1. Introduction
Over the last decade, outcomes have significantly improved in ovarian and endometrial cancers with the introduction of new treatment regimens, however, many patients will relapse within a median time of 18-24 months suggesting that effective biomarkers to predict relapse and facilitate escalated therapy are needed.[1, 2] The standard practice after completing primary therapy is surveillance with serial follow up of CA-125 and/or imaging, however, the ability to predict or detect relapse early is limited.[
3] Circulating tumor DNA (ctDNA) is tumor-derived DNA detectable in circulation via next generation sequencing technologies, providing a minimally-invasive approach without requiring invasive tissue biopsy, leading to clinical adoption and regulatory approval for several assays.[4, 5] Prior work has demonstrated the utility of quantifying the amount of tumor-derived DNA, or tumor fraction (TFx), for prognostic value in breast, lung, prostate, colon and other cancers.[6, 7] However, there is a paucity of data regarding their role in the treatment and surveillance of gynecologic malignancies, thus the objective of this study was to investigate the use of ctDNA and TFx in a cohort of patients with ovarian and endometrial cancer.
2. Methods
2.1. Patient Identification and Clinicopathologic Data
Patients with pathologic diagnosis of ovarian or endometrial cancers at a single tertiary cancer center were consented for collection of plasma for ctDNA analyses. The study was approved by the Cleveland Clinic Institutional Review Board and patients signed informed consent for the study. Patient characteristics and clinical and treatment data of our patients are summarized in
Table 1. Patients were non-consecutive and represented a diverse array of stages, grades, and other factors. Patients with a partial or complete response by RECIST 1.1 when evaluated by CT scan per judgment of their treating physician were classified as ‘responders’, while patients with best response as stable disease or progression were considered ‘non-responders’. Progression-free survival was defined as the time from diagnosis (for primary setting) or start of treatment (for recurrent tumors) to disease progression, last follow up, or death. Overall survival was calculated from the time of diagnosis for primary setting or start of treatment in recurrent setting to death or last follow up.
2.2. Sample Processing and Ultra Low-Pass Whole-Genome Sequencing
Venous blood samples were collected in EDTA (BD) or Cell-Free DNA BCT (Streck) tubes. Blood processing to component parts, cell-free DNA extraction from plasma, and DNA quantification was performed as described previously.[
8] Library construction of cell-free DNA was performed using the Kapa HyperPrep kit with custom adapters (IDT). 5-50 ng of cfDNA input (1000-20,000 haploid genome equivalents) was used for ultra low-pass whole-genome sequencing (ULP-WGS). Constructed sequencing libraries were pooled (2 uL of each x 96 per pool) and sequenced using 100bp paired-end runs over 1 x lane on a HiSeq2500 (Illumina) for ULP-WGS. ULP-WGS of cfDNA was performed to average genome-wide fold coverage of 0.1X. Segment copy number and TFx were derived via ichorCNA.[
8] Samples were excluded if the median absolute deviation (MAD) of the copy ratios (2
log2 ratio) between adjacent bins, genome-wide, was greater than 0.20 suggesting poor quality sequence data. Genome-wide copy number plots were generated by ichorCNA.
2.3. Statistical Analyses and Data Visualization
All statistical analyses and data visualizations were performed in R version 3.3.1. The association of TFx to continuous and categorical clinicopathologic factors was evaluated using Wilcoxon rank-sum and chi-square test or analysis of variance, respectively. Multiple linear regression models were constructed using the ‘lm’ function in R. Association with progression-free survival was assessed via log-rank test and Kaplan-Meier visualization constructed using the ‘packHV’ package in R.
3. Results
A total of 210 plasma samples from 78 patients with biopsy-proven ovarian cancer or endometrial cancer collected between 04/2018 and 04/2020 under IRB-approved protocols at a single institution and abstracted detailed clinicopathologic information were collected (
Table 1). Among ovarian cancers, 90.2% (46/51) were serous histology and 94.1% (48/51) high grade. Among endometrial cancers, 55% (11/20) were endometrioid and 50% (10/20) high grade. Among both cancer types, there were samples collected in both the primary (ovarian 62.7%, 32/51; endometrial 55%, 11/20) and recurrent (ovarian 37.3%, 19/51; endometrial 45%, 9/20) settings. At the time of data freeze, 17.6% (9/51) ovarian patients and 35% (7/20) endometrial patients had died of disease. Median first progression-free survival (PFS1) was 15 months for ovarian and 9 months for endometrial cancer.
The association of the first ‘sentinel’ blood draw with cancer and clinicopathologic characteristics were assessed (
Supplementary Figure S1;
Table 2). The mean tumor fraction for ovarian cancer was 5.5% and for endometrial cancer was 2.4%, not significantly different between cancer types (t-test p=0.065). We evaluated the characteristics of ‘sentinel’ TFx by sites of metastatic disease (
Table 2). For metastatic sites with at least two representative patients, mean TFx ranged from 1.6% (lung/pleural effusion metastases in endometrial cancer) to 5.2% (peritoneal metastases in ovarian cancer). When evaluating association with clinicopathologic features, there was no significant difference in primary versus recurrent settings within each individual cancer type (ovarian t-test p=0.84; endometrial p=0.64), tumor histology within each cancer type (ovarian Wilcoxon rank-sum p=0.24 endometrial p=0.62), or best overall therapy response (ovarian cancer ANOVA p=0.40; endometrial p=0.99). Interestingly, grade was associated with significant difference in ‘sentinel’ TFx among ovarian cancers (t-test p=0.01) but not endometrial cancers (ANOVA p=0.48). No association between clinical and pathologic variables in endometrial and ovarian cancer was significant after multiple test correction for either endometrial or ovarian cancer (
Supplementary Table S1).
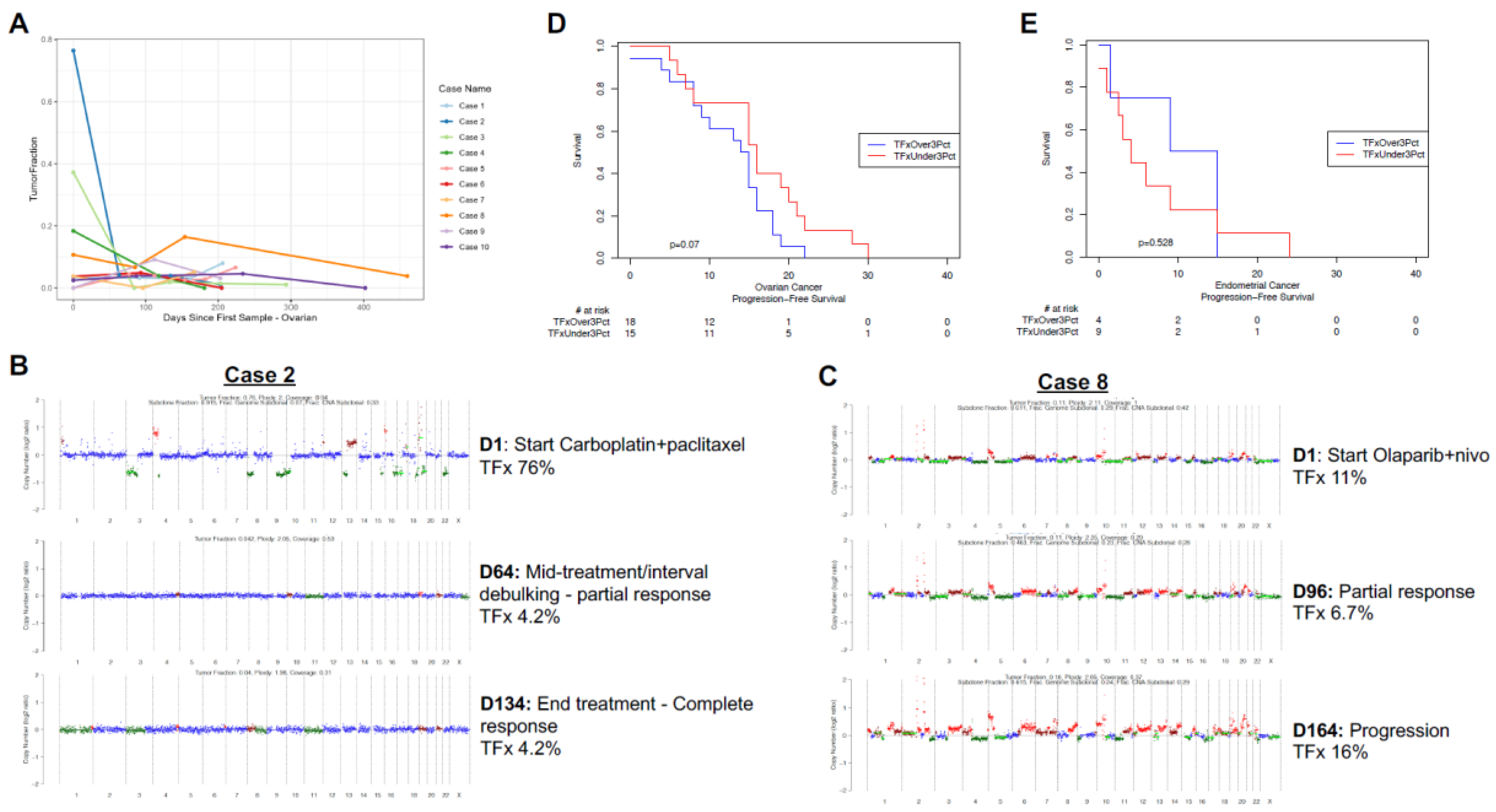
A unique aspect of this cohort was the rich collection of serial samples, with average of nearly three samples per individual (range 1 – 13 samples per patient). In the majority of cases, most samples had persistently low TFx, often near or below the threshold of ‘detectable’ ctDNA of 3%.[
8] We visualized the dynamics of ctDNA TFx for the ten patients with the greatest dynamic range and at least three samples (
Figure 1A,
Supplementary Figure S1G). For ovarian cancer, multiple patients demonstrate very high initial TFx, associated with rapid decline then persistently low subsequent TFx values (e.g., Cases 2/3/4
Figure 1A). Alternatively, for endometrial cancer there did not appear to be a consistent trend, with modest increases and decreases all at relatively low TFx<6% (
Supplementary Figure S1G).
Circulating tumor DNA TFx or similar metrics (e.g., highest variant allele fraction/VAF of detectable mutations) has been associated with prognosis across a variety of cancers.[6, 7, 8] Given the heterogeneity of the sample cohort, we investigated the association of ‘sentinel’ TFx with progression-free survival for patients with a sample collected at initiation of therapy for either primary or recurrent ovarian (
Figure 1B) or endometrial (
Figure 1C) cancers. While various TFx thresholds have been investigated, given the low overall TFx among the cohort we
a priori established the 3% TFx ‘detectable’ level as our threshold. For ovarian cancer, there was a non-significant trend (log-rank p=0.07) toward shorter PFS for patients with detectable ctDNA (TFx>3%;
Figure 1B). For endometrial cancer, with a limited number of evaluable patients there was no appreciable trend (log-rank p=0.53;
Figure 1C).
We investigated example cases in greater detail: a patient with newly diagnosed stage IIIC high grade serous ovarian cancer, germline BRCA mutated with high baseline TFx and dramatic decline with initial therapy, without evidence of relapse to date (
Figure 1B); and a patient with recurrent high grade serous ovarian cancer, germline BRCA wild-type initiating salvage therapy with Olaparib and nivolumab who demonstrated initial decline the rebound of ctDNA TFx (
Figure 1C).
4. Discussion
The treatment of advanced gynecologic malignancies has significantly improved patient outcomes, however, the treatment of platinum refractory and resistant disease remains challenging. Better diagnostic markers that can identify high-risk patients with molecular residual cancer at the time of primary treatment completion or at the time of recurrence are needed.
The present study evaluated the feasibility and utility of ctDNA TFx in ovarian and endometrial cancer as a prognostic biomarker. To screen a large number of samples, we used a low-cost/high throughput ULP-WGS assay that can enumerate TFx for <$200/sample. A strength of this study was the large number of samples overall and, in addition, serial samples from individual patients over time. Interestingly, most clinicopathologic features were not significantly associated with TFx – which may reflect the low overall TFx values. While our data suggests that ctDNA is potentially detectable, it rather dramatically reinforces that ctDNA TFx is overall very low in ovarian and endometrial cancers.
Limitations of the ULP-WGS technique include the potential for small ctDNA yield particularly in the setting of low tumor burden.[
9] Further, there is evidence that ctDNA TFx may be lower in peritoneal-based malignancies (such as appendiceal, colorectal, and ovarian cancers) than other solid tumors.[10-13] Interestingly, we were able to detect ctDNA in patients with primarily peritoneal disease, as well as metastases to other sites. This suggests that ‘low shed’ of ctDNA may be more a feature of ovarian and endometrial cancers themselves, rather than specific to the location (e.g., within the peritoneum).
Multiple patients with ovarian cancer demonstrated a high TFx at the time of initial collection followed by rapid decline and persistently low TFx values, corresponding to treatment response. We did not see a similar trend in endometrial cancer, however, Feng et al. assessed ctDNA as a biomarker of disease relapse in high-risk endometrial cancers by target panel of 78 known cancer genes and found an initial decrease in ctDNA from baseline to surgery in detectable cases, and subsequent increase in conjunction with disease relapse.[
14]
There are several limitations to this study. First, this study is limited by its small sample size. Future studies evaluating the use of TFx in gynecologic malignancies using larger cohorts are necessary to validate these findings. Further, direct comparison of cases with uneven number and/or times of sampling events is difficult, and subsequent investigations will benefit from standardization of TFx sampling across cases. Additionally, the use of TFx in ovarian and endometrial cancer will need to be correlated with targeted panel or whole exome sequencing, which will be the focus of a forthcoming study.
5. Conclusions
This study demonstrates that collection of ctDNA TFx by ULP-WGS is feasible in ovarian and endometrial cancers, yet reinforces that both ovarian and endometrial cancers appear to have low TFx irrespective of disease cancer type, stage (limited vs. advanced), or other clinicopathologic features. Based on these findings, future studies should focus on high sensitivity assays
Supplementary Materials
The following supporting information can be downloaded at the website of this paper posted on Preprints.org.
Author Contributions
Conceptualization, DGS, HM.; Methodology, MV, DGS, HM.; Formal Analysis, MV, DGS, HM; Data Curation, AG, PR, HM; Writing – Original Draft Preparation, MV, AG, AS, KC, DGS, HM; Writing – Review & Editing, all authors.; Visualization, MV, DGS.; Supervision, HM; Funding Acquisition, DGS, HM.
Funding
This research was funded by Cleveland Clinic Philanthropy.
Institutional Review Board Statement
The study was conducted according to the guidelines of the Declaration of Helsinki, and approved by the Institutional Review Board of Cleveland Clinic.
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study
Data Availability Statement
The data presented in this study are available on request from the corresponding author. The data are not publicly available due to restrictions secondary to information that could compromise the privacy of research participants.
Acknowledgments
The authors would like to acknowledge the Broad Institute Genomics Platform for sequencing support.
Conflicts of Interest
DGS has served on an advisory board for Novartis.
References
- Wang, Q.; Zheng, Y.; Wang, P.; Zhang, J.; Liu, H.; Li, Q.; et al. The prognostic factor for recurrence in advanced-stage high-grade serous ovarian cancer after complete clinical remission: a nested case-control study. J Ovarian Res. 2021, 14, 179. [Google Scholar] [CrossRef] [PubMed]
- Piedimonte, S.; Kim, R.; Bernardini, M.Q.; Atenafu, E.G.; Clark, M.; Lheureux, S.; et al. Validation of the KELIM score as a predictor of response to neoadjuvant treatment in patients with advanced high grade serous ovarian cancer. Gynecol Oncol. 2022, 167, 417–22. [Google Scholar] [CrossRef] [PubMed]
- Zachou, G.; El-Khouly, F.; Dilley, J. Evaluation of follow-up strategies for women with epithelial ovarian cancer following completion of primary treatment. Cochrane Database Syst Rev. 2023, 8, CD006119. [Google Scholar] [PubMed]
- FDA FoundationOne Liquid CDx - P190032 | FDA. 2020.
- FDA Guardant360 CDx – P200010/S001. 2020.
- Stover, D.G.; Parsons, H.A.; Ha, G.; Freeman, S.S.; Barry, W.T.; Guo, H.; et al. Association of Cell-Free DNA Tumor Fraction and Somatic Copy Number Alterations With Survival in Metastatic Triple-Negative Breast Cancer. J Clin Oncol. 2018, 36, 543–53. [Google Scholar] [CrossRef] [PubMed]
- Reichert, Z.R.; Morgan, T.M.; Li, G.; Castellanos, E.; Snow, T.; Dall’Olio, F.G.; et al. Prognostic value of plasma circulating tumor DNA fraction across four common cancer types: a real-world outcomes study. Ann Oncol. 2023, 34, 111–20. [Google Scholar] [CrossRef] [PubMed]
- Adalsteinsson, V.A.; Ha, G.; Freeman, S.S.; Choudhury, A.D.; Stover, D.G.; Parsons, H.A.; et al. Scalable whole-exome sequencing of cell-free DNA reveals high concordance with metastatic tumors. Nature communications. 2017, 8, 1324. [Google Scholar] [CrossRef] [PubMed]
- Asante, D.B.; Calapre, L.; Ziman, M.; Meniawy, T.M.; Gray, E.S. Liquid biopsy in ovarian cancer using circulating tumor DNA and cells: Ready for prime time? Cancer Lett. 2020, 468, 59–71. [Google Scholar] [CrossRef] [PubMed]
- Singh, H.; Klempner, S.J.; Melnitchouk, N.; Chander, D.P.; Negrea, O.G.; Patel, A.K.; et al. Highly Sensitive Circulating Tumor DNA Assay Aids Clinical Management of Radiographically Occult Isolated Peritoneal Metastases in Patients With GI Cancer. JCO Precision Oncology. 2023, e2200572. [Google Scholar] [CrossRef] [PubMed]
- Baumgartner, J.M.; Botta, G.P. Role of Circulating Tumor DNA Among Patients with Colorectal Peritoneal Metastases. J Gastrointest Cancer. 2023. [CrossRef] [PubMed]
- Bando, H.; Nakamura, Y.; Taniguchi, H.; Shiozawa, M.; Yasui, H.; Esaki, T.; et al. Effects of Metastatic Sites on Circulating Tumor DNA in Patients With Metastatic Colorectal Cancer. JCO Precision Oncology. 2022, e2100535. [Google Scholar] [CrossRef] [PubMed]
- Oikkonen, J.; Zhang, K.; Salminen, L.; Schulman, I.; Lavikka, K.; Andersson, N.; et al. Prospective Longitudinal ctDNA Workflow Reveals Clinically Actionable Alterations in Ovarian Cancer. JCO Precision Oncology. 2019, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Feng, W.; Jia, N.; Jiao, H.; Chen, J.; Chen, Y.; Zhang, Y.; et al. Circulating tumor DNA as a prognostic marker in high-risk endometrial cancer. Journal of Translational Medicine. 2021, 19, 51. [Google Scholar] [CrossRef] [PubMed]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).