Submitted:
18 September 2024
Posted:
20 September 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
1.1. Information Extraction
1.2. Symptomatology
1.3. Objective
2. Materials and Methods
2.1. Data Sources
2.2. Prevalence Analysis
2.3. Dataset Curation for Model Fine-Tuning and Validation
2.4. Model Validation
3. Results
3.1. Prevalence Analysis
3.2. Performance Evaluation
4. Discussion
4.1. Prevalence Analysis
4.2. Error Analysis
4.3. Performance Evaluation
4.4. Limitations
4.5. Future Work
5. Conclusions
Acknowledgments
Appendix A
Prevalence Analysis
| UMN PASC | UMN COVID | N3C COVID |
|---|---|---|
| Pain (0.131) | Fever (0.119) | Pain (0.090) |
| Shortness of Breath (0.079) | Shortness of Breath (0.118) | Fever (0.059) |
| Cough (0.077) | Cough (0.100) | Shortness of Breath (0.084) |
| Myalgia (0.053) | Myalgia (0.053) | Nausea or Vomiting (0.064) |
| Fever (0.049) | Nausea or Vomiting (0.058) | Cough (0.053) |
| Rash (0.029) | Rash (0.009) | Diarrhea (0.040) |
| Anxiety (0.025) | Fatigue (0.041) | Abdominal Pain (0.039) |
| Nausea or Vomiting (0.054) | Abdominal Pain (0.014) | Respiratory Depression (0.037) |
| Fatigue (.060) | Diarrhea (0.031) | Fatigue (0.034) |
| Headaches (0.052) | Headaches (0.023) | Mental Status Change (0.031) |
Appendix B
BioMedICUS/Microservice Text Analysis Platform (MTAP) Concept Extraction
Appendix C
Lexica
Appendix D
LLM Model Development
| Question | Answer |
| “What are the positive and negative symptoms of the patient given the following clinical text:\n Clinical Text: note_text\n” | “Positive symptoms are [symptomA, symptomB]. Negative symptoms are [symptomP, symptomQ].” |
References
- Pakhomov S, Finley GP, McEwan R, et al. Corpus domain effects on distributional semantic modeling of medical terms. Bioinformatics. 2016;32 23:3635–44. [CrossRef]
- Silverman GM, Sahoo HS, Ingraham NE, et al. NLP Methods for Extraction of Symptoms from Unstructured Data for Use in Prognostic COVID-19 Analytic Models. Journal of Artificial Intelligence Research. 2021;72:429–74. [CrossRef]
- National Center for Biotechnology Information. UMLS® Reference Manual. National Library of Medicine (US) 2009. https://www.ncbi.nlm.nih.gov/books/NBK9676/ (accessed 5 October 2021).
- Bodenreider O. The UMLS Semantic Network. The UMLS Semantic Network. 2020. https://semanticnetwork.nlm.nih.gov (accessed 3 February 2020).
- He Z, Perl Y, Elhanan G, et al. Auditing the assignments of top-level semantic types in the UMLS semantic network to UMLS concepts. 2017 IEEE International Conference on Bioinformatics and Biomedicine (BIBM). 2017;1262–9.
- Wang L, Foer D, MacPhaul E, et al. PASCLex: A comprehensive post-acute sequelae of COVID-19 (PASC) symptom lexicon derived from electronic health record clinical notes. J Biomed Inform. 2022;125:103951. [CrossRef]
- Knoll BC, Lindemann EA, Albert AL, et al. Recurrent Deep Network Models for Clinical NLP Tasks: Use Case with Sentence Boundary Disambiguation. Stud Health Technol Inform. 2019;264:198–202.
- Knoll BC, McEwan R, Finzel R, et al. MTAP - A Distributed Framework for NLP Pipelines. 2022 IEEE 10th International Conference on Healthcare Informatics (ICHI). Rochester, MN, USA: IEEE 2022:537–8. [CrossRef]
- Knoll BC, Gunderson M, Rajamani G, et al. Advanced Care Planning Content Encoding with Natural Language Processing. Stud Health Technol Inform. 2024;310:609–13.
- Xu D, Chen W, Peng W, et al. Large Language Models for Generative Information Extraction: A Survey. Published Online First: 2023. [CrossRef]
- Yang Z, Dai Z, Yang Y, et al. XLNet: Generalized Autoregressive Pretraining for Language Understanding. Published Online First: 2019. [CrossRef]
- Beltagy I, Peters ME, Cohan A. Longformer: The Long-Document Transformer. Published Online First: 2020. [CrossRef]
- Hoogendoorn M, Szolovits P, Moons L, et al. Utilizing uncoded consultation notes from electronic medical records for predictive modeling of colorectal cancer. Artificial intelligence in medicine. 2016;69:53–61. [CrossRef]
- Stephens KA, Au MA, Yetisgen M, et al. Leveraging UMLS-driven NLP to enhance identification of influenza predictors derived from electronic medical record data. Bioinformatics 2020. [CrossRef]
- Skube SJ, Hu Z, Simon GJ, et al. Accelerating Surgical Site Infection Abstraction with a Semi-automated Machine-learning Approach. Ann Surg. Published Online First: 14 October 2020. [CrossRef]
- Sahoo HS, Silverman GM, Ingraham NE, et al. A fast, resource efficient, and reliable rule-based system for COVID-19 symptom identification. JAMIA Open. 2021;4:ooab070. [CrossRef]
- Silverman GM, Rajamani G, Ingraham NE, et al. A Symptom-Based Natural Language Processing Surveillance Pipeline for Post-COVID-19 Patients. In: Bichel-Findlay J, Otero P, Scott P, et al., eds. Studies in Health Technology and Informatics. IOS Press 2024. [CrossRef]
- Devlin J, Chang M-W, Lee K, et al. BERT: Pre-training of Deep Bidirectional Transformers for Language Understanding. NAACL-HLT. 2019.
- Jahan I, Laskar MTR, Peng C, et al. A comprehensive evaluation of large Language models on benchmark biomedical text processing tasks. Computers in Biology and Medicine. 2024;171:108189. [CrossRef]
- Fleiss JL. Measuring nominal scale agreement among many raters. Psychological Bulletin. 1971;76:378–82. [CrossRef]
- He Y, Yu H, Ong E, et al. CIDO, a community-based ontology for coronavirus disease knowledge and data integration, sharing, and analysis. Sci Data. 2020;7:181. [CrossRef]
- N3C. OHNLP/N3C-NLP-Documentation Wiki. 2022. https://github.com/OHNLP/N3C-NLP-Documentation/wiki (accessed 30 April 2024).
- Nagamine T, Gillette B, Kahoun J, et al. Data-driven identification of heart failure disease states and progression pathways using electronic health records. Sci Rep. 2022;12:17871. [CrossRef]
- Pojanapunya P, Watson Todd R. Log-likelihood and odds ratio: Keyness statistics for different purposes of keyword analysis. Corpus Linguistics and Linguistic Theory. 2018;14:133–67. [CrossRef]
- Patra BG, Lepow LA, Kumar PKRJ, et al. Extracting Social Support and Social Isolation Information from Clinical Psychiatry Notes: Comparing a Rule-based NLP System and a Large Language Model. 2024. [CrossRef]
- OpenAI, Achiam J, Adler S, et al. GPT-4 Technical Report. 2023. [CrossRef]
- Lewis M, Liu Y, Goyal N, et al. BART: Denoising Sequence-to-Sequence Pre-training for Natural Language Generation, Translation, and Comprehension. 2019. [CrossRef]
- Chen Q, Du J, Hu Y, et al. Large language models in biomedical natural language processing: benchmarks, baselines, and recommendations. 2023. [CrossRef]
- University of Minnesota NLP/IE Program. BioMedICUS Section Header Detector. GitHub. 2024. https://github.com/nlpie/biomedicus/blob/main/java/src/main/java/edu/umn/biomedicus/sections/RuleBasedSectionHeaderDetector.java (accessed 23 April 2024).
- University of Minnesota NLP/IE Program. BioMedICUS UMLS Concept Detection Algorithm. GitHub. 2024. https://github.com/nlpie/biomedicus/tree/main/java/src/main/java/edu/umn/biomedicus/concepts (accessed 23 April 2024).
- Chapman WW, Bridewell W, Hanbury P, et al. A Simple Algorithm for Identifying Negated Findings and Diseases in Discharge Summaries. Journal of Biomedical Informatics. 2001;34:301–10. [CrossRef]
- CDC. Symptoms of Coronavirus. Centers for Disease Control and Prevention. 2020. https://www.cdc.gov/coronavirus/2019-ncov/symptoms-testing/symptoms.html (accessed 12 December 2020).
- Davis HE, Assaf GS, McCorkell L, et al. Characterizing long COVID in an international cohort: 7 months of symptoms and their impact. EClinicalMedicine. 2021;38:101019. [CrossRef]
- Touvron H, Martin L, Stone K, et al. Llama 2: Open Foundation and Fine-Tuned Chat Models. Published Online First: 2023. [CrossRef]
- Paszke A, Gross S, Massa F, et al. PyTorch: An Imperative Style, High-Performance Deep Learning Library. Published Online First: 2019. [CrossRef]
- Dettmers T, Pagnoni A, Holtzman A, et al. QLoRA: Efficient Finetuning of Quantized LLMs. Published Online First: 2023. [CrossRef]
- Loshchilov I, Hutter F. Decoupled Weight Decay Regularization. Published Online First: 2017. [CrossRef]
| Corpus | Source | Note Count |
|---|---|---|
| UMN PASC | Outpatient (OP) Emergency Department (ED) |
387 84 |
| UMN COVID | Emergency Department (ED) | 46 |
| N3C COVID | Not Available (NA) | 148 |
| Corpus | Number of Patients | Median age in years, IQR (Q1, Q3) | Male % | Racial Distribution & Ethnicity % |
|---|---|---|---|---|
| UMN PASC | 476 | 53.10, (38.00, 67.90) | 58.00 % | 5.00% Asian 11.90% Black 3.36% Hispanic 15.74% Others* 64.00% White |
| UMN COVID | 46 | 54.68, (39.00, 66.64) | 54.00 % | 9.09% Asian 27.27% Black 4.54% Hispanic 7.10% Others* 52.00% White |
| N3C COVID** | 148 | Not Available (NA) | NA | NA |
| Corpora | Number of Patients | Median age in years, IQR (Q1, Q3) | Male % | Racial Distribution & Ethnicity % |
|---|---|---|---|---|
| Training | 351 | 53.89, (38.00, 67.00) | 55.00 % | 7.00% Asian 19.50% Black 3.90% Hispanic 11.60% Others* 58.00% White |
| Validation | 20 | 53.50, (38.00, 67.00) |
56.00 % | 6.00% Asian 18.00% Black 4.00% Hispanic 12.00% Others* 60.00% White |
| Hold-out Test** | 299 | 53.00, (38.00, 67.00) |
56.00 % | 6.5% Asian 20% Black 4.00% Hispanic 11.5% Others* 58% White |
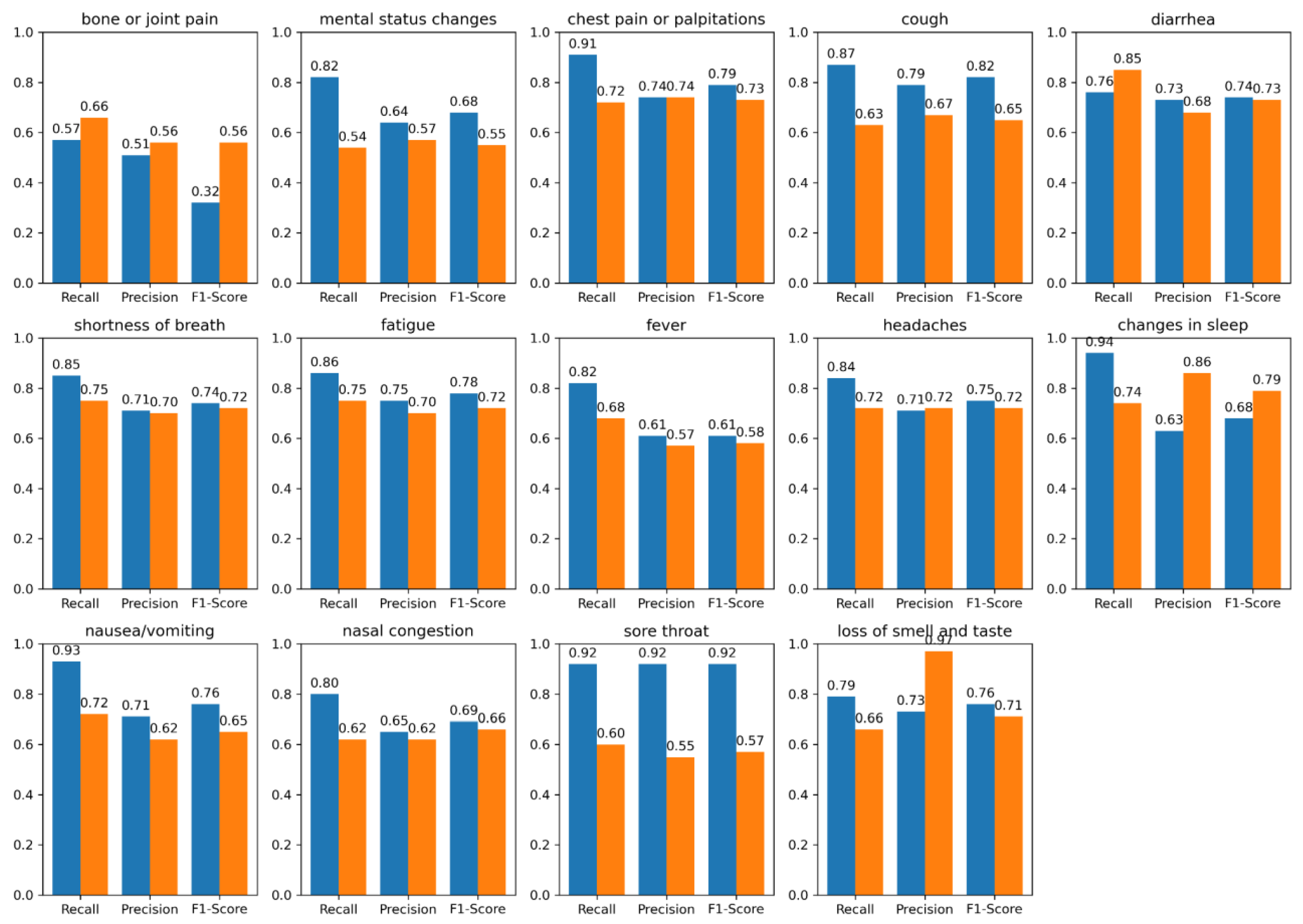
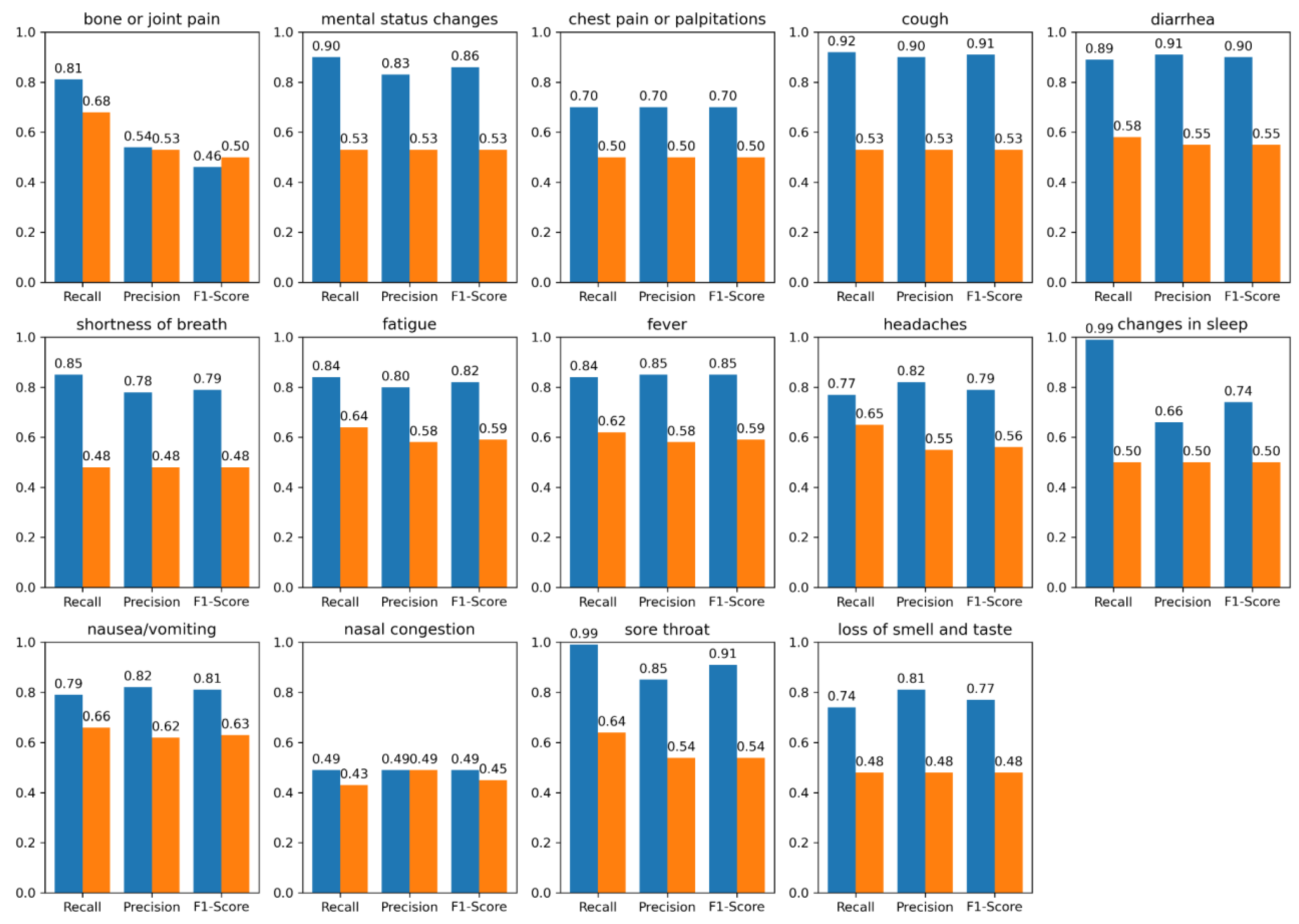
| Symptom | Model | UMN PASC (+) r*, p*, f1* | UMN PASC (-) r, p, f1 | N3C COVID (+) r, p, f1 | N3C COVID (-) r, p, f1 |
|---|---|---|---|---|---|
| bone or joint pain | BioMedICUS LLM |
0.57, 0.51, 0.32 0.66, 0.56, 0.56 |
0.64, 0.58, 0.46 0.53, 0.54, 0.53 |
0.81, 0.54, 0.46 0.68,0.53,0.50 |
0.56, 0.50, 0.47 0.45, 0.48, 0.47 |
| mental status changes | BioMedICUS LLM |
0.82, 0.64, 0.68 0.54, 0.57, 0.55 |
0.86, 0.62, 0.66 0.48, 0.48, 0.48 |
0.90, 0.83, 0.86 0.53, 0.53,0.53 |
0.65, 0.59, 0.61 0.48, 0.48, 0.48 |
| chest pain or palpitations | BioMedICUS LLM |
0.91, 0.74, 0.79 0.72, 0.74, 0.73 |
0.95, 0.91, 0.92 0.62, 0.77, 0.64 |
0.70, 0.70, 0.70 0.50, 0.50, 0.50 |
0.83, 0.71, 0.76 0.46, 0.47, 0.47 |
| cough | BioMedICUS LLM |
0.87, 0.79, 0.82 0.63, 0.67, 0.65 |
0.81, 0.73, 0.74 0.66, 0.69, 0.67 |
0.92, 0.90, 0.91 0.53, 0.53, 0.53 |
0.65, 0.68, 0.66 0.44, 0.47, 0.45 |
| diarrhea | BioMedICUS LLM |
0.76 ,0.73, 0.74 0.85, 0.68, 0.73 |
0.89, 0.86, 0.87 0.63, 0.64, 0.64 |
0.89, 0.91, 0.90 0.58, 0.55, 0.55 |
0.87, 0.83, 0.85 0.59, 0.55, 0.56 |
| shortness of breath | BioMedICUS LLM |
0.85, 0.71, 0.74 0.75, 0.70, 0.72 |
0.86, 0.82, 0.83 0.61, 0.65, 0.62 |
0.85, 0.78, 0.79 0.48, 0.48, 0.48 |
0.72, 0.69, 0.71 0.50, 0.50, 0.50 |
| fatigue | BioMedICUS LLM |
0.86, 0.75, 0.78 0.75, 0.70, 0.72 |
0.81, 0.63, 0.67 0.67, 0.59, 0.61 |
0.84, 0.80, 0.82 0.64, 0.58, 0.59 |
0.98, 0.62, 0.69 0.47, 0.49, 0.48 |
| fever | BioMedICUS LLM |
0.82, 0.61, 0.61 0.68, 0.57, 0.58 |
0.85, 0.86, 0.86 0.61, 0.62, 0.61 |
0.84, 0.85, 0.85 0.62, 0.58, 0.59 |
0.88, 0.91, 0.89 0.50, 0.50, 0.49 |
| headaches | BioMedICUS LLM |
0.84, 0.71, 0.75 0.72, 0.72, 0.72 |
0.91, 0.81, 0.85 0.66, 0.66, 0.66 |
0.77, 0.82, 0.79 0.65, 0.55, 0.56 |
0.84, 0.80, 0.82 0.45, 0.47, 0.46 |
| changes in sleep | BioMedICUS LLM |
0.94, 0.63, 0.68 0.74, 0.86, 0.79 |
0.48, 0.49, 0.48 0.50, 0.50, 0.50 |
0.99, 0.66, 0.74 0.50, 0.50, 0.50 |
0.75, 0.99, 0.83 0.50, 0.49, 0.49 |
| nausea/vomiting | BioMedICUS LLM |
0.93, 0.71, 0.76 0.72, 0.62, 0.65 |
0.91, 0.89, 0.90 0.67, 0.68, 0.68 |
0.79, 0.82, 0.81 0.66, 0.62, 0.63 |
0.78, 0.77, 0.77 0.56, 0.58, 0.57 |
| nasal congestion and obstruction | BioMedICUS LLM |
0.80, 0.65, 0.69 0.62, 0.62, 0.66 |
0.86, 0.88, 0.87 0.63, 0.68, 0.65 |
0.49, 0.49, 0.49 0.43, 0.49, 0.45 |
0.99, 0.75, 0.83 0.47, 0.49, 0.48 |
| sore throat | BioMedICUS LLM |
0.92, 0.92, 0.92 0.60, 0.55, 0.57 |
0.90, 0.84, 0.87 0.60, 0.63, 0.61 |
0.99, 0.85, 0.91 0.64, 0.54, 0.54 |
0.75, 0.99, 0.82 0.47, 0.48, 0.47 |
| loss of smell and taste | BioMedICUS LLM |
0.79, 0.73, 0.76 0.66, 0.97, 0.71 |
0.65, 0.65, 0.65 0.66, 0.74, 0.70 |
0.74, 0.81, 0.77 0.48, 0.48, 0.48 |
0.65, 0.65, 0.65 0.49, 0.48, 0.49 |
| BioMedICUS LLM |
0.81, 0.69, 0.70 0.68, 0.68, 0.68 |
0.79, 0.75, 0.74 0.61, 0.63, 0.62 |
0.82, 0.76, 0.77 0.56, 0.53, 0.54 |
0.77, 0.74, 0.74 0.48, 0.50, 0.49 |
| Model | Category | r*, p*, f1* (+) | r, p , f1 (-) |
|---|---|---|---|
| LLM | Male Female |
0.69, 0.71, 0.70 0.67, 0.66, 0.66 |
0.62, 0.65, 0.63 0.61, 0.62, 0.63 |
| Asian Black Other** White |
0.64, 0.65, 0.64 0.67, 0.66, 0.66 0.70, 0.70, 0.70 0.70, 0.72, 0.71 |
0.58, 0.62, 0.60 0.60, 0.64, 0.61 0.62, 0.65, 0.63 0.63, 0.66, 0.64 |
|
| BioMedICUS | Male Female |
0.82, 0.71, 0.76 0.80, 0.67, 0.73 |
0.80, 0.76, 0.77 0.78, 0.74, 0.76 |
| Asian Black Other** White |
0.75, 0.65, 0.69 0.80, 0.68, 0.73 0.84, 0.70, 0.76 0.85, 0.73, 0.78 |
0.75, 0.70, 0.72 0.79, 0.74, 0.76 0.81, 0.76, 0.78 0.83, 0.80, 0.81 |
| Corpus/ Symptom | Ground Truth | BioMedICUS | LLM | Issues |
|---|---|---|---|---|
| N3C COVID/Fever | 1 | 0 | 0 | both UMLS and LLM did not pick up 38 °C’ following heading of ‘Temp’ for positive mention; other similar |
| 1 | 1 | 0 | contextual : “had been sick with fever... over 12 days prior to his admission.”, etc. | |
| 1 | 0 | 1 | UMLS did not pick up “increase in temperature” for positive mention; “highest temp 37. 5,” etc. | |
| UMN PASC/Diarrhea | 1 | 0 | 0 | classified as negation: “Mild diarrhea today, kaopectate resolved” |
| 1 | 1 | 0 | none | |
| 1 | 0 | 1 | improper negation: “Constipation - senna not doing well, still either rabbit turds or diarrhea” | |
| N3C COVID/Chest Pain | 1 | 0 | 0 | missed lexical synonym: “Persistent pain or pressure in the chest,” etc. |
| 1 | 1 | 0 | missed case of both positive and negative mention due to "possible" symptom | |
| 1 | 0 | 1 | none |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





