Submitted:
14 September 2024
Posted:
18 September 2024
You are already at the latest version
Abstract
Keywords:
Introduction
Genetic of Meningioma:
Molecular Signaling Pathways
Objectives
Material and Methods
Whole Exome Sequencing (WES)
Sample Preparation
DNA Extraction Method
Exploratory Analysis
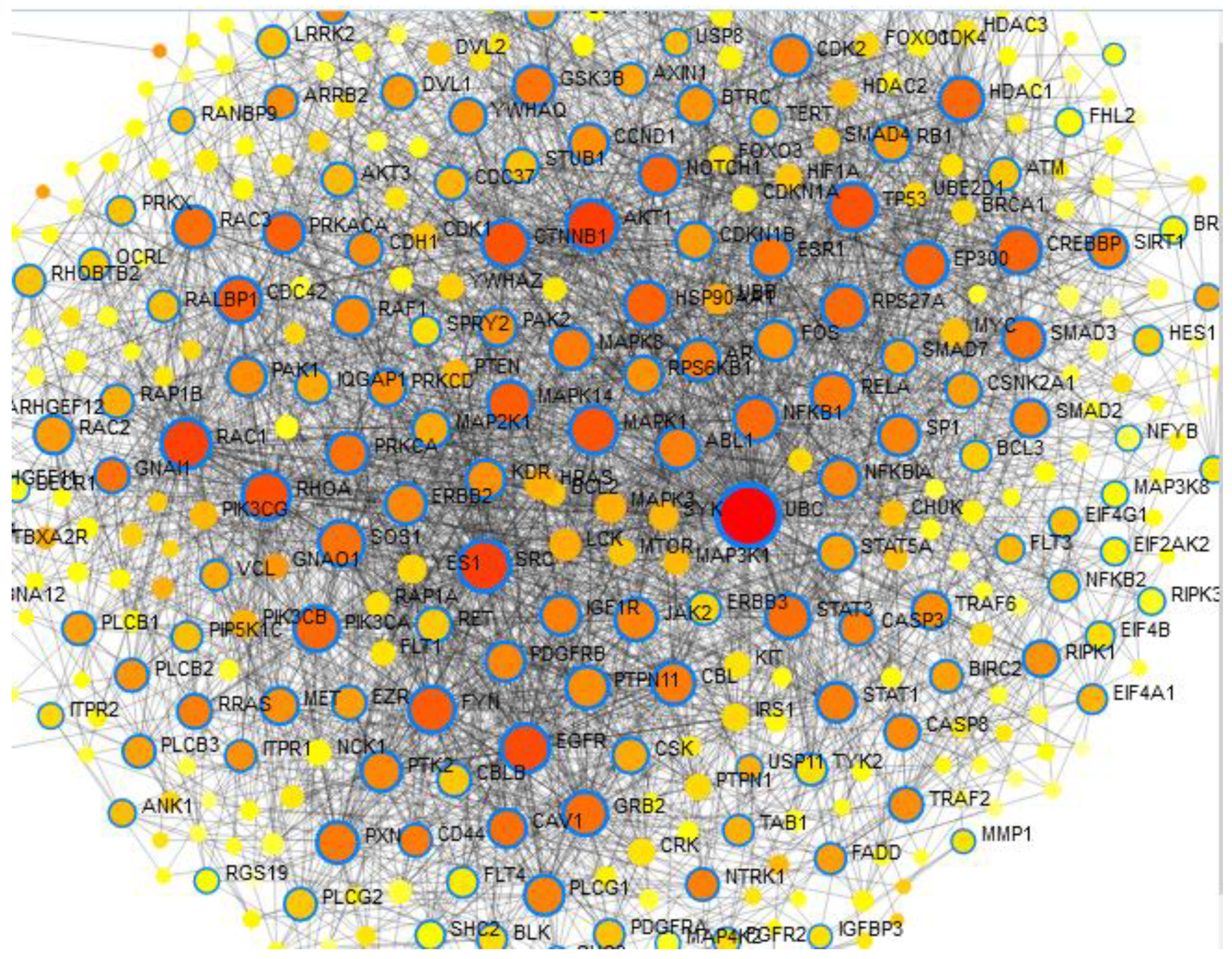
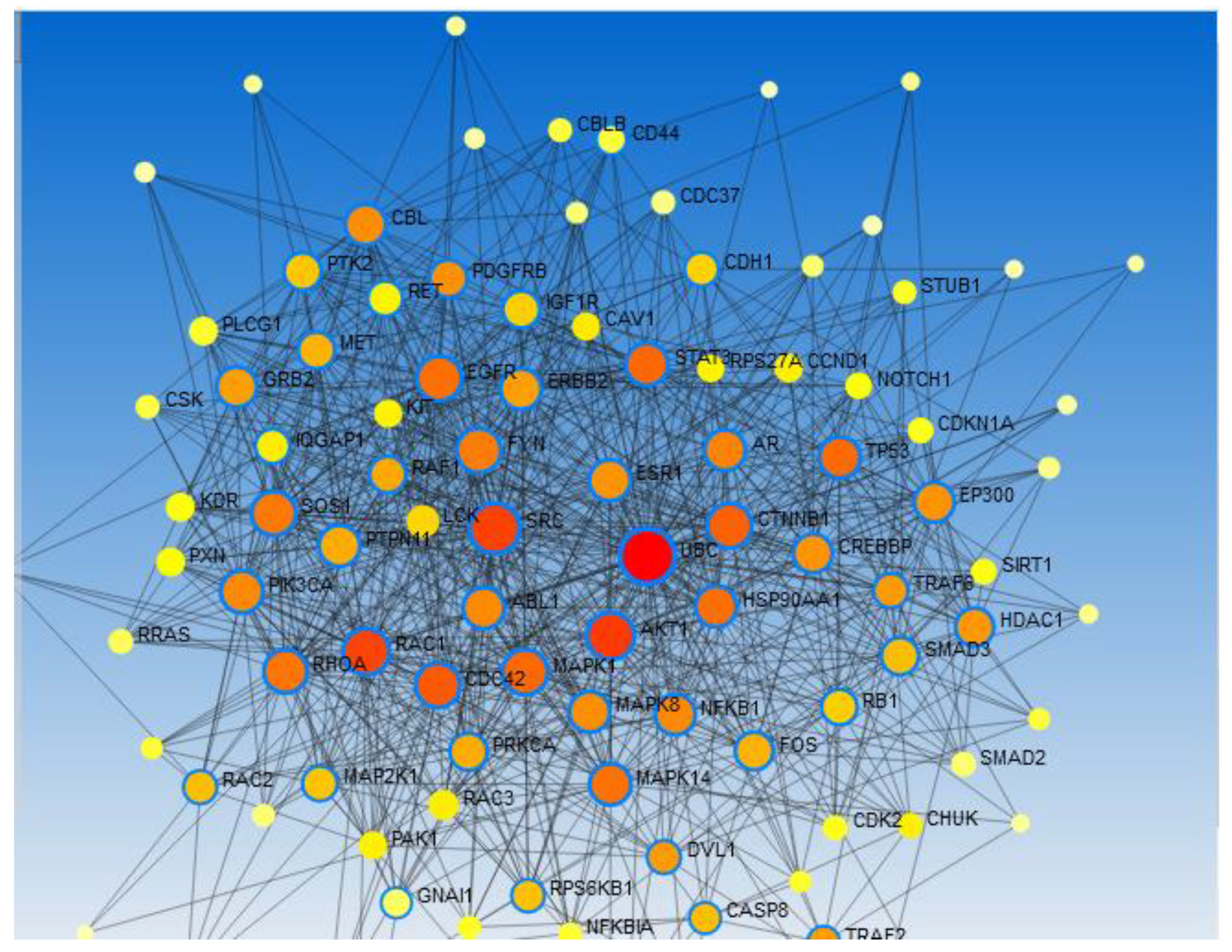
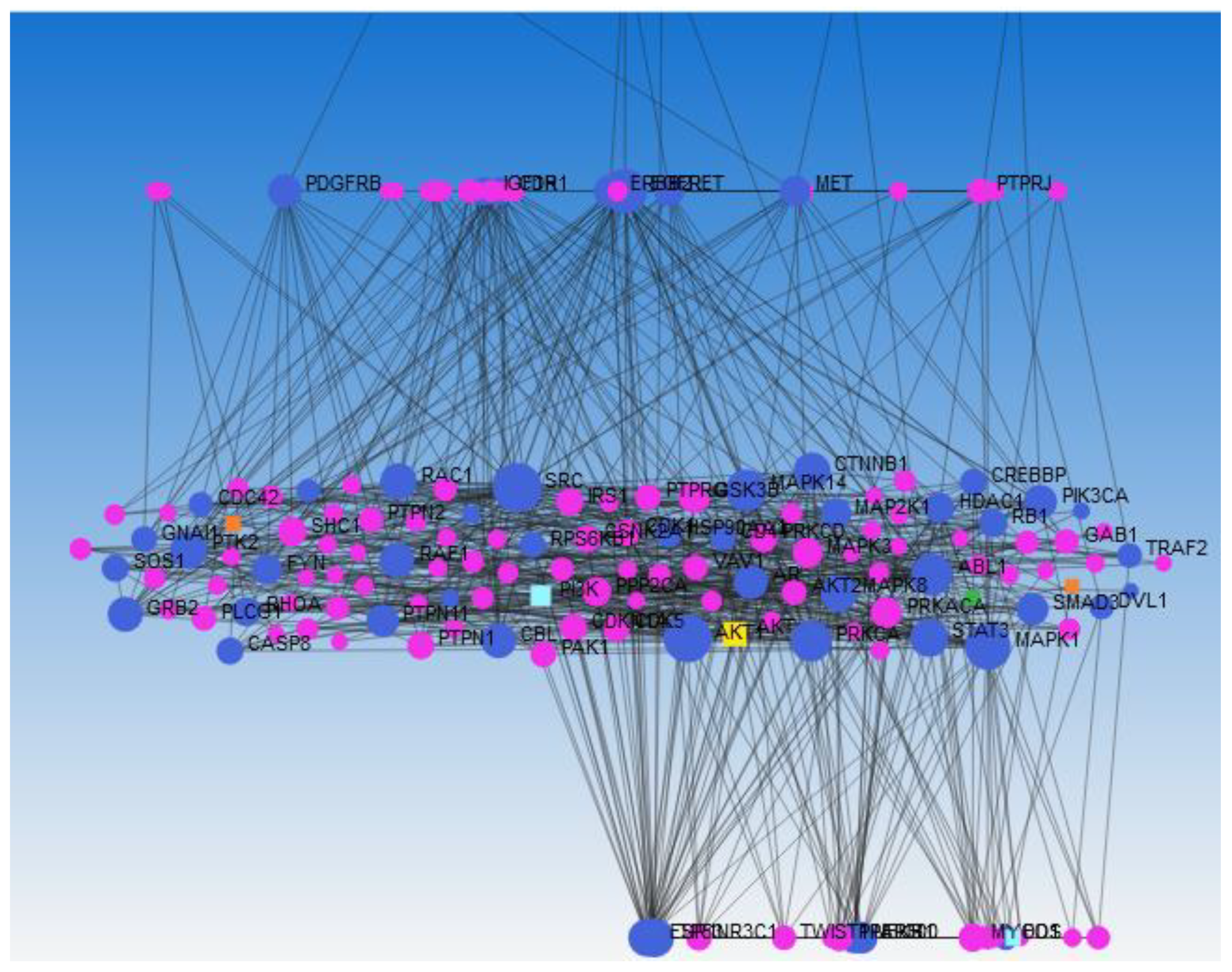
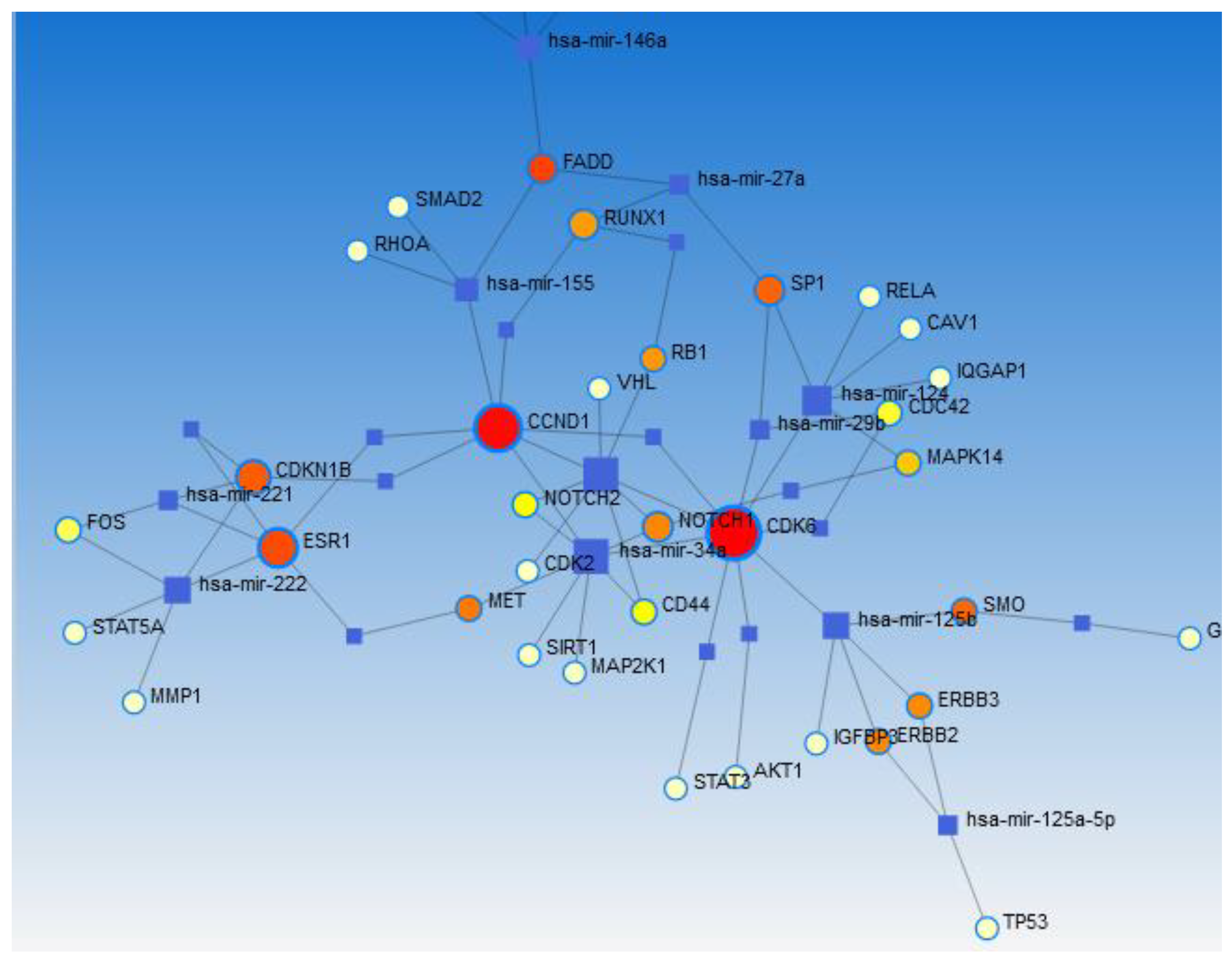
Network Analysis Using Network Analyst 2021
Pathway and GO Enrichment Analysis
| AKT1 | F | CCCCTCAGATGATCTCTCCA |
| R | TGAAGAATTTGGAGGGAAGG | |
| CD44 | F | AGGAACAGTGGTTTGGCAAC |
| R | AACTGGCTTGTATCCATTCCT |
Results
Result of Exploratory Analysis
| Class | Sample | All | Novel | Miss | Intronic | 3UTR | 5UTR | Splicing |
|---|---|---|---|---|---|---|---|---|
| SNVs | 1B | 116490 | 930 | 11323 | 74276 | 3481 | 2125 | 2560 |
| 2B | 119465 | 6032 | 11512 | 75940 | 3734 | 379 | 2386 | |
| 4A | 118987 | 5930 | 10719 | 77996 | 3689 | 369 | 2157 | |
| INDEL | Frs | |||||||
| 1B | 15103 | 4884 | 662 | 11638 | 554 | 282 | 660 | |
| 2B | 15579 | 5007 | 94 | 3743 | 173 | 76 | 278 | |
| 4A | 17114 | 5745 | 627 | 13269 | 660 | 336 | 677 |
| Sample | Total high impact SNVs/INDEL | Total moderate impact SNVs/INDEL | P value |
|---|---|---|---|
| 1B | 17 | 253 | <0.001 |
| 2B | 17 | 267 | <0.001 |
| 4A | 11 | 242 | <0.001 |
| Pathway | P value |
|---|---|
| Pathways in cancer | 1.61E-70 |
| Proteoglycans in cancer | 2.83E-40 |
| ErbB signaling pathway | 2.14E-28 |
| Focal adhesion | 6.22E-28 |
| MAPK signaling pathway | 2.03E-27 |
| Prostate cancer | 1.71E-26 |
| Pancreatic cancer | 0.02E-25 |
| Breast cancer | 1.21E-25 |
| Colorectal cancer | 2.4E-25 |
| Glioma | 4.72E-20 |
| Wnt signaling pathway | 8.79E-18 |
| MicroRNAs in cancer | 2.03E-17 |
| PI3K-Akt signaling pathway | 9.42E-17 |
| Signaling pathways regulating pluripotency of stem cells | 5.32E-11 |
| mTOR signaling pathway | 9.57E-08 |
| Hedgehog signaling pathway | 4.05E-07 |
| Phosphatidylinositol signaling system | 5.85E-06 |
| Disease Name | P value |
|---|---|
| Malignant neoplasm of thyroid | 2.060e-20 |
| Ovarian Carcinoma | 2.770e-26 |
| Transitional Meningioma | 0.00001274 |
| Rhabdomyosarcoma | 1.382e-18 |
| Neuroectodermal Tumors | 9.353e-8 |
| GlioblastomaMultiforme | 2.024e-22 |
| Endometrial Neoplasms | 1.741e-15 |
| Sphenoid Wing Meningioma | 0.00001588 |
| Clear Cell Meningioma | 0.00001588 |
| Squamous cell carcinoma of the head and neck | 1.458e-22 |
| Carcinoma of bladder | 2.235e-23 |
| Malignant Peripheral Nerve Sheath Tumor | 4.794e-14 |
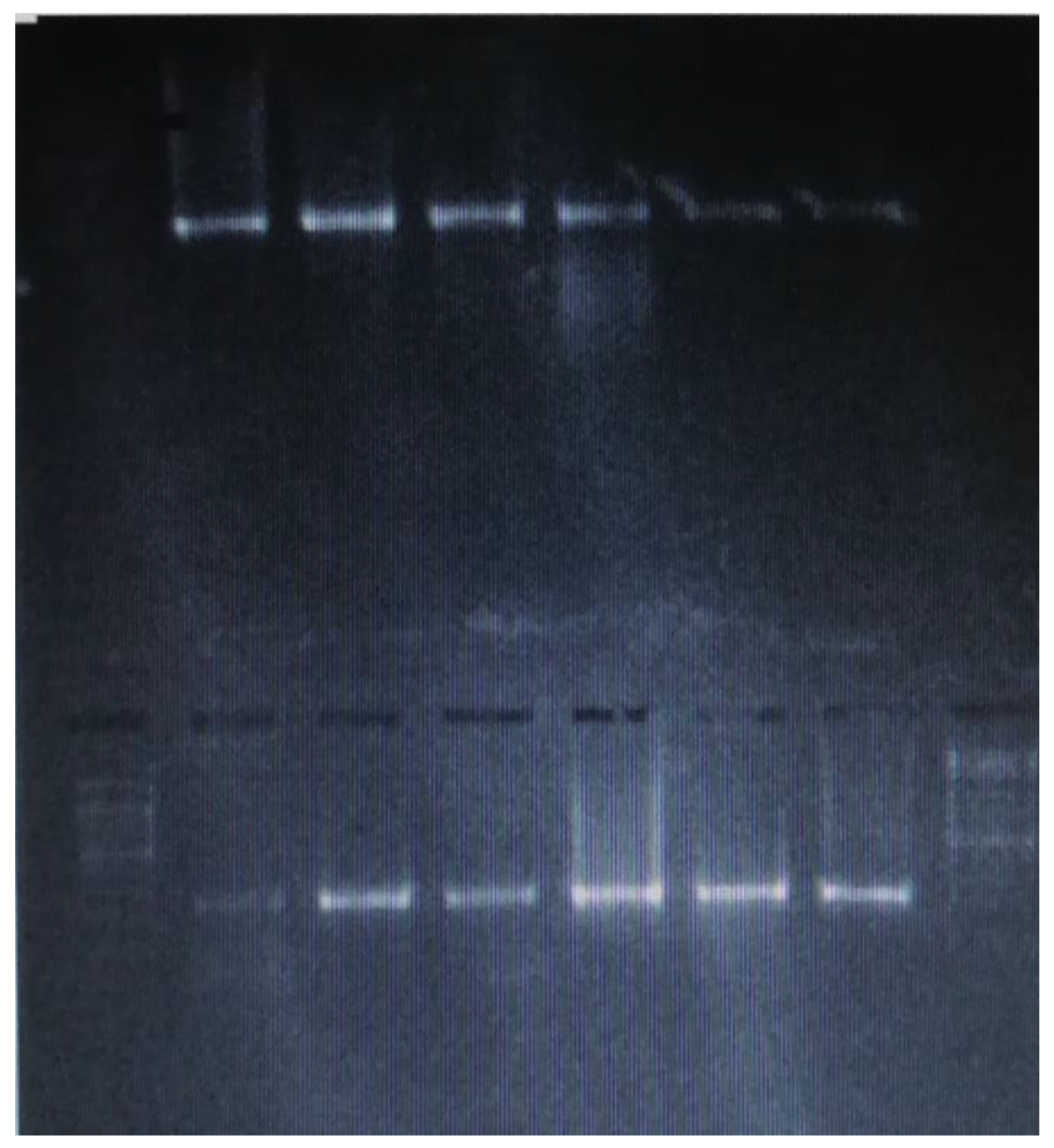
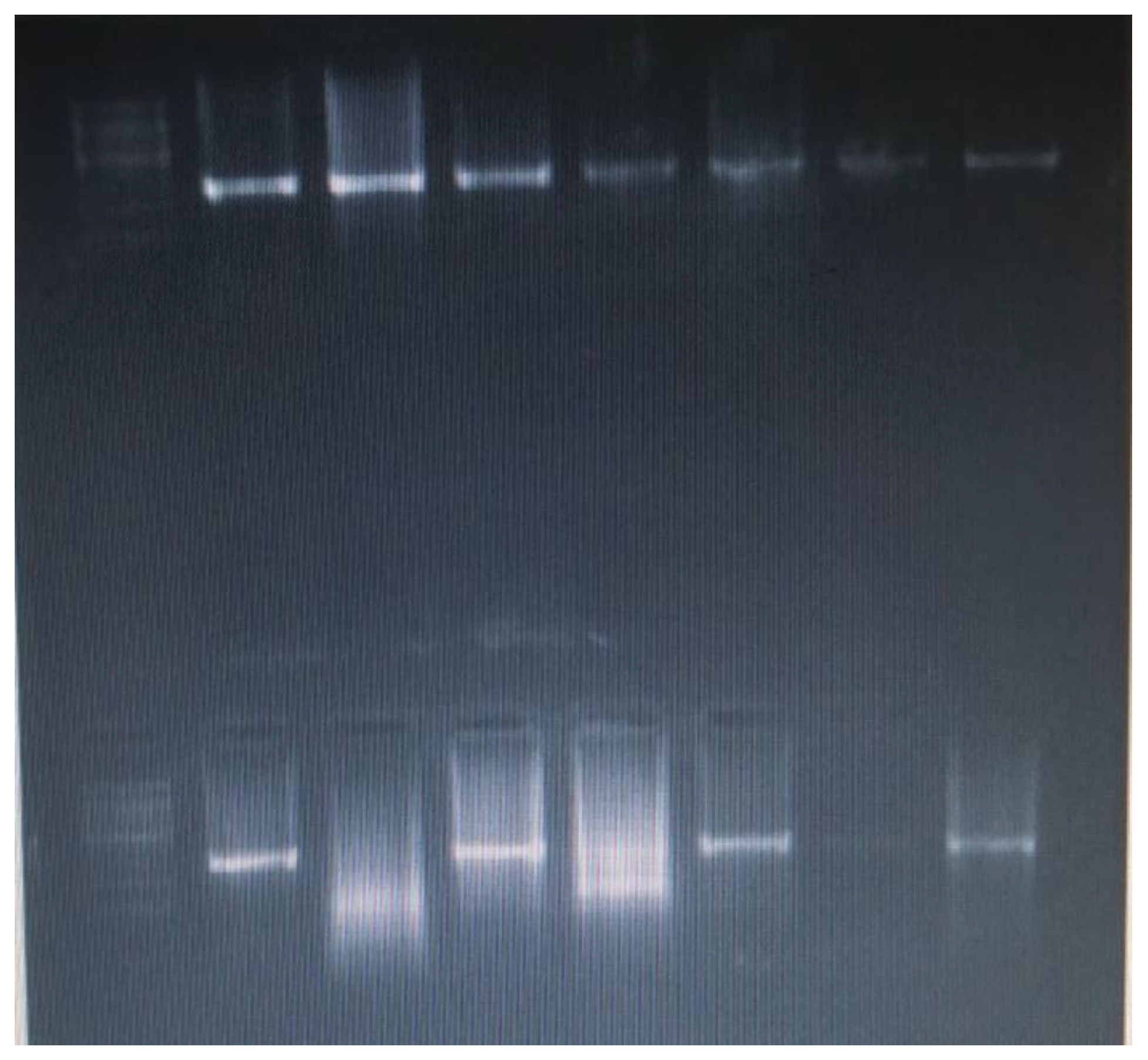
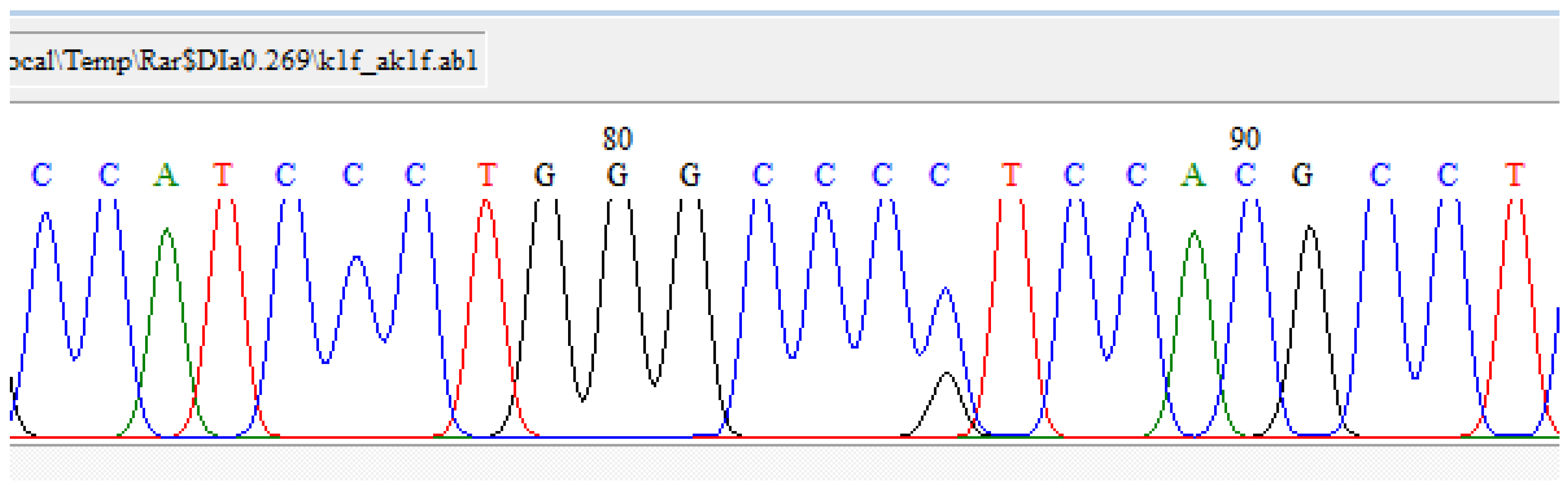
PCR and Sanger Sequencing Results of Genes of Centrality (CD44 and Akt1)
Discussion:
Conclusion
Funding
Conflict of interest
References
- Baldi I, Engelhardt J, Bonnet C, Bauchet L, Berteaud E, Grüber A, Loiseau H. Epidemiology of meningiomas. Neurochirurgie. 2018 Mar 1;64(1):5-14. [CrossRef]
- Holleczek B, Zampella D, Urbschat S, Sahm F, von Deimling A, Oertel J, Ketter R. Incidence, mortality and outcome of meningiomas: a population-based study from Germany. Cancer epidemiology. 2019 Oct 1;62:101562. [CrossRef]
- Mohamed Abdel RahmanArbab, Sawsan.A.H.Aldeaf, Lamyaa. A.Elhasan, AhmedElhasan, Cranial meningioma in Sudanese patients: clinical and pathological characteristic. International Journal of Research (IJR) 2014; Vol-1, Issue-7 ISSN 2348-684.
- van Alkemade H, de Leau M, Dieleman EM, Kardaun JW, van Os R, Vandertop WP, van Furth WR, Stalpers LJ. Impaired survival and long-term neurological problems in benign meningioma.Neuro-oncology. 2012 May 1;14(5):658-66. [CrossRef]
- Wen PY, Quant E, Drappatz J, Beroukhim R, Norden AD. Medical therapies for meningiomas.JNeurooncol.2010;99(3):365–378. [CrossRef]
- Moazzam AA, Wagle N, Zada G. Recent developments in chemotherapy for meningiomas: a review. Neurosurg Focus. 2013; 35(6):E18. [CrossRef]
- Choy W, Kim W, Nagasawa D, Stramotas S, Yew A, Gopen Q, Parsa AT, Yang I. The molecular genetics and tumor pathogenesis of meningiomas and the future directions of meningioma treatments.Neurosurgical focus. 2011 May 1;30(5):E6. [CrossRef]
- AbdElmontalab FY, Fadl EI, Abushama HM, Kreskowski K, Liehr T. Molecular cytogenetic study of the NF2 gene deletion in meningioma in sudanese patients. Balkan journal of medical genetics: BJMG. 2013 Dec;16(2):29. [CrossRef]
- Evans JJ, Jeun SS, Lee JH, Harwalkar JA, Shoshan Y, Cowell JK, Golubic M. Molecular alterations in the neurofibromatosis type 2 gene and its protein rarely occurring in meningothelialmeningiomas. Journal of neurosurgery. 2001 Jan 1;94(1):111-7.
- Schmitz U, Mueller W, Weber M, Sevenet N, Delattre O, vonDeimling A. INI1 mutations in meningiomas at a potential hotspotin exon 9. Br J Cancer 2001;84:199–201.
- Lamszus K. Meningioma pathology, genetics, and biology. Journal of Neuropathology & Experimental Neurology. 2004 Apr 1;63(4):275-86.
- Malmer BS, Feychting M, Lönn S, Lindström S, Grönberg H, Ahlbom A, Schwartzbaum J, Auvinen A, Collatz-Christensen H, Johansen C, Kiuru A. Genetic variation in p53 and ATM haplotypes and risk of glioma and meningioma. Journal of neuro-oncology. 2007 May;82(3):229-37. [CrossRef]
- Gassoum A, Arbab MA, Aldeaf SA, Elhassan LA, Elhassan AM. P53 Gene Allele Frequency in Linguistic Affiliation of Different Sudanese Tribes.International Journal of Research.2014;1:880-5.
- Hilton DA, Shivane A, Kirk L, Bassiri K, Enki DG, Hanemann CO. Activation of multiple growth factor signalling pathways is frequent in meningiomas. Neuropathology. 2016; Jun;36(3):250-61. [CrossRef]
- Mu J, Hui T, Shao B, Li L, Du Z, Lu L, Ye L, Li S, Li Q, Xiao Q, Qiu Z. Dickkopf-related protein 2 induces G0/G1 arrest and apoptosis through suppressing Wnt/β-catenin signaling and is frequently methylated in breast cancer. Oncotarget. 2017 Jun 13;8(24):39443.
- Ali MM. Aberrant expression of β-catenin in invasive ductal breast carcinomas.Journal of the Egyptian National Cancer Institute. 2009 Jun 1;21(2):185-95.
- Mao J, Fan S, Ma W, Fan P, Wang B, Zhang J, Wang H, Tang B, Zhang Q, Yu X, Wang L. Roles of Wnt/β-catenin signaling in the gastric cancer stem cells proliferation and salinomycin treatment. Cell death & disease. 2014 Jan;5(1):e1039-. [CrossRef]
- Wu K, Ma L, Zhu J. miR-483-5p promotes growth, invasion and self-renewal of gastric cancer stem cells by Wnt/β-catenin signaling. Molecular medicine reports. 2016 Oct 1;14(4):3421-8.
- Nagarajan N, Bertrand D, Hillmer AM, Zang ZJ, Yao F, Jacques PÉ, et al. Whole-genome reconstruction and mutational signatures in gastric cancer. Genome Biol. 2012; 13: R115. [CrossRef]
- Davila-Velderrain J, Servin-Marquez A, Alvarez-Buylla ER. Molecular evolution constraints in the floral organ specification gene regulatory network module across 18 angiosperm genomes. Molecular biology and evolution. 2014 Mar 1;31(3):560-73. [CrossRef]
- Mathivanan S, Periaswamy B, Gandhi TK, Kandasamy K, Suresh S, Mohmood R, Ramachandra YL, Pandey A. An evaluation of human protein-protein interaction data in the public domain.BMC bioinformatics. 2006 Dec;7(5):1-4. [CrossRef]
- Carpten JD, Faber AL, Horn C, Donoho GP, Briggs SL, Robbins CM, Hostetter G, Boguslawski S, Moses TY, Savage S, Uhlik M. A transforming mutation in the pleckstrin homology domain of AKT1 in cancer.Nature. 2007 Jul;448(7152):439-44. [CrossRef]
- Bianchi M, Giacomini E, Crinelli R, Radici L, Carloni E, Magnani M. Dynamic transcription of ubiquitin genes under basal and stressful conditions and new insights into the multiple UBC transcript variants. Gene. 2015 Nov 15;573(1):100-9. [CrossRef]
- Clague MJ, Heride C, Urbé S. The demographics of the ubiquitin system.Trends in cell biology. 2015 Jul 1;25(7):417-26. [CrossRef]
- Husnjak, K. &Dikic, I. Ubiquitin-binding proteins: decoders of ubiquitin-mediated cellular functions. Annu. Rev. Biochem.2012; 81, 291–322.
- Jewell JL, Russell RC, Guan KL. Amino acidsignaling upstream of mTOR.Nature reviews Molecular cell biology. 2013 Mar;14(3):133-9.
- Saxton RA, Sabatini DM. mTOR signaling in growth, metabolism, and disease. Cell. 2017 Mar 9;168(6):960-76.
- Buckley SM, Aranda-Orgilles B, Strikoudis A, Apostolou E, Loizou E, Moran-Crusio K, Farnsworth CL, Koller AA, Dasgupta R, Silva JC, Stadtfeld M. Regulation of pluripotency and cellular reprogramming by the ubiquitin-proteasome system. Cell stem cell. 2012 Dec 7;11(6):783-98. [CrossRef]
- Buckley SM, Aranda-Orgilles B, Strikoudis A, Apostolou E, Loizou E, Moran-Crusio K, Farnsworth CL, Koller AA, Dasgupta R, Silva JC, Stadtfeld M. Regulation of pluripotency and cellular reprogramming by the ubiquitin-proteasome system. Cell stem cell. 2012 Dec 7;11(6):783-98. [CrossRef]
- Malta TM, Sokolov A, Gentles AJ, Burzykowski T, Poisson L, Weinstein JN, Kamińska B, Huelsken J, Omberg L, Gevaert O, Colaprico A. Machine learning identifies stemness features associated with oncogenic dedifferentiation. Cell. 2018 Apr 5;173(2):338-54. [CrossRef]
- Shivapathasundram, G.;Wickremesekera, A.C.; Brasch, H.D.; Marsh, R.; Tan, S.T.; Itinteang, T. Expression of Embryonic StemCell Markers on the Microvessels of WHO Grade I Meningioma. Front. Surg. 2018; 5, 65.
- Li M, Dai FR, Du XP, Yang QD, Zhang X, Chen Y. Infusion of BDNF into the nucleus accumbens of aged rats improves cognition and structural synaptic plasticity through PI3K-ILK-Akt signaling. Behavioural brain research. 2012 May 16;231(1):146-53. [CrossRef]
- Balu DT, Carlson GC, Talbot K, Kazi H, Hill-Smith TE, Easton RM, Birnbaum MJ, Lucki I. Akt1 deficiency in schizophrenia and impairment of hippocampal plasticity and function. Hippocampus. 2012 Feb;22(2):230-40. [CrossRef]
- .Brastianos PK, Horowitz PM, Santagata S, Jones RT, McKenna A, Getz G, Ligon KL, Palescandolo E, Van Hummelen P, Ducar MD, Raza A. Genomic sequencing of meningiomas identifies oncogenic SMO and AKT1 mutations. Nature genetics. 2013 Mar;45(3):285-9. [CrossRef]
- Yesilöz Ü, Kirches E, Hartmann C, Scholz J, Kropf S, Sahm F, Nakamura M, Mawrin C. Frequent AKT1 E17K mutations in skull base meningiomas are associated with mTOR and ERK1/2 activation and reduced time to tumor recurrence. Neuro-oncology. 2017 Aug 1;19(8):1088-96.
- Weller M, Roth P, Sahm F, Burghardt I, Schuknecht B, Rushing EJ, Regli L, Lindemann JP, Von Deimling A. Durable control of metastatic AKT1-mutant WHO grade 1 meningothelialmeningioma by the AKT inhibitor, AZD5363. Journal of the National Cancer Institute. 2017 Mar 1;109(3):djw320.
- Boehm M and Slack FJ: microRNA control of lifespan and metabolism. Cell Cycle. 5:837–840. 2006.
- Freedman JA, Wang Y, Li X, Liu H, Moorman PG, George DJ, Lee NH, Hyslop T, Wei Q, Patierno SR. Single-nucleotide polymorphisms of stemness genes predicted to regulate RNA splicing, microRNA and oncogenic signaling are associated with prostate cancer survival. Carcinogenesis. 2018 Jul 3;39(7):879-88. [CrossRef]
- Iozzo RV, Sanderson RD. Proteoglycans in cancer biology, tumour microenvironment and angiogenesis.Journal of cellular and molecular medicine. 2011 May;15(5):1013-31.
- Venselaar, H., teBeek, T.A.H., Kuipers, R.K.P., Hekkelman, M.L. and Vriend, G. Protein Structure Analysis of Mutations Causing Inheritable Diseases. An e-Science Approach with Life Scientist Friendly Interfaces. BMC Bioinformatics.2010 11, 548. [CrossRef]


| Sample | SIFT/Total deleterious effect |
PolyPhen-2/total D | PolyPhen-2/total P/D | PolyPhen-2/total/B |
|---|---|---|---|---|
| 1B/253SNVs | 54 | 16 | 10 | 21 |
| 2B/267SNVs | 63 | 18 | 18 | 20 |
| 4A/242SNVs | 48 | 30 | 16 | 25 |
| NO | Histopathology variant | WHO Grades | Recurrence | Mutation Akt1 | Mutation CD44 |
|---|---|---|---|---|---|
| 1 | Meningiothelial | 1 | NO | Del A | A>G (rs9666607) |
| 2 | Clear cell | 11 | NO | A>G (rs9666607) D419Tfs*61 InsC |
|
| 3 | Meningiothelial | 1 | NO | Del A | A>G (rs9666607) |
| 4 | Meningiothelial | 1 | NO | Del A/ InsC |
A>G (rs9666607) |
| 5 | Fibrous | 1 | NO | G>A (rs17846829) | A>G (rs9666607) D419Tfs*61 |
| 6 | Clear cell | 11 | Yes | Del A | A>G (rs9666607) |
| 7 | Fibrous | 1 | Yes | Del A G>A(rs17846829) | A>G (rs9666607) D419Tfs*61 InsC |
| 8 | Fibrous | 1 | Yes | Del A | A>G (rs9666607) D419Tfs*61 |
| 9 | Fibrous | 1 | Yes | Del A | A>G (rs9666607) |
| 10 | Atypical | 11 | Yes | Del A G>A (rs17846829) |
A>G (rs9666607) |
| 11 | Fibrous | 1 | Yes | Del A InsC |
A>G (rs9666607) InsC |
| 12 | Fibrous | 1 | NO | A>G (rs9666607) D419Tfs*61 |
|
| 13 | Fibrous | 1 | Yes | Del A | A>G (rs9666607) |
| 14 | Fibrous | 1 | Yes | Del A | A>G (rs9666607) D419Tfs*61 |
| 15 | Fibrous | 1 | NO | Del A | A>G (rs9666607) InsC |
| 16 | Fibrous | 1 | NO | A>G (rs9666607) D419Tfs*61 |
|
| 17 | Clear cell | 11 | Yes | Del A | A>G (rs9666607) D419Tfs*61 |
| 18 | Fibrous | 1 | Yes | Del A | A>G (rs9666607) |
| 19 | Atypical | 11 | Yes | Del A G>A(rs17846829) | A>G (rs9666607) |
| 20 | Meningiothelial | 1 | Yes | Del A | A>G (rs9666607) D419Tfs*61 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




