supplied by EY, contains fundamental details for each crop, including District Name, Latitude, Longitude, Crop Season, Crop Frequency, Harvest Date, Crop Area, and Yield as given in
Table 1.
Every entry in the primary dataset represents an individual crop and is characterised by eight features, including three categorical variables (District name, Crop Season [WS=Winter Spring; SA = Summer Autumn], and Crop Frequency of the specified farm [D = Twice; T = Thrice]) and five numerical variables (Latitude, Longitude, Harvest Date, Area [Hectares], and Yield Rate [Kg/Ha]). The harvesting dates of the crops in question extend from March 18, 2022, to August 9, 2022, covering two significant crop seasons, namely Summer-Autumn and Winter-Spring.
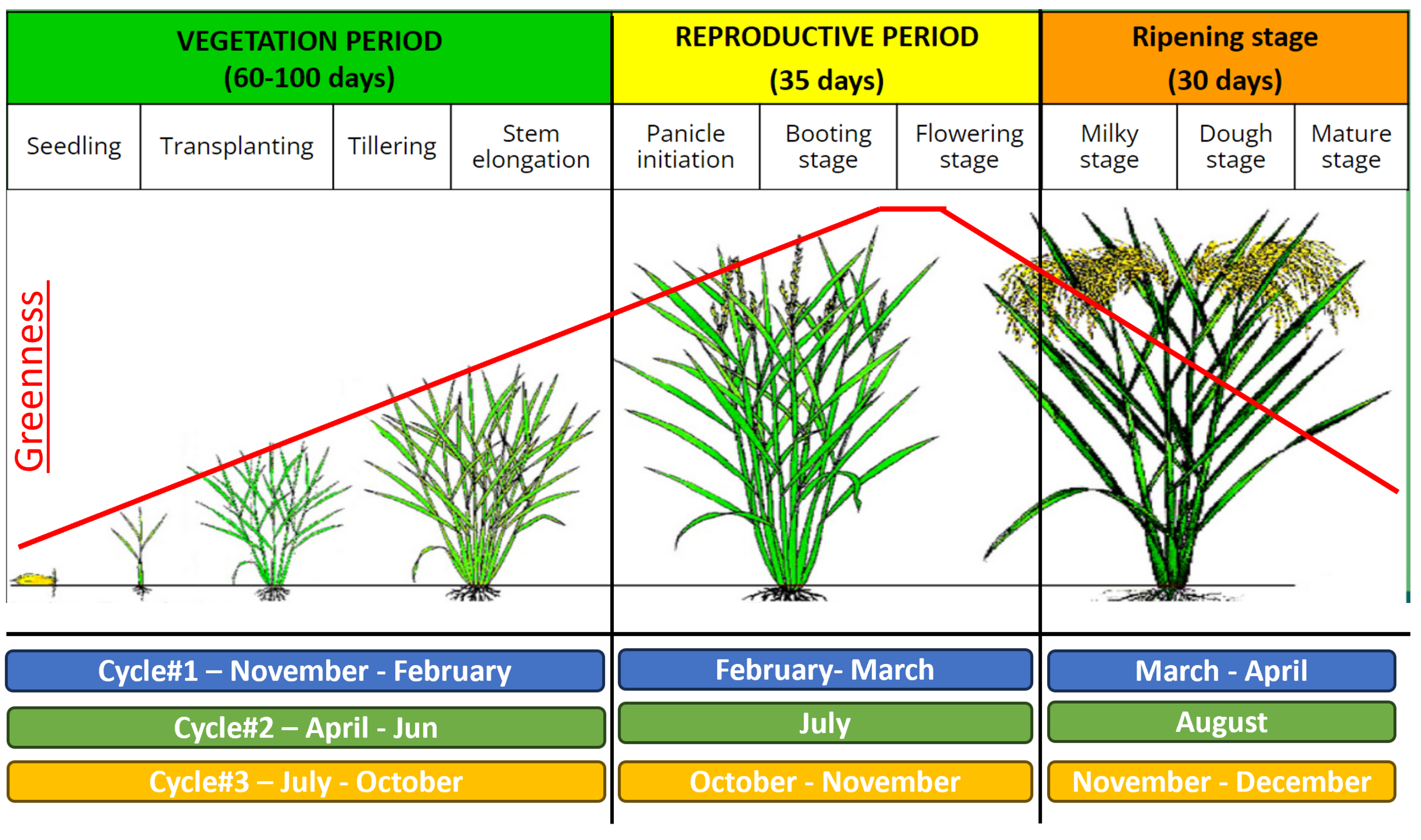
While the overall duration of the crop cycle spans approximately 5-6 months, contingent on the season, the period from transplanting to harvesting typically ranges from 90 to 130 days. The growth trajectory can be categorised into three principal phases: Vegetative, Reproductive, and Ripening stages, illustrated in
Figure 2. During the Reproductive stage, rice plants attain their maximum greenness, marking the culmination of this phase. Subsequently, the crop transitions into the Ripening stage, characterised by the transformation of the plants from green to yellow, coinciding with the maturation of rice grains. In the context of Vietnam, rice cultivation occurs biannually or tri-annually within two seasons: Winter-Spring (Nov-Apr), Summer-Autumn (Apr-Aug), and Autumn-Winter (Jul-Dec).
The necessary geospatial information can be obtained from either Landsat or Sentinel satellites, taking into account the designated location and harvest time outlined in the dataset. After careful consideration of technical details such as Ground Spatial Resolution (GSD) and revisit frequency, the decision is made to opt for data sourced from Sentinel satellites. Additionally, there is anticipation in leveraging the complete spectrum of SAR, MSI, and Meteorological data. To gather remote sensing data for a specified area, it is essential to finalise two key parameters: the time window and the geographical bounding box or crop window.
We intend to gather data during the phase when the crop is nearing complete maturation. The progress of the crop’s growth is discerned through the intensity of the green hue in the data, with the period from transplantation to maturity spanning 60-100 days, contingent on the prevailing season. Following full maturity, the crop progresses into the ripening stage, which typically lasts around 30 days. To determine the timeframe for data collection, we use the harvest date as a reference point. The initiation date is established as 60-90 days before the harvest date, while the conclusion date is set at 30 days before the harvest date (refer to
Figure 2). This designated time window encapsulates the entire duration from the crop’s transplantation to the conclusion of the maturity stage, ensuring that the remote sensing data aligns with the various growth phases of the crop.
The primary input data exclusively provides information about the field’s area without specifying the precise bounding region for each field. This lack of detail hinders our ability to extract data for the exact boundaries of individual crop fields. To address this limitation, we proceed by identifying the precise location of each field in the MSI data and cropping the image to obtain a set of pixels surrounding that specific location. Given that the visual bands of the MSI data have a spatial resolution of 10 meters per pixel, a 3x3 pixel collection corresponds to an area of 900 square meters, equivalent to 0.222 acres or 0.09 hectares. Notably, the SWIR16 and SWIR22 bands of Sentinel 2’s MSI exhibit a spatial resolution of 20 meters per pixel, meaning a 3x3 pixel bounding box represents an area of 3600 square meters, or 0.89 acres, or 0.36 hectares.
3.2. Data/Feature Extraction
3.2.1. Sentinel-1
Sentinel 1 delivers SAR images featuring two polarisations, vertical (VV) and horizontal (VH), characterised by the difference in the polarisation of their transmitted and received signals. The polarisation of radar signals plays a crucial role in deciphering the structure and orientation of surface elements on the land. The radar signal experiences scattering and depolarisation due to the randomly oriented structure of plant leaves as it undergoes multiple bounces. By comparing the vertical (VV) and horizontal (VH) components, the degree of scattering by the land surface can be discerned. In our model, we incorporate this technique through the feature. The data was collected by defining a geographical bounding box with a size of 0.001 degree, resulting in an output array size of 3x3. However, some location data did not conform to this shape and required trimming to achieve a 3x3 box size. Given Sentinel 1’s spatial resolution of 10 meters, each box corresponds to an area of 30 meters by 30 meters. The subsequent features were derived from Sentinel 1 SAR data
Set 1 (4 variables) →
where the radar vegetation index (RVI) is given as
3.2.2. Sentinel-2
Sentinel 2 data furnishes spectral intensities across 13 bands, encompassing Visual-NIR (VNIR) to Short-Wave Infra-Red (SWIR) regions. Notably, there are four spectral bands, namely Red, Green, Blue, and NIR (B04, B03, B02, and B08), offering a ground resolution of 10 meters. Additionally, six bands exhibit a 20-meter ground resolution, comprising four Red Edge bands (B05, B06, B07, and B08A) and two SWIR bands with distinct wavelengths (B11 and B12). The remaining three bands, with a 60-meter ground resolution, serve specific purposes: B01 for aerosol detection (0.443 m), B09 for water vapour observation (0.945 m), and B10 for cirrus detection (1.374 m). Notably, the Sentinel 2 mission boasts a revisit frequency of 5 days.
When acquiring MSI data, it is crucial to account for the potential impact of cloud cover in the targeted area. With a revisit frequency of 5 days, there are only 6-8 chances to capture images during the period when crops reach full growth before maturation. Given Vietnam’s tropical monsoon climate, these image opportunities are prone to cloudiness. The dataset at hand reveals median cloud coverage values of 16% and 21% during the Winter-Spring and Summer-Autumn seasons, respectively.
In order to prevent the occurrence of unclear or cloudy images, it is necessary to eliminate those with a high level of cloud coverage. Simultaneously, we aim to capture comprehensive crop data when the plants are at their full growth and exhibit maximum greenness. To achieve optimal outcomes, we conducted experiments with various values for maximum cloud coverage and time windows. Based on the findings detailed in
Table 2, we concluded that setting the maximum cloud coverage to 60% and collecting Sentinel-2 MSI data during the 50 days preceding the crop maturation provides favourable results. The objective is to secure a minimum of 4-5 images for each specific location.
To ensure that the spectral intensity trends are captured, we identify the minimum, maximum, mean and variance of 9 MSI bands based on all the MSI images available for each location.
Set 2 (36 variables) → is in a format of “” where , , , , , , , , and , , , .
MSI data has been used to create transformational features known as
Vegetation Indices given in
Table A1 like NDVI, SR, EVI, EVI2, SAVI, RGVI, DVI, MSR, NIRv, kNDVI, NDVIre, NDRE1, NDRE2 to indicate the volume of vegetation on the land surface.
Set 3 (26 variables) → is in a format of “” where , , , , , , , , , , , , and , .
Utilising MSI data, various features, such as NDWI, BSI, and LSWI, as outlined in
Table A1, have been generated to depict soil and water content. This application is particularly advantageous in the context of rice cultivation, where the crop is submerged in water.
Set 4 (6 variables) → is in a format of “” where , , and , .
Additional features have been generated using MSI spectral data, incorporating information derived from the biochemical properties of the plants (refer to
Table A1).
3.2.3. Sentinel-3
Sentinel 3 data was acquired to obtain meteorological information related to environmental variables such as ambient air temperature (), land surface temperature (LST), solar radiation (), and specific humidity (). These data sets have been integrated into the model as the following features:
Set 6 (8 variables) → is in a format of “” where , , , and , .
3.2.4. NASA GES DISC
Rainfall information was acquired from NASA’s Goddard Earth Sciences (GES) Data and Information Services Centre (DISC) through the utilisation of the Google Earth Engine API. The data retrieval involved the utilisation of the precipitationCal parameter, which denotes rainfall in mm per hour. We organise this data into two distinct features: Rainfall-Totalgrowth and Rainfall-Totalmaturity, encompassing three statistical measures—mean, maximum, and sum as
Set 7 (6 variables) → is in a format of “” where , and , , .
3.3. Correlation Analysis
After meticulously gathering all potentially valuable engineered features from multi-modal remote sensing data, we proceed to examine their statistical and predictive capabilities for subsequent feature selection. This phase, delineated in this sub-section, initiates with a correlation analysis.
Concerning the relationship between SAR features and , all four data features exhibit a strong correlation with . As anticipated, the feature effectively captures the backscattering of the SAR signal by the rice plant leaves, resulting in a higher correlation (0.32) with compared to (0.25). The serves as a transformative feature, demonstrating an enhanced correlation (0.45) in comparison to both and individually. Notably, the Radar Vegetation Index (RVI) shows an a similar positive correlation (0.41) with the Yield Rate.
As previously indicated regarding Sentinel-2 data, we derive spectral statistics from 9 bands: B02 Blue, B03 Green, B04 Red, B05-B07 Red Edge, B08 NIR, and B11-B12 SWIR. These statistics, namely min, max, mean, and variance, are incorporated into the model. Upon analysing observations across all bands, it is noteworthy that variance features exhibit a relatively low correlation () with the target variable, whereas other statistical measures demonstrate correlation coefficient values surpassing 0.3.
Spectral data is employed to generate transformative characteristics known as Vegetative Indices (VI). These indices serve as a more efficient measure for discerning and monitoring variations in plant phenology. In our approach, we utilise VIs such as NDVI, NDVIre, NDRE1, NDRE2, SR, DVI, MSR, EVI, EVI2, SAVI and RGVI, NIRv, and kNDVI. These features are integrated into the model in the form of their respective mean and variance features. However, the variance feature is omitted from the model due to its limited correlation with the target variable. Notably, kNDVI exhibits one of the highest correlations with Yield, while features like DVI, EVI, and NIRv demonstrate some of the lowest correlations.
Similar to the vegetation indices, we can employ optical data to compute additional indices that precisely quantify the environmental conditions of the crop’s cultivation. NDWI, LSWI16, and LSWI20 specifically indicate the water or moisture content in the soil, which is crucial for rice cultivation, requiring flooded fields. Conversely, BSI reflects the soil condition. We incorporate these attributes into the model as their respective Mean and Variance features. Similar to the approach with vegetation indices, we have excluded the variance feature from the models due to its limited correlation with Yield. Notably, all water indices exhibit high correlations with each other and share a similar correlation with Yield.
Lastly, concerning meteorological characteristics, the average ambient air temperature (refers to variable “”) and specific humidity () exhibit the strongest correlation with crop yield, and they also demonstrate a high degree of correlation between themselves. Solar radiation () emerges as a significant predictor due to its notable correlation with yield and comparatively lower correlation with other meteorological features.
3.4. Feature Selection
Up to this point, all the engineered features, totalling 94 in number, have undergone various stages of processing. These stages include (i) grouping, involving the arrangement of data types and condensation into categorical, numerical, and object types; (ii) scaling, which entails MinMax scaling; and (iii) splitting through a train-test split with a ratio of 3:1.
As outlined in the preceding sections, the subset of the 94 features exhibits significant correlation, and incorporating all these features in the models would lead to computationally intensive experiments. To mitigate this, during the final processing stage, we execute multiple rounds of feature selection, including Pairwise Feature Independence Check using the
test, statistical significance tests based on p-values, outlier removal, and thresholding for correlation and variance. Following these stages, the outcome is a refined set of 15 predictive (11 numerical & 4 categorical) features and 1 target feature all of which are shown in
Table 3.
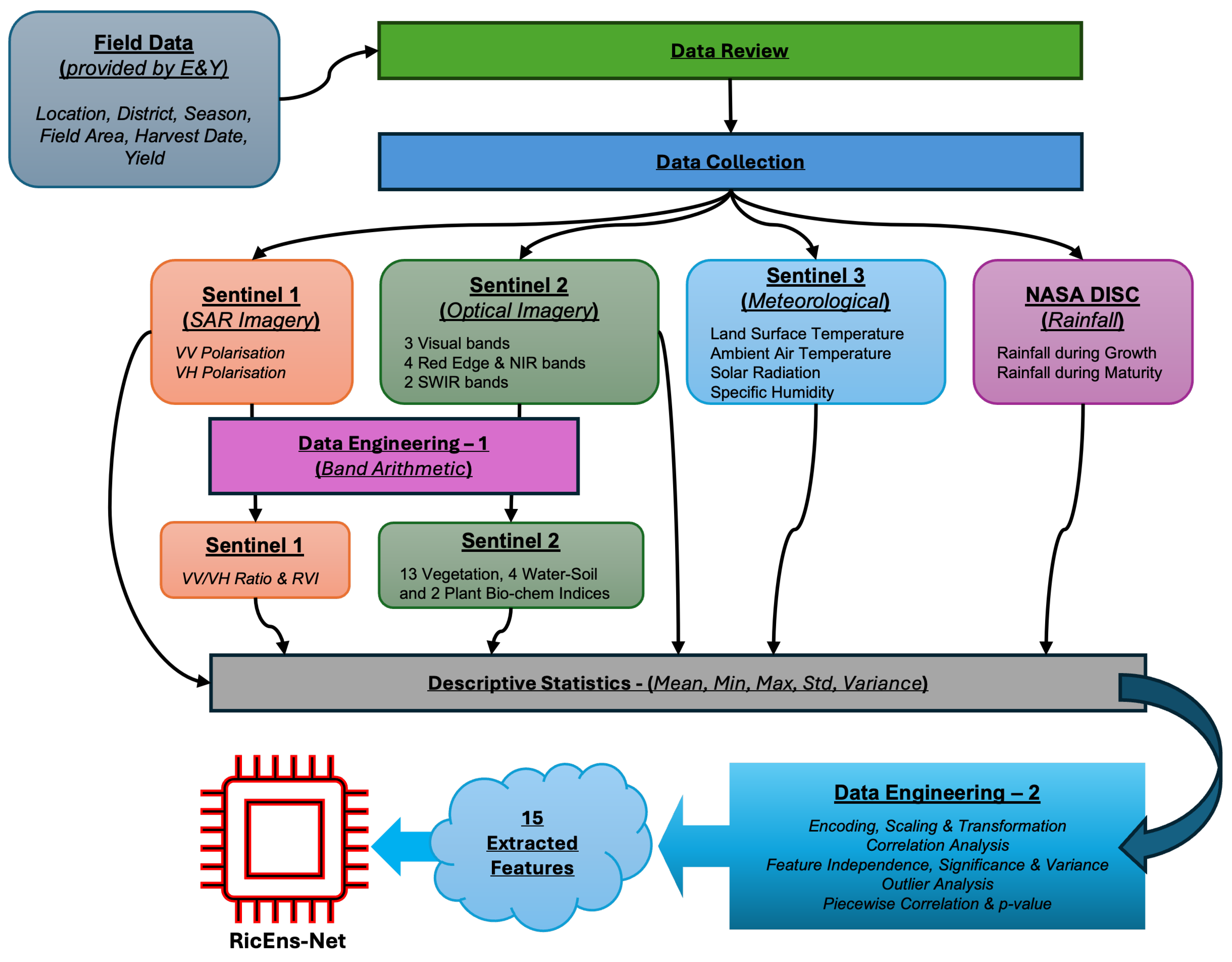
The whole data collection, processing and engineering stages are plotted in
Figure 3.
4. Methodology
After the extensive stages of data engineering, pre-processing, and preparation mentioned earlier, the 15 most informative and significant data features from 5 different data modalities will be employed to predict rice yield for the specified locations. This section provides a comprehensive introduction to the proposed deep ensemble model, RicEns-Net, and explains the reasoning behind the selection of the advanced techniques used in this study. The section will conclude with a description of the computational environment for implementation and definitions of the performance metrics.
4.1. The Proposed Model - RicEns-Net
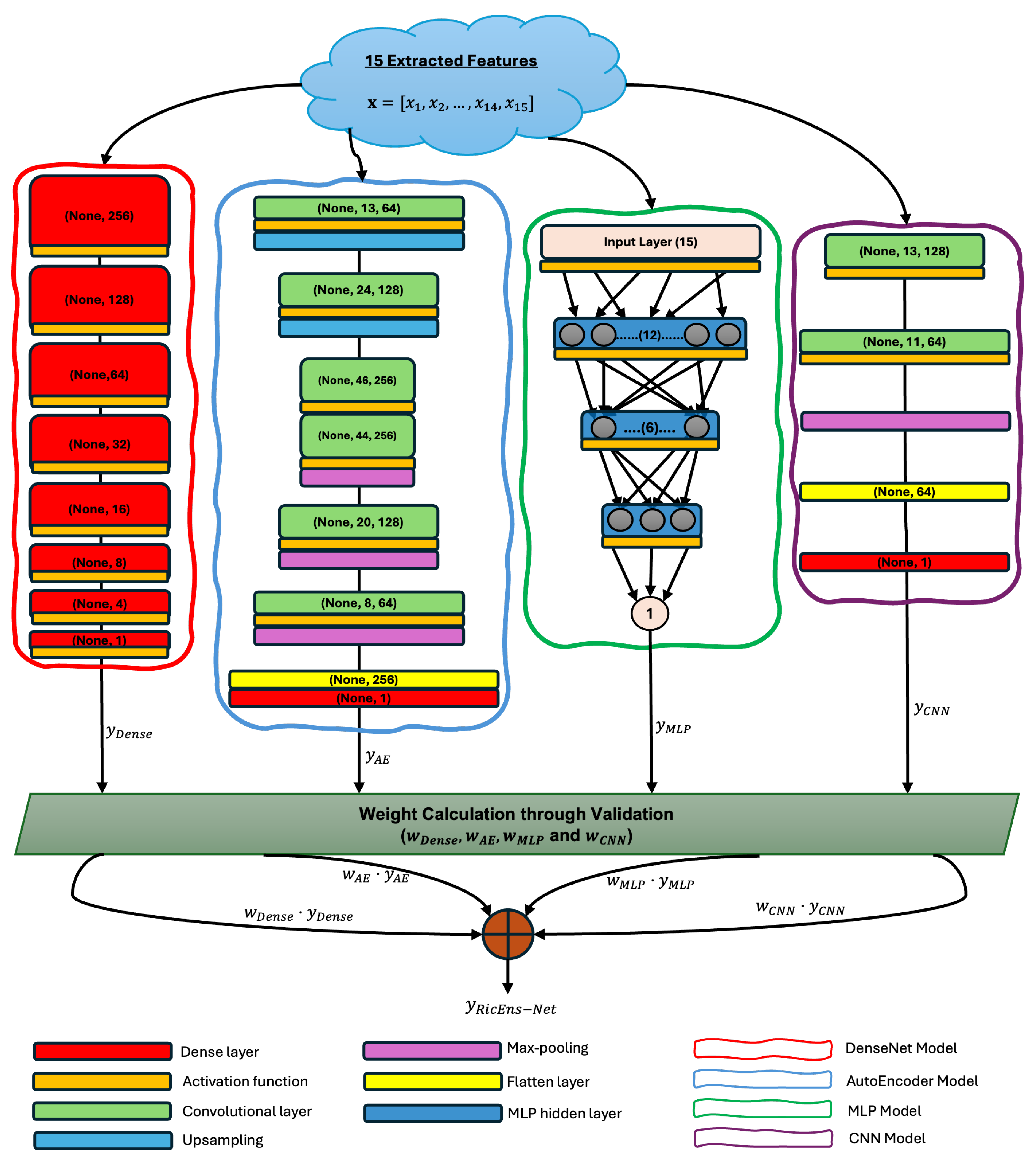
As mentioned in the earlier stages, deep learning models have currently been dominating yield prediction studies. This paper stands parallel with these advances in the literature but tries to explore complementary advantages of different deep learning regression techniques under a deep ensemble architecture named RicEns-Net. The details of RicEns-Net model architectures are presented in
Figure 4.
RicEns-Net utilises two cutting-edge deep learning architectures, CNN and MLP. Despite the various designed versions of these models in the literature, no single architecture has emerged as a general solution. This motivated us to create our architecture tailored to achieve optimal performance for the rice yield prediction dataset. In addition to CNN and MLP, RicEns-Net incorporates two significant deep learning architectures: DenseNet and AutoEncoder (AE). While these architectures are scarcely used in yield prediction literature, they are prevalent in crucial remote sensing applications, such as semantic segmentation and classification.
Our motivation for incorporating DenseNet and AE architectures into our ensemble model stems from their respective advantages in handling complex data. AE architectures are valuable for rice yield prediction as they efficiently reduce dimensionality, extracting essential features while minimizing noise. Conversely, DenseNet architectures enhance deep learning by ensuring robust gradient flow through dense connections, thereby improving feature propagation and enabling the model to learn intricate patterns within agricultural data. These combined capabilities make the ensemble model more effective in predicting rice yield.
In developing the RicEns-Net ensemble model, extensive testing was conducted to determine the optimal architectures for each individual deep learning model. This rigorous process involved systematically evaluating various configurations, including the number of layers, types of activation functions, and loss functions. For each model, we experimented with different depths ranging from shallow to deep architectures to find the optimal balance between complexity and performance. We also tested various activation functions, such as ReLU, sigmoid, SiLU, and tanh, to identify the most effective function for each model. Additionally, we compared multiple loss functions, including mean squared error, mean absolute error, and Huber loss, to select the one that minimized prediction errors most effectively. Furthermore, each model’s architecture was refined through a series of experiments, incorporating cross-validation and hyperparameter tuning, to ensure the best performance for predicting rice yield. This exhaustive testing process ensured that each model within the ensemble was optimally configured to contribute to the overall predictive power of RicEns-Net. The details of these architectures, including the final selected configurations, are illustrated in
Figure 4. These configurations were chosen based on comprehensive performance evaluations, ensuring that each component model enhances the proposed deep ensemble’s accuracy and robustness.
In order to create the RicEns-Net’s ensemble output, we use a weighted average approach. Let
be the output of the
i-th model, and let
be the weight assigned to the
i-th model. The ensemble output
is then given by:
where
N is 4 - the total number of models in the ensemble - and
i refers to each model with keywords
.
The weights
are determined based on the validation errors of each model. Let
be the validation error of the
i-th model. We assign the weights such that a lower validation error corresponds to a higher weight. Specifically, we can define the weight
as:
This choice ensures that the weights are normalised to sum to 1:
Thus, the final ensemble output is a weighted average of the individual model outputs, where the weights are inversely proportional to the validation errors, normalised to sum to 1. This approach prioritises models with lower validation errors, giving them a higher influence on the ensemble output.
4.2. On Deciding State-of-the-Art
Despite significant advancements in computer vision research through the development of novel deep learning architectures, yield prediction research predominantly relies on traditional deep learning methods like CNN and MLP, as well as state-of-the-art machine learning algorithms such as RF, XGBoost, and SVR. Additionally, traditional time series deep learning techniques, including Long Short-Term Memory (LSTM) and its variants, have been extensively utilized in the literature. Given this context, selecting the appropriate state-of-the-art models for the comparison study in this paper is of paramount importance. This section is dedicated to explaining the rationale behind the choice of each comparison model.
As previously discussed, this paper utilizes a dataset from the EY Open Science Challenge 2023. This global data science competition, sponsored by EY, a leading consulting firm, is notable for its innovative application of Data Science and Analytics to real-world business scenarios. The EY Open Science Challenge 2023 ran for two months, from January 31, 2023, to March 31, 2023, attracting over 13,000 participants and more than 7,500 submissions. The competition awarded USD 10,000 to the winner and USD 5,000 to the runner-up. EY has provided performance results for the winning teams (global and employees) on the same test dataset, making these two models ideal candidates for comparison in this study. The global winner used a CatBoost regression model (referred to as CatBoost-EY in this paper), while the employee winner’s model was based on Extremely Randomized Trees (referred to as ExtRa-EY). We present these models’ performance metrics without infringing on their copyrights.
Considering that our ensemble model incorporates four deep learning architectures— DenseNet, CNN, MLP, and AE— we also evaluate their individual performance in the comparison study. This approach aligns with the systematic review paper [
32], which notes that 78 out of 102 proposed models in the literature up to 2023 include these four architectures. This validation supports our choice of comparison models, ensuring that the proposed RicEns-Net is benchmarked against state-of-the-art standards. Additionally, we incorporate advanced machine learning algorithms such as XGBoost, RF, SVR, AdaBoost, ElasticNet, and Gradient Boosting into the comparison pool, along with their Voting and Stacked ensemble models.
4.3. Implementation of Models
We conducted our research using Python version 3.10.9, leveraging the rich ecosystem of libraries available for data science and machine learning. Our desktop workstation, featuring a robust 20-core processor and ample 32GB of RAM, provided the computational power necessary for handling large datasets and complex modelling tasks efficiently. The widely-used scikit-learn (sklearn) module served as the cornerstone for implementing traditional and state-of-the-art machine learning algorithms, offering a comprehensive suite of tools for data preprocessing, model selection, and evaluation. Additionally, for the implementation of deep learning architectures, we employed the widespread Python libraries tensorFlow and keras, which provide powerful abstractions and efficient computation frameworks tailored specifically for neural network development. These Python modules enabled us to explore a diverse set of modelling techniques and methodologies, ultimately facilitating the realisation of our research objectives with precision and scalability.
4.4. Evaluation Metrics
All the models will undergo assessment utilising regression metrics such as RMSE (Root Mean Square Error), MAE (Mean Absolute Error),
Score (Coefficient Of Determination), and Adjusted
(Adjusted Coefficient Of Determination). While MAE stands as the most straightforward performance metric, RMSE poses a more rigorous criterion by squaring the prediction error before calculating its mean and taking the square root. MAE and RMSE exhibit differences in their sensitivity to outliers.
Another critical metric is
which is anticipated to fall within the 0 to 1 range, although it can dip below 0 for specific models. In simpler terms, the
value gauges the model’s capacity to elucidate the variance of the target variable. To provide a more precise definition, as outlined in the SkLearn user guide, it signifies the proportion of variance (of
y) explained by the independent variables in the model. It offers insight into the goodness of fit and serves as an indicator of how effectively the model is likely to predict unseen samples through the explained variance proportion [
33].
Additionally, we employ a modified version of
known as Adjusted
, which factors in the impact of an elevated number of predictors contributing to a higher
value. Furthermore, we compute the
score for the input training data to compare it with the testing
score. The disparity between them signals the potential for overfitting. The evaluation of models based on disparities in training and testing
scores aids in the identification of models that may not generalise effectively with unseen data.
where
n refers to number of observations and
k is the number of predictors.
5. Experimental Analysis
We start our experimental analysis with a general comparison of all state-of-the-art models mentioned above to the proposed RicEns-Net model. The analysis compares the MAE and RMSE errors in predicting the rice yield along with goodness of fit measures of train/test and adjusted
values which are presented in
Table 4.
Analyzing the performance metrics presented in
Table 4, it is evident that the RMSE values for most models range between 430 and 470 Kg/Ha, while the MAE values fall between 340 and 380 Kg/Ha. The proposed Deep Ensemble model, RicEns-Net, stands out as the top-performing model in both RMSE and MAE, showing at least a 2% (and up to 10%) improvement over the next best models, CNN and SVR. When considering the adjusted
values of the test results, it is clear that the traditional model ensemble - Voting regressor - along with CNN, demonstrate excellent performance, while RicEns-Net surpasses all 12 models. Besides the leading performance of the Deep Ensemble model, SVR, CNN, and Voting Regressor, the DenseNet deep learning model, as well as traditional machine learning models such as AdaBoost and Gradient Boosting, also exhibit strong performance. Their MAE and RMSE values are within 5% of those of the best-performing models.
Examining
Table 4 with a comparison between the EY contest winners and the proposed RicEns-Net, the findings indicate that the outcomes of this study demonstrate comparable or superior performance metrics relative to the global competition winners. As shown in
Table 4, RicEns-Net consistently outperforms both EY-winning models in terms of MAE and RMSE. Additionally, among the reference models, the CNN, SVR, DenseNet, and Voting regressor models specifically achieved performance values that are competitive with those of the EY winner models.
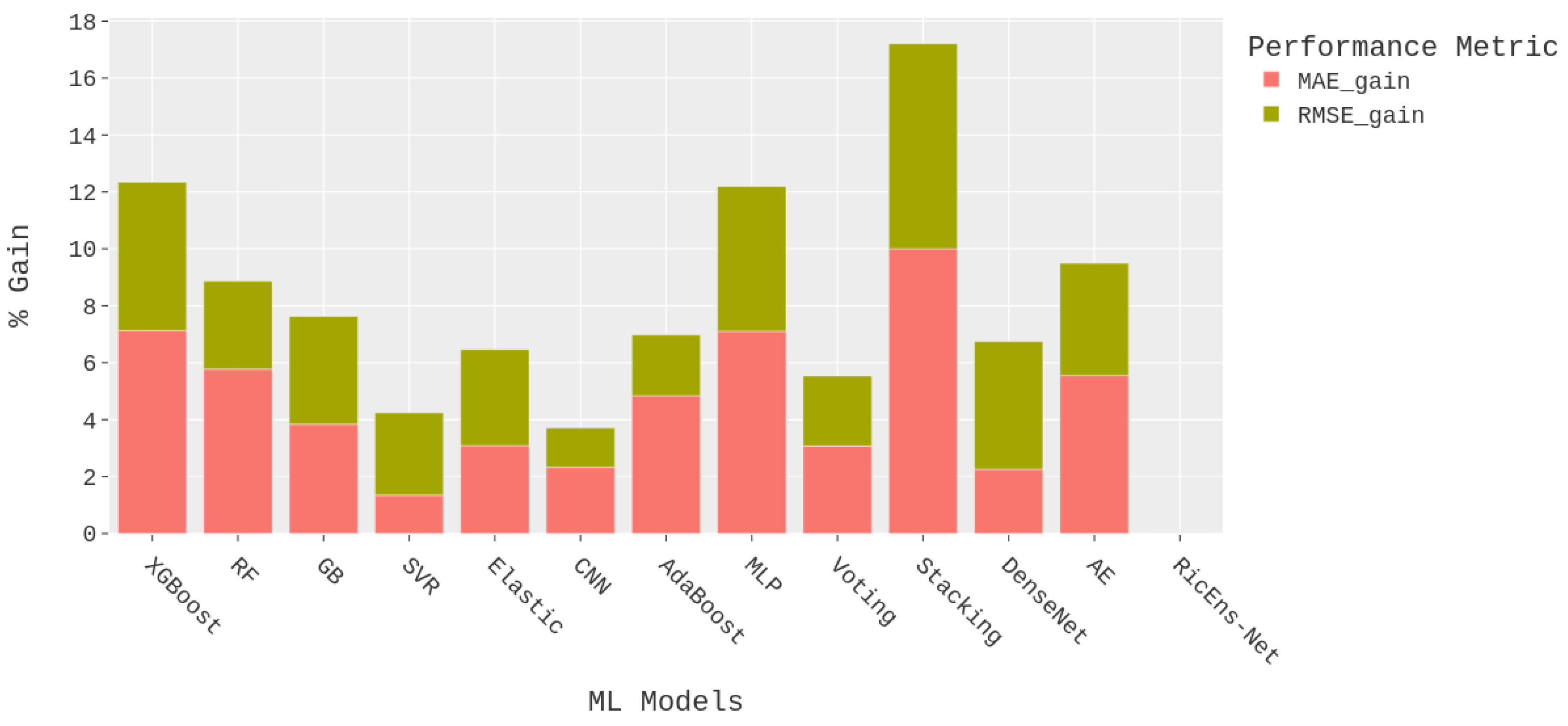
Figure 5 illustrates the results presented in
Table 4 from the perspective of performance improvement, focusing on the proposed RicEns-Net model as the best-performing model. The performance gains of Ricens-Net for MAE and RMSE metrics were computed as percentages and visualised as a stacked bar chart in
Figure 5. It can be observed from
Figure 5 that CNN, SVR and Voting Regressor models exhibit performance closest to that of RicEns-Net, with a cumulative performance degradation of approximately 5%. Conversely, Stacking regressor, MLP and XGBoost techniques demonstrate the poorest performance among all referenced models with more than 10% cumulative errors.
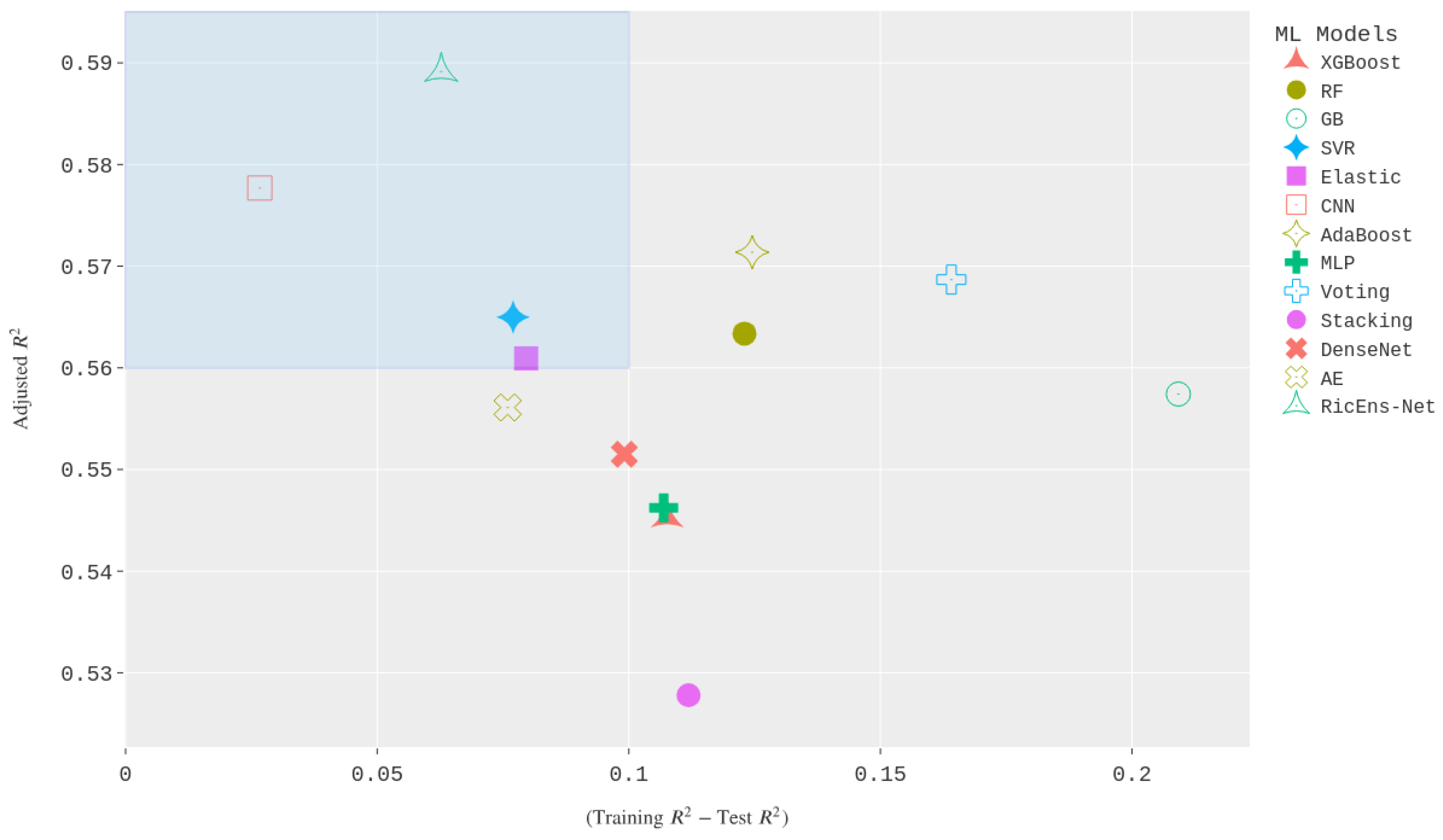
Figure 6 illustrates a scatter plot in two dimensions, showcasing the relationship between the
Difference and the Adjusted Test
values.
Figure 6 serves as a visual representation for best-performing models in terms of goodness of fit measures, where the optimal performance is achieved when the
Difference is minimised, and the Adjusted Test
is maximised. Therefore, models positioned nearer to the top-left corner of the plot represent the most effective ones in terms of
metrics. The rectangular area depicted in
Figure 6 serves to highlight a region for best-performing models. Subsequently, the top-performing models selected within this region include the proposed RicEns-Net, CNN, SVR, and ElasticNet models. Notably, certain models that showed favourable RMSE and MAE values were excluded from selection due to unsatisfactory
performance during either the test or training phases. For example, even though the Voting and Adaboost regressors ranked in the top 5 for cumulative MAE/RMSE, they did not make it into the top left rectangle because of a significant disparity between their training and test performance which can be seen as an indicator that these methods are likely to overfit during training.
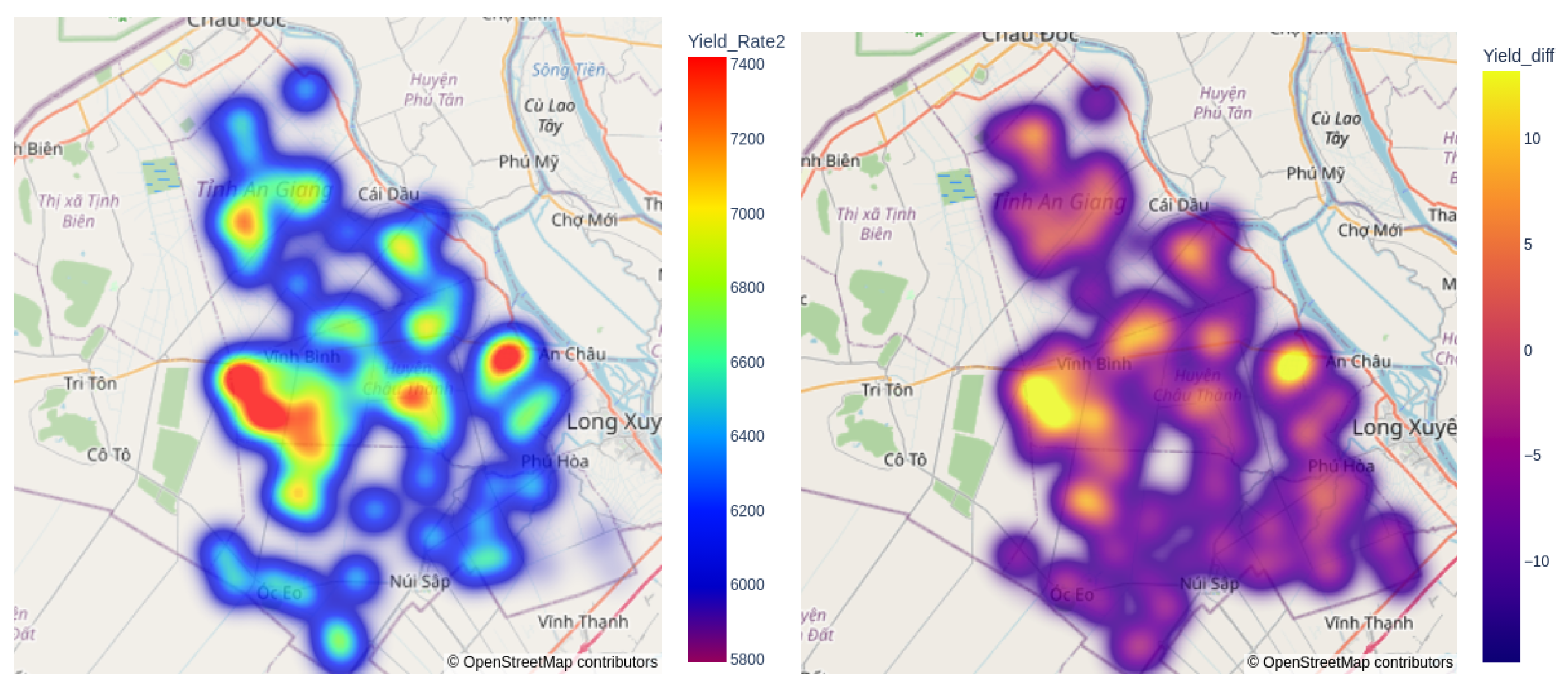
The final experimental analysis includes a visualisation output displaying the predictions of the RicEns-Net model for
, alongside a separate subplot illustrating the discrepancies between these predictions and the actual ground truth values, as shown in
Figure 7. The analysis reveals that the maximum absolute error exceeds 10% in only a small portion of the region of interest, with consistently low errors observed across most regions in the test dataset. Furthermore, it is evident that areas with higher yield rates tend to show higher errors, while the RicEns-Net model generally underestimates the
values.
6. Conclusions
In this study, we proposed a novel deep ensemble model called RicEns-Net for predicting rice crop yields. Leveraging multi-modal data sourced from Ernst & Young as part of their annual data science challenge, our predictive model incorporated radar and optical-type remote sensing data along with meteorological information to enhance forecasting accuracy. Through the creation of a comprehensive set of data features derived from spectral indices and advanced feature engineering techniques, we addressed the complex interplay of natural factors influencing crop yield. Employing an extensive feature selection process, we curated a refined set of 15 features from 5 different modalities to mitigate the curse of dimensionality. Our evaluation of deep ensemble methodology and various state-of-the-art machine learning regression models revealed their capability to predict crop yield rates with acceptable margins of error. These findings underscore the practical utility and effectiveness of the proposed data features in real-world agricultural applications, offering promising avenues for enhancing crop yield forecasting methodologies.
The MSI remote sensing data gathered from various geographical locations in this study are significantly influenced by cloud presence and shadows. Moreover, there is a lack of input data regarding the shape of individual crop fields. To address these limitations in future research, it is suggested to employ cloud and cloud shadow detection algorithms alongside algorithms for accessing location-specific spectral statistics. This approach would allow for the establishment of a higher threshold for cloud coverage across the entire MSI image, as well as the consideration of location-specific cloud and shadow coverage before deciding on further image processing for spectral statistics extraction. Although the development of a cloud detection algorithm utilising Sentinel 2’s B09 Band input for water vapour detection is feasible, it has not been implemented in this study and is proposed as a future endeavour.
Another area for enhancement involves integrating an algorithm capable of delineating the geographical perimeters of individual crop fields. This enhancement would allow for the gathering of comprehensive data across entire fields, contrasting with the current method of assessing spectral statistics within a limited 3 x 3-pixel area (equivalent to 30 x 30 meters in Sentinel 1/2 imagery) centred around designated coordinates.
In recent years, there have been several technological advancements that hold substantial promise for enhancing the scope and goals of this study. As previously mentioned, there is growing interest in employing user-centric remote sensing techniques for “Precision Farming,” aiming not only to optimise crop yields but also to efficiently manage resources such as water, fertilisers, insecticides, and pesticides. Additionally, there has been notable progress in the availability of high-resolution multi-spectral optical sensors equipped with LiDAR and Radar capabilities. When coupled with UAVs/drones, these sensors offer farmers unprecedented advantages in crop monitoring, enabling them to schedule flights flexibly at low altitudes to circumvent cloud cover. This integration promises to revolutionise crop health monitoring by enabling the measurement of biochemical and physiological parameters of crops more efficiently and effectively.
Author Contributions
Conceptualization, A.D.Y and O.K.; methodology, A.D.Y. and O.K.; software, A.D.Y.; validation, A.D.Y. and O.K.; formal analysis, A.D.Y. and O.K.; investigation, A.D.Y.; resources, A.D.Y.; data curation, A.D.Y.; writing—original draft preparation, A.D.Y. and O.K.; writing—review and editing, O.K.; visualization, A.D.Y. and O.K.; supervision, O.K. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding. The APC was funded by Cardiff University.
Data Availability Statement
The data used in this study belongs to EY and cannot be shared by the authors. Access to the data is subject to EY’s policies and restrictions.
Acknowledgments
We would like to express our sincere gratitude to EY for providing the data that made this research possible. Special thanks go to Dr. Brian Killough (Program Director, EY Open Science Data Challenge), Anna Biel (Program Manager, EY), and Saurabh Agarwal (AI Manager and Design Lead) for their invaluable assistance in obtaining the data and sharing the winner team codes. Their support and contributions were instrumental in the success of this study.
Conflicts of Interest
The authors declare no conflicts of interest.
Appendix A. Vegetation, Soil and Plant Biochemical Indices
This appendix section presents important vegetation, soil and plant biochemical indices in
Table A1.
Table A1.
Vegetation, Soil, Water and Plant Biochemical Indices.
Table A1.
Vegetation, Soil, Water and Plant Biochemical Indices.
| Index |
Reference |
| Normalized Difference Vegetation Index (NDVI) |
|
| Transformed Vegetation Index (TVI) |
|
| Simple Ratio (SR) |
|
| Enhanced Vegetation Index (EVI) |
|
| EVI - 2-Bands (EVI2) |
|
| Soil adjusted vegetation index (SAVI) |
|
| Rice Growth Vegetation Index (RGVI) |
|
| Difference Vegetation Index (DVI) |
|
| Modified Simple Ratio (MSR) |
|
| Near Infra-Red Reflectance Of Vegetation (NIRv) |
|
| Kernelized NDVI (kNDVI) |
|
| NDVI-Red Edge (NNDVIre) |
|
| Normalized Difference Red Edge 1 (NDRE1) |
|
| Normalized Difference Red Edge 2 (NDRE2) |
|
| Normalized Difference Water Index (NDWI) |
|
| Bare Soil Index (BSI) |
|
| Land Surface Water Index (1.6 m) (LSWI16) |
|
| Land Surface Water Index (2.2 m) (LSWI22) |
|
| Chlorophyll Carotenoid Index (CCI) |
|
| Green Chromatic Coordinate (GCC) |
|
References
- United Nations. United Nations Sustainable Development Goals, 2023. Accessed: 20 August 2023.
- United Nations. Goal 2: Zero Hunger, no date. Accessed: 20 August 2023.
- Food and Agriculture Organization (FAO). Rice market monitor, 2022. Accessed: January 18, 2024.
- Rowley, A.; Karakuş, O. Predicting air quality via multimodal AI and satellite imagery. Remote Sensing of Environment 2023, 293, 113609. [CrossRef]
- Fan, X.; Zhou, W.; Qian, X.; Yan, W. Progressive adjacent-layer coordination symmetric cascade network for semantic segmentation of multimodal remote sensing images. Expert Systems with Applications 2024, 238, 121999. [CrossRef]
- Ma, W.; Karakuş, O.; Rosin, P.L. AMM-FuseNet: Attention-based multi-modal image fusion network for land cover mapping. Remote Sensing 2022, 14, 4458. [CrossRef]
- Roy, S.K.; Deria, A.; Hong, D.; Rasti, B.; Plaza, A.; Chanussot, J. Multimodal fusion transformer for remote sensing image classification. IEEE Transactions on Geoscience and Remote Sensing 2023. [CrossRef]
- Baier, W.; Robertson, G.W. The performance of soil moisture estimates as compared with the direct use of climatological data for estimating crop yields. Agricultural Meteorology 1968, 5, 17–31. [CrossRef]
- Bouman, B. Crop modelling and remote sensing for yield prediction. Netherlands Journal of Agricultural Science 1995, 43, 143–161. [CrossRef]
- Baret, F.; Guyot, G. Potentials and limits of vegetation indices for LAI and APAR assessment. Remote sensing of environment 1991, 35, 161–173. [CrossRef]
- Uno, Y.; Prasher, S.; Lacroix, R.; Goel, P.; Karimi, Y.; Viau, A.; Patel, R. Artificial neural networks to predict corn yield from Compact Airborne Spectrographic Imager data. Computers and electronics in agriculture 2005, 47, 149–161. [CrossRef]
- Li, A.; Liang, S.; Wang, A.; Qin, J. Estimating crop yield from multi-temporal satellite data using multivariate regression and neural network techniques. Photogrammetric Engineering & Remote Sensing 2007, 73, 1149–1157.
- Bala, S.; Islam, A. Correlation between potato yield and MODIS-derived vegetation indices. International Journal of Remote Sensing 2009, 30, 2491–2507. [CrossRef]
- Li, R.; Li, C.; Xu, X.; Wang, J.; Yang, X.; Huang, W.; Pan, Y.; others. Winter wheat yield estimation based on support vector machine regression and multi-temporal remote sensing data. Transactions of the Chinese Society of Agricultural Engineering 2009, 25, 114–117.
- Stojanova, D.; Panov, P.; Gjorgjioski, V.; Kobler, A.; Džeroski, S. Estimating vegetation height and canopy cover from remotely sensed data with machine learning. Ecological Informatics 2010, 5, 256–266. [CrossRef]
- Mosleh, M.K.; Hassan, Q.K.; Chowdhury, E.H. Application of remote sensors in mapping rice area and forecasting its production: A review. Sensors 2015, 15, 769–791. [CrossRef] [PubMed]
- Johnson, M.D.; Hsieh, W.W.; Cannon, A.J.; Davidson, A.; Bédard, F. Crop yield forecasting on the Canadian Prairies by remotely sensed vegetation indices and machine learning methods. Agricultural and forest meteorology 2016, 218, 74–84. [CrossRef]
- Pantazi, X.E.; Moshou, D.; Alexandridis, T.; Whetton, R.L.; Mouazen, A.M. Wheat yield prediction using machine learning and advanced sensing techniques. Computers and electronics in agriculture 2016, 121, 57–65. [CrossRef]
- Ramos, A.P.M.; Osco, L.P.; Furuya, D.E.G.; Gonçalves, W.N.; Santana, D.C.; Teodoro, L.P.R.; da Silva Junior, C.A.; Capristo-Silva, G.F.; Li, J.; Baio, F.H.R.; others. A random forest ranking approach to predict yield in maize with uav-based vegetation spectral indices. Computers and Electronics in Agriculture 2020, 178, 105791. [CrossRef]
- Li, X.; Yuan, W.; Dong, W. A machine learning method for predicting vegetation indices in China. Remote Sensing 2021, 13, 1147. [CrossRef]
- Zhang, L.; Zhang, Z.; Luo, Y.; Cao, J.; Xie, R.; Li, S. Integrating satellite-derived climatic and vegetation indices to predict smallholder maize yield using deep learning. Agricultural and Forest Meteorology 2021, 311, 108666. [CrossRef]
- Son, N.T.; Chen, C.F.; Cheng, Y.S.; Toscano, P.; Chen, C.R.; Chen, S.L.; Tseng, K.H.; Syu, C.H.; Guo, H.Y.; Zhang, Y.T. Field-scale rice yield prediction from Sentinel-2 monthly image composites using machine learning algorithms. Ecological informatics 2022, 69, 101618. [CrossRef]
- Perich, G.; Turkoglu, M.O.; Graf, L.V.; Wegner, J.D.; Aasen, H.; Walter, A.; Liebisch, F. Pixel-based yield mapping and prediction from Sentinel-2 using spectral indices and neural networks. Field Crops Research 2023, 292, 108824. [CrossRef]
- Khan, H.R.; Gillani, Z.; Jamal, M.H.; Athar, A.; Chaudhry, M.T.; Chao, H.; He, Y.; Chen, M. Early Identification of Crop Type for Smallholder Farming Systems Using Deep Learning on Time-Series Sentinel-2 Imagery. Sensors 2023, 23, 1779. [CrossRef] [PubMed]
- Shahhosseini, M.; Hu, G.; Khaki, S.; Archontoulis, S.V. Corn yield prediction with ensemble CNN-DNN. Frontiers in plant science 2021, 12, 709008. [CrossRef]
- Gavahi, K.; Abbaszadeh, P.; Moradkhani, H. DeepYield: A combined convolutional neural network with long short-term memory for crop yield forecasting. Expert Systems with Applications 2021, 184, 115511. [CrossRef]
- Zare, H.; Weber, T.K.; Ingwersen, J.; Nowak, W.; Gayler, S.; Streck, T. Within-season crop yield prediction by a multi-model ensemble with integrated data assimilation. Field Crops Research 2024, 308, 109293. [CrossRef]
- Gopi, P.; Karthikeyan, M. Red fox optimization with ensemble recurrent neural network for crop recommendation and yield prediction model. Multimedia Tools and Applications 2024, 83, 13159–13179. [CrossRef]
- Boppudi, S.; others. Deep ensemble model with hybrid intelligence technique for crop yield prediction. Multimedia Tools and Applications 2024, pp. 1–21.
- EY. Open Science Data Challenge, 2023. Accessed: 20 August 2023.
- e-Extension. Major Insect Pests at Each Growth Stage of the Rice Plant, 2024. Accessed: 20 January 2024.
- Joshi, A.; Pradhan, B.; Gite, S.; Chakraborty, S. Remote-sensing data and deep-learning techniques in crop mapping and yield prediction: A systematic review. Remote Sensing 2023, 15, 2014. [CrossRef]
- scikit-learn contributors. scikit-learn - R2 Score Documentation, 2023. Accessed: 3 October 2023.
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).