Submitted:
20 September 2024
Posted:
23 September 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
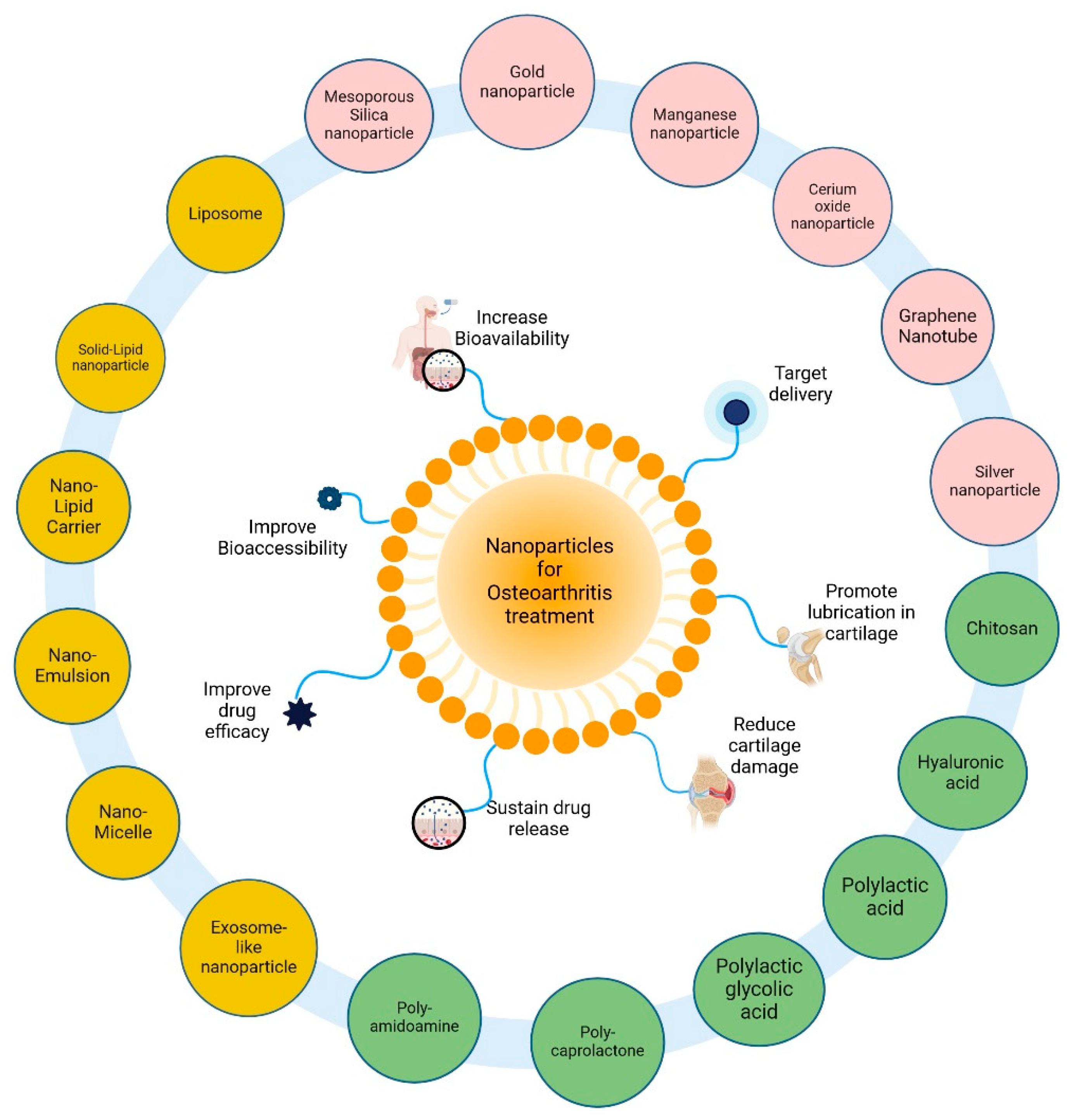
2. Nanotherapeutics in Osteoarthritis
2.1. Nanoformulation and Nanoemulsion as Bioactive Carriers
2.2. Polyphenolic Nanoformulation and Osteoarthritis
2.3. Epigallocatechin in Osteoarthritis
2.4. Resveratrol in Osteoarthritis
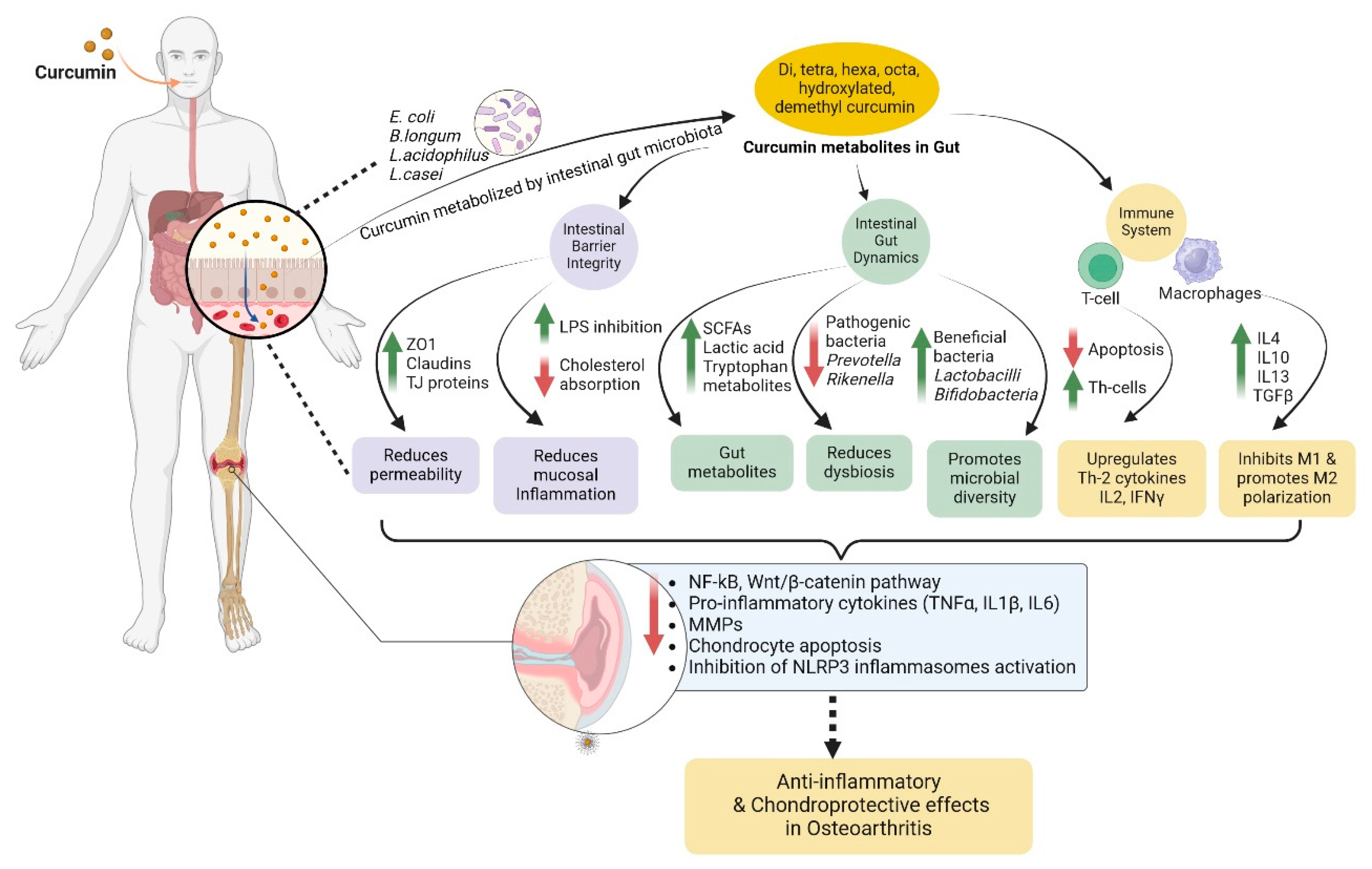
2.5. Curcumin in Osteoarthritis
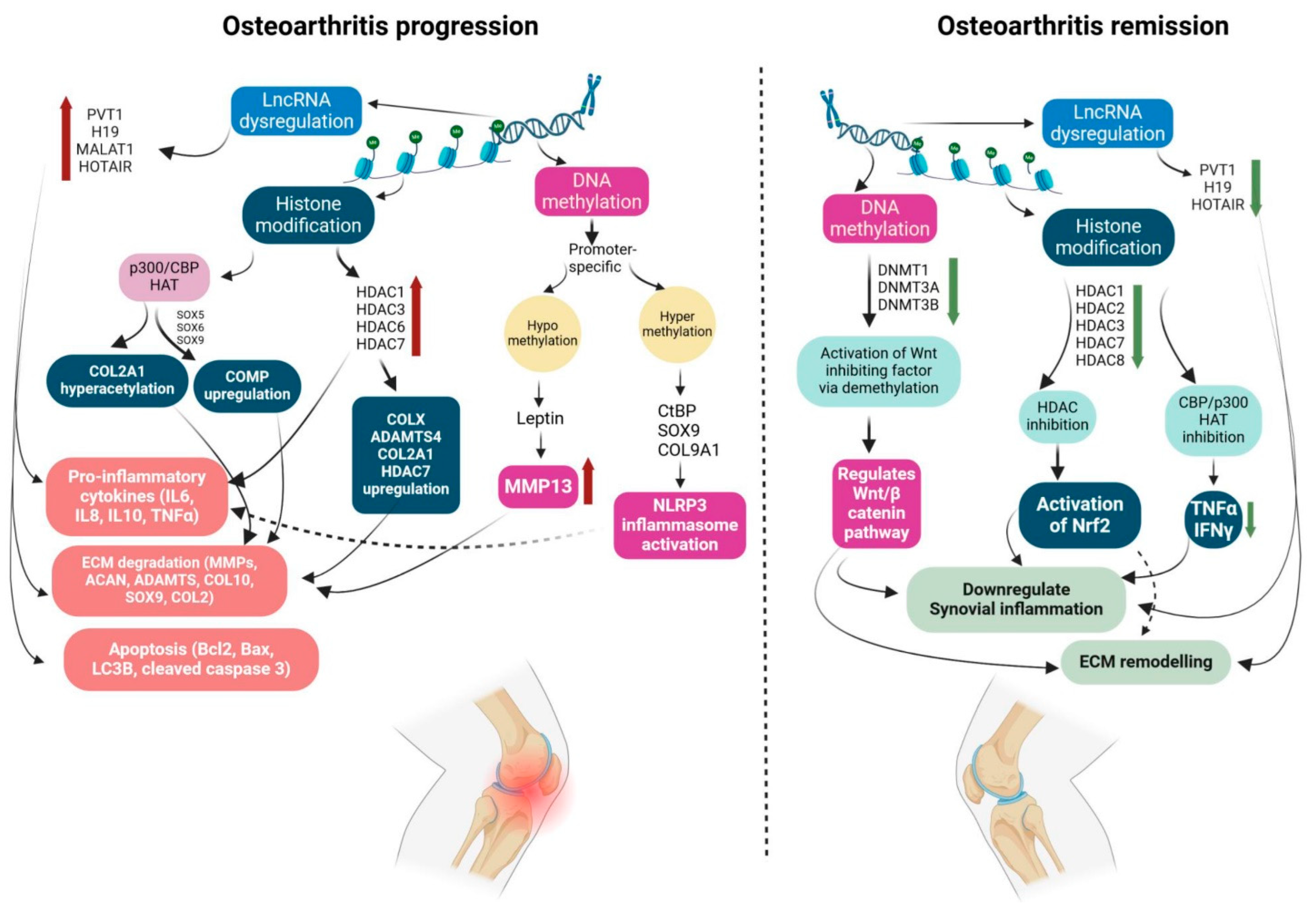
3. Epigenetic Modulations in Osteoarthritis Pathogenesis
3.1. Role of Histone Modifications in Osteoarthritis Pathology
3.2. Modulation of Promoter DNA Methylation in Osteoarthritis
3.3. Role of Non-Coding RNAs in Osteoarthritis Pathogenesis
3.4. Polyphenol as a Potential Epigenetic Modulator
4. Interplay of Gut Microbiome, Epigenetics, and Osteoarthritis: Mechanism of Bioactive Actions
4.1. Target Osteoarthritis by Epigenetic Modulation
4.2. Gut Microbiome-Mediated Epigenetic Modulation
5. Future Perspectives and Conclusions
Author Contributions
Funding
Conflicts of Interest
Acknowledgement
References
- Olansen, J.; Dyke, J.P.; Aaron, R.K. Is Osteoarthritis a Vascular Disease? Front. Biosci. (Landmark Ed. ) 2024, 29, 113. [Google Scholar] [CrossRef] [PubMed]
- Steinmetz, J.D.; Culbreth, G.T.; Haile, L.M.; Rafferty, Q.; Lo, J.; Fukutaki, K.G.; Cruz, J.A.; Smith, A.E.; Vollset, S.E.; Brooks, P.M.; et al. Global, regional, and national burden of osteoarthritis, 1990–2020 and projections to 2050: a systematic analysis for the Global Burden of Disease Study 2021. Lancet Rheumatol. 2023, 5, e508–e522. [Google Scholar] [CrossRef]
- Yao, Q.; Wu, X.; Tao, C.; Gong, W.; Chen, M.; Qu, M.; Zhong, Y.; He, T.; Chen, S.; Xiao, G. Osteoarthritis: pathogenic signaling pathways and therapeutic targets. Signal Transduct. Target. Ther. 2023, 8, 56. [Google Scholar] [CrossRef]
- Marcum, Z.A.; Hanlon, J.T. Recognizing the Risks of Chronic Nonsteroidal Anti-Inflammatory Drug Use in Older Adults. Ann. Long-Term Care Off. J. Am. Med. Dir. Assoc. 2010, 18, 24–27. [Google Scholar]
- Rodriguez-Merchan, E.C. The Current Role of Disease-modifying Osteoarthritis Drugs. Arch. Bone Jt. Surg. 2023, 11, 11–22. [Google Scholar] [CrossRef]
- Pulido, L.; Parvizi, J.; Macgibeny, M.; Sharkey, P.F.; Purtill, J.J.; Rothman, R.H.; Hozack, W.J. In hospital complications after total joint arthroplasty. J. Arthroplast. 2008, 23, 139–145. [Google Scholar] [CrossRef]
- Sukhikh, S.; Noskova, S.; Ivanova, S.; Ulrikh, E.; Izgaryshev, A.; Babich, O. Chondroprotection and Molecular Mechanism of Action of Phytonutraceuticals on Osteoarthritis. Molecules 2021, 26. [Google Scholar] [CrossRef]
- Crofford, L.J. Use of NSAIDs in treating patients with arthritis. Arthritis Res Ther 2013, 15 (Suppl. S3), S2. [Google Scholar] [CrossRef]
- Anwar, A.; Anwar, I.J.; Delafontaine, P. Elevation of cardiovascular risk by non-steroidal anti-inflammatory drugs. Trends Cardiovasc. Med. 2015, 25, 726–735. [Google Scholar] [CrossRef]
- Maniar, K.H.; Jones, I.A.; Gopalakrishna, R.; Vangsness, C.T., Jr. Lowering side effects of NSAID usage in osteoarthritis: recent attempts at minimizing dosage. Expert Opin. Pharmacother. 2018, 19, 93–102. [Google Scholar] [CrossRef]
- Basak, S.; Hridayanka, K.S.N.; Duttaroy, A.K. Bioactives and their roles in bone metabolism of osteoarthritis: evidence and mechanisms on gut-bone axis. Front Immunol 2024, 14, 1323233. [Google Scholar] [CrossRef] [PubMed]
- Mileo, A.M.; Nisticò, P.; Miccadei, S. Polyphenols: Immunomodulatory and Therapeutic Implication in Colorectal Cancer. Front Immunol 2019, 10, 729. [Google Scholar] [CrossRef] [PubMed]
- Tomar, R.; Das, S.S.; Balaga, V.K.R.; Tambe, S.; Sahoo, J.; Rath, S.K.; Ruokolainen, J.; Kesari, K.K. Therapeutic Implications of Dietary Polyphenols-Loaded Nanoemulsions in Cancer Therapy. ACS Appl. Bio Mater. 2024, 7, 2036–2053. [Google Scholar] [CrossRef] [PubMed]
- Yusuf, A.; Almotairy, A.R.Z.; Henidi, H.; Alshehri, O.Y.; Aldughaim, M.S. Nanoparticles as Drug Delivery Systems: A Review of the Implication of Nanoparticles’ Physicochemical Properties on Responses in Biological Systems. Polymers 2023, 15. [Google Scholar] [CrossRef] [PubMed]
- Soares, S.; Sousa, J.; Pais, A.; Vitorino, C. Nanomedicine: Principles, Properties, and Regulatory Issues. Front. Chem. 2018, 6. [Google Scholar] [CrossRef] [PubMed]
- Tong, L.; Yu, H.; Huang, X.; Shen, J.; Xiao, G.; Chen, L.; Wang, H.; Xing, L.; Chen, D. Current understanding of osteoarthritis pathogenesis and relevant new approaches. Bone Res. 2022, 10, 60. [Google Scholar] [CrossRef]
- Krishnan, Y.; Grodzinsky, A.J. Cartilage diseases. Matrix Biol. : J. Int. Soc. Matrix Biol. 2018, 71-72, 51–69. [Google Scholar] [CrossRef]
- Lu, H.; Zhang, S.; Wang, J.; Chen, Q. A Review on Polymer and Lipid-Based Nanocarriers and Its Application to Nano-Pharmaceutical and Food-Based Systems. Front. Nutr. 2021, 8, 783831. [Google Scholar] [CrossRef]
- Lv, Z.; Wang, P.; Li, W.; Xie, Y.; Sun, W.; Jin, X.; Jiang, R.; Fei, Y.; Liu, Y.; Shi, T.; et al. Bifunctional TRPV1 Targeted Magnetothermal Switch to Attenuate Osteoarthritis Progression. Res. (Wash. D.C.) 2024, 7, 0316. [Google Scholar] [CrossRef]
- Wang, H.; Wang, C.; Zou, Y.; Hu, J.; Li, Y.; Cheng, Y. Natural polyphenols in drug delivery systems: Current status and future challenges. Giant 2020, 3, 100022. [Google Scholar] [CrossRef]
- Pontes, A.P.; Welting, T.J.M.; Rip, J.; Creemers, L.B. Polymeric Nanoparticles for Drug Delivery in Osteoarthritis. Pharmaceutics 2022, 14. [Google Scholar] [CrossRef] [PubMed]
- Mohammed, M.A.; Syeda, J.T.M.; Wasan, K.M.; Wasan, E.K. An Overview of Chitosan Nanoparticles and Its Application in Non-Parenteral Drug Delivery. Pharmaceutics 2017, 9. [Google Scholar] [CrossRef] [PubMed]
- Rao, N.V.; Rho, J.G.; Um, W.; Ek, P.K.; Nguyen, V.Q.; Oh, B.H.; Kim, W.; Park, J.H. Hyaluronic Acid Nanoparticles as Nanomedicine for Treatment of Inflammatory Diseases. Pharmaceutics 2020, 12. [Google Scholar] [CrossRef]
- Danhier, F.; Ansorena, E.; Silva, J.M.; Coco, R.; Le Breton, A.; Préat, V. PLGA-based nanoparticles: An overview of biomedical applications. J. Control. Release 2012, 161, 505–522. [Google Scholar] [CrossRef]
- Puri, A.; Loomis, K.; Smith, B.; Lee, J.H.; Yavlovich, A.; Heldman, E.; Blumenthal, R. Lipid-based nanoparticles as pharmaceutical drug carriers: from concepts to clinic. Crit. Rev. Ther. Drug Carr. Syst. 2009, 26, 523–580. [Google Scholar] [CrossRef] [PubMed]
- Guimarães, D.; Cavaco-Paulo, A.; Nogueira, E. Design of liposomes as drug delivery system for therapeutic applications. Int. J. Pharm. 2021, 601, 120571. [Google Scholar] [CrossRef]
- Lei, Y.; Wang, X.; Liao, J.; Shen, J.; Li, Y.; Cai, Z.; Hu, N.; Luo, X.; Cui, W.; Huang, W. Shear-responsive boundary-lubricated hydrogels attenuate osteoarthritis. Bioact. Mater. 2022, 16, 472–484. [Google Scholar] [CrossRef]
- Bishnoi, M.; Jain, A.; Hurkat, P.; Jain, S.K. Aceclofenac-loaded chondroitin sulfate conjugated SLNs for effective management of osteoarthritis. J. Drug Target. 2014, 22, 805–812. [Google Scholar] [CrossRef]
- Müller, R.H.; Shegokar, R.; Keck, C.M. 20 years of lipid nanoparticles (SLN and NLC): present state of development and industrial applications. Curr. Drug Discov. Technol. 2011, 8, 207–227. [Google Scholar] [CrossRef]
- Sütő, B.; Berkó, S.; Kozma, G.; Kukovecz, Á.; Budai-Szűcs, M.; Erős, G.; Kemény, L.; Sztojkov-Ivanov, A.; Gáspár, R.; Csányi, E. Development of ibuprofen-loaded nanostructured lipid carrier-based gels: characterization and investigation of in vitro and in vivo penetration through the skin. Int J Nanomed. 2016, 11, 1201–1212. [Google Scholar] [CrossRef]
- Jaiswal, M.; Dudhe, R.; Sharma, P.K. Nanoemulsion: an advanced mode of drug delivery system. 3 Biotech 2015, 5, 123–127. [Google Scholar] [CrossRef]
- Leite, C.B.; Coelho, J.M.; Ferreira-Nunes, R.; Gelfuso, G.M.; Durigan, J.L.; Azevedo, R.B.; Muehlmann, L.A.; Sousa, M.H. Phonophoretic application of a glucosamine and chondroitin nanoemulsion for treatment of knee chondropathies. Nanomed. (Lond. Engl. ) 2020, 15, 647–659. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.R.; Desu, P.K.; Nakkala, R.K.; Kondi, V.; Devi, S.; Alam, M.S.; Hamid, H.; Athawale, R.B.; Kesharwani, P. Nanotechnology-based approaches applied to nutraceuticals. Drug Delivery and Translational Research 2021 12:3 2021, 12, 485–499. [Google Scholar] [CrossRef] [PubMed]
- Ansari, M.Y.; Ahmad, N.; Haqqi, T.M. Oxidative stress and inflammation in osteoarthritis pathogenesis: Role of polyphenols. Biomed Pharmacother 2020, 129, 110452. [Google Scholar] [CrossRef]
- Ashruf, O.S.; Ansari, M.Y. Natural Compounds: Potential Therapeutics for the Inhibition of Cartilage Matrix Degradation in Osteoarthritis. Life (Basel Switz. ) 2022, 13. [Google Scholar] [CrossRef]
- Suzuki, T.; Ohishi, T.; Tanabe, H.; Miyoshi, N.; Nakamura, Y. Anti-Inflammatory Effects of Dietary Polyphenols through Inhibitory Activity against Metalloproteinases. Molecules 2023, 28. [Google Scholar] [CrossRef]
- Wu, Z.; Yang, Z.; Liu, L.; Xiao, Y. Natural compounds protect against the pathogenesis of osteoarthritis by mediating the NRF2/ARE signaling. Front Pharmacol 2023, 14, 1188215. [Google Scholar] [CrossRef]
- Valsamidou, E.; Amerikanou, C.; Tzavara, C.; Skarpas, G.; Mariolis-Sapsakos, T.D.; Zoumpoulakis, P.; Kaliora, A.C. A standardized nutraceutical supplement contributes to pain relief, improves quality of life and regulates inflammation in knee osteoarthritis patients; A randomized clinical trial. Heliyon 2023, 9, e20143. [Google Scholar] [CrossRef]
- Li, Y.; Shen, B.; Lv, C.; Zhu, X.; Naren, Q.; Xu, D.; Chen, H.; Wu, F. Methyl gallate prevents oxidative stress induced apoptosis and ECM degradation in chondrocytes via restoring Sirt3 mediated autophagy and ameliorates osteoarthritis progression. Int Immunopharmacol 2023, 114, 109489. [Google Scholar] [CrossRef]
- Permatasari, D.A.; Karliana, D.; Iskandarsyah, I.; Arsianti, A.; Bahtiar, A. Quercetin prevent proteoglycan destruction by inhibits matrix metalloproteinase-9, matrix metalloproteinase-13, a disintegrin and metalloproteinase with thrombospondin motifs-5 expressions on osteoarthritis model rats. J. Adv. Pharm. Technol. Res. 2019, 10, 2–8. [Google Scholar] [CrossRef]
- Eo, S.H.; Kim, S.J. Rosmarinic acid induces rabbit articular chondrocyte differentiation by decreases matrix metalloproteinase-13 and inflammation by upregulating cyclooxygenase-2 expression. J. Biomed. Sci. 2017, 24, 75. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.P.; Jin, G.J.; Xiong, Y.; Hu, P.F.; Bao, J.P.; Wu, L.D. Rosmarinic acid down-regulates NO and PGE(2) expression via MAPK pathway in rat chondrocytes. J Cell Mol Med 2018, 22, 346–353. [Google Scholar] [CrossRef] [PubMed]
- Valentino, A.; Conte, R.; De Luca, I.; Di Cristo, F.; Peluso, G.; Bosetti, M.; Calarco, A. Thermo-Responsive Gel Containing Hydroxytyrosol-Chitosan Nanoparticles (Hyt@tgel) Counteracts the Increase of Osteoarthritis Biomarkers in Human Chondrocytes. Antioxid. (Basel Switz. ) 2022, 11. [Google Scholar] [CrossRef] [PubMed]
- Cai, J.; Liu, L.F.; Qin, Z.; Liu, S.; Wang, Y.; Chen, Z.; Yao, Y.; Zheng, L.; Zhao, J.; Gao, M. Natural Morin-Based Metal Organic Framework Nanoenzymes Modulate Articular Cavity Microenvironment to Alleviate Osteoarthritis. Res. (Wash. D.C.) 2023, 6, 0068. [Google Scholar] [CrossRef]
- Chen, Y.; Xu, W.; Shafiq, M.; Song, D.; Wang, T.; Yuan, Z.; Xie, X.; Yu, X.; Shen, Y.; Sun, B.; et al. Injectable nanofiber microspheres modified with metal phenolic networks for effective osteoarthritis treatment. Acta Biomater. 2023, 157, 593–608. [Google Scholar] [CrossRef]
- Craciunescu, O.; Icriverzi, M.; Florian, P.E.; Roseanu, A.; Trif, M. Mechanisms and Pharmaceutical Action of Lipid Nanoformulation of Natural Bioactive Compounds as Efficient Delivery Systems in the Therapy of Osteoarthritis. Pharmaceutics 2021, 13. [Google Scholar] [CrossRef]
- Li, Y.; Chen, M.; Yan, J.; Zhou, W.; Gao, S.; Liu, S.; Li, Q.; Zheng, Y.; Cheng, Y.; Guo, Q. Tannic acid/Sr(2+)-coated silk/graphene oxide-based meniscus scaffold with anti-inflammatory and anti-ROS functions for cartilage protection and delaying osteoarthritis. Acta Biomater. 2021, 126, 119–131. [Google Scholar] [CrossRef]
- Xiong, F.; Qin, Z.; Chen, H.; Lan, Q.; Wang, Z.; Lan, N.; Yang, Y.; Zheng, L.; Zhao, J.; Kai, D. pH-responsive and hyaluronic acid-functionalized metal-organic frameworks for therapy of osteoarthritis. J Nanobiotechnology 2020, 18, 139. [Google Scholar] [CrossRef]
- Li, G.; Liu, S.; Chen, Y.; Zhao, J.; Xu, H.; Weng, J.; Yu, F.; Xiong, A.; Udduttula, A.; Wang, D. , et al. An injectable liposome-anchored teriparatide incorporated gallic acid-grafted gelatin hydrogel for osteoarthritis treatment. Nat. Commun. 2023, 14, 3159. [Google Scholar] [CrossRef]
- Yang, H.; Yan, R.; Chen, Q.; Wang, Y.; Zhong, X.; Liu, S.; Xie, R.; Ren, L. Functional nano drug delivery system with dual lubrication and immune escape for treating osteoarthritis. J. Colloid Interface Sci. 2023, 652, 2167–2179. [Google Scholar] [CrossRef]
- Yan, R.; Yang, H.; Liu, Y.; Wang, Y.; Liu, S.; Xie, R.; Ren, L. A Dual Functional Bioinspired Lubricant for Osteoarthritis Treatment and Potential Prevention. ACS Appl. Mater. Interfaces, 2024. [Google Scholar] [CrossRef]
- Ouyang, Z.; Tan, T.; Liu, C.; Duan, J.; Wang, W.; Guo, X.; Zhang, Q.; Li, Z.; Huang, Q.; Dou, P.; et al. Targeted delivery of hesperetin to cartilage attenuates osteoarthritis by bimodal imaging with Gd(2)(CO(3))(3)@PDA nanoparticles via TLR-2/NF-κB/Akt signaling. Biomaterials 2019, 205, 50–63. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Zhong, Y.; He, S.; Liang, R.; Liao, C.; Zheng, L.; Zhao, J. Application of the pH-Responsive PCL/PEG-Nar Nanofiber Membrane in the Treatment of Osteoarthritis. Front Bioeng Biotechnol 2022, 10, 859442. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Chen, X.; Zhou, Z.; Fang, B.; Chen, Z.; Huang, Y.; Hu, Y.; Liu, H. Berberine oleanolic acid complex salt grafted hyaluronic acid/silk fibroin (BOA-g-HA/SF) composite scaffold promotes cartilage tissue regeneration under IL-1β caused stress. Int. J. Biol. Macromol. 2023, 250, 126104. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.; Chang, L.; Zhao, X.; Hu, Y.; Lin, Y.; Chen, Z.; Ren, X.; Mei, X. Preparation of epigallocatechin gallate decorated Au-Ag nano-heterostructures as NIR-sensitive nano-enzymes for the treatment of osteoarthritis through mitochondrial repair and cartilage protection. Acta Biomater. 2022, 144, 168–182. [Google Scholar] [CrossRef]
- Lee, C.Y.; Chang, Y.C.; Yang, K.C.; Lin, Y.F.; Wu, A.T.H.; Tseng, C.L. Development and functional evaluation of a hyaluronic acid coated nano-formulation with kaempferol as a novel intra-articular agent for Knee Osteoarthritis treatment. Biomed Pharmacother 2024, 175, 116717. [Google Scholar] [CrossRef]
- Guo, J.; Su, K.; Wang, L.; Feng, B.; You, X.; Deng, M.; Toh, W.S.; Wu, J.; Cheng, B.; Xia, J. Poly(p-coumaric acid) nanoparticles alleviate temporomandibular joint osteoarthritis by inhibiting chondrocyte ferroptosis. Bioact. Mater. 2024, 40, 212–226. [Google Scholar] [CrossRef]
- Shaban, N.S.; Radi, A.M.; Abdelgawad, M.A.; Ghoneim, M.M.; Al-Serwi, R.H.; Hassan, R.M.; Mohammed, E.T.; Radi, R.A.; Halfaya, F.M. Targeting Some Key Metalloproteinases by Nano-Naringenin and Amphora coffeaeformis as a Novel Strategy for Treatment of Osteoarthritis in Rats. Pharm. (Basel Switz. ) 2023, 16. [Google Scholar] [CrossRef]
- Nagle, D.G.; Ferreira, D.; Zhou, Y.D. Epigallocatechin-3-gallate (EGCG): chemical and biomedical perspectives. Phytochemistry 2006, 67, 1849–1855. [Google Scholar] [CrossRef]
- Akhtar, N.; Haqqi, T.M. Epigallocatechin-3-gallate suppresses the global interleukin-1beta-induced inflammatory response in human chondrocytes. Arthritis Res Ther 2011, 13, R93. [Google Scholar] [CrossRef]
- Rasheed, Z.; Rasheed, N.; Al-Shaya, O. Epigallocatechin-3-O-gallate modulates global microRNA expression in interleukin-1β-stimulated human osteoarthritis chondrocytes: potential role of EGCG on negative co-regulation of microRNA-140-3p and ADAMTS5. Eur. J. Nutr. 2018, 57, 917–928. [Google Scholar] [CrossRef]
- Rasheed, Z.; Rasheed, N.; Al-Shobaili, H.A. Epigallocatechin-3-O-gallate up-regulates microRNA-199a-3p expression by down-regulating the expression of cyclooxygenase-2 in stimulated human osteoarthritis chondrocytes. J Cell Mol Med 2016, 20, 2241–2248. [Google Scholar] [CrossRef] [PubMed]
- Wei, H.; Qin, J.; Huang, Q.; Jin, Z.; Zheng, L.; Zhao, J.; Qin, Z. Epigallocatechin-3-gallate (EGCG) based metal-polyphenol nanoformulations alleviates chondrocytes inflammation by modulating synovial macrophages polarization. Biomed Pharmacother 2023, 161, 114366. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Song, Z.; Yu, J.; Ren, B.; Dong, Y.; You, Y.; Zhang, Z.; Jia, C.; Zhao, Y.; Zhou, X.; et al. Supramolecular self-assembly of EGCG-selenomethionine nanodrug for treating osteoarthritis. Bioact. Mater. 2024, 32, 164–176. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Xiao, L.; Yu, C.; Jin, P.; Qin, D.; Xu, Y.; Yin, J.; Liu, Z.; Du, Q. Enhanced Antiarthritic Efficacy by Nanoparticles of (-)-Epigallocatechin Gallate-Glucosamine-Casein. J. Agric. Food Chem. 2019, 67, 6476–6486. [Google Scholar] [CrossRef] [PubMed]
- Jin, Y.; Koh, R.H.; Kim, S.H.; Kim, K.M.; Park, G.K.; Hwang, N.S. Injectable anti-inflammatory hyaluronic acid hydrogel for osteoarthritic cartilage repair. Mater. Sci. Eng.. C Mater. Biol. Appl. 2020, 115, 111096. [Google Scholar] [CrossRef]
- Li, H.; Xiang, D.; Gong, C.; Wang, X.; Liu, L. Naturally derived injectable hydrogels with ROS-scavenging property to protect transplanted stem cell bioactivity for osteoarthritic cartilage repair. Front Bioeng Biotechnol 2022, 10, 1109074. [Google Scholar] [CrossRef]
- Dave, M.; Attur, M.; Palmer, G.; Al-Mussawir, H.E.; Kennish, L.; Patel, J.; Abramson, S.B. The antioxidant resveratrol protects against chondrocyte apoptosis via effects on mitochondrial polarization and ATP production. Arthritis Rheum 2008, 58, 2786–2797. [Google Scholar] [CrossRef]
- Wang, J.; Gao, J.S.; Chen, J.W.; Li, F.; Tian, J. Effect of resveratrol on cartilage protection and apoptosis inhibition in experimental osteoarthritis of rabbit. Rheumatol. Int. 2012, 32, 1541–1548. [Google Scholar] [CrossRef]
- Singh, K.; Gupta, J.K.; Kumar, S. The Pharmacological Potential of Resveratrol in Reducing Soft Tissue Damage in Osteoarthritis Patients. Curr Rheumatol Rev 2024, 20, 27–38. [Google Scholar] [CrossRef]
- Kann, B.; Spengler, C.; Coradini, K.; Rigo, L.A.; Bennink, M.L.; Jacobs, K.; Offerhaus, H.L.; Beck, R.C.; Windbergs, M. Intracellular Delivery of Poorly Soluble Polyphenols: Elucidating the Interplay of Self-Assembling Nanocarriers and Human Chondrocytes. Anal. Chem. 2016, 88, 7014–7022. [Google Scholar] [CrossRef]
- Le Clanche, S.; Cheminel, T.; Rannou, F.; Bonnefont-Rousselot, D.; Borderie, D.; Charrueau, C. Use of Resveratrol Self-Emulsifying Systems in T/C28a2 Cell Line as Beneficial Effectors in Cellular Uptake and Protection Against Oxidative Stress-Mediated Death. Front. Pharmacol. 2018, 9. [Google Scholar] [CrossRef]
- Wei, L.; Pan, Q.; Teng, J.; Zhang, H.; Qin, N. Intra-articular administration of PLGA resveratrol sustained-release nanoparticles attenuates the development of rat osteoarthritis. Mater. Today. Bio 2024, 24, 100884. [Google Scholar] [CrossRef] [PubMed]
- Zhou, M.; Wang, D.; Tang, J. Identification of the Resveratrol Potential Targets in the Treatment of Osteoarthritis. Evid Based Complement Altern. Med 2021, 2021, 9911286. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Zhang, H.; You, W.; Tang, X.; Li, X.; Gong, Z. Therapeutic effect of Resveratrol in the treatment of osteoarthritis via the MALAT1/miR-9/NF-κB signaling pathway. Exp Ther Med 2020, 19, 2343–2352. [Google Scholar] [CrossRef]
- Hussain, S.A.; Marouf, B.H.; Ali, Z.S.; Ahmmad, R.S. Efficacy and safety of co-administration of resveratrol with meloxicam in patients with knee osteoarthritis: a pilot interventional study. Clin. Interv. Aging 2018, 13, 1621–1630. [Google Scholar] [CrossRef] [PubMed]
- Marouf, B.H.; Hussain, S.A.; Ali, Z.S. Correlation between serum pro inflammatory cytokines and clinical scores of knee osteoarthritic patients using resveratrol as a supplementary therapy with meloxicam. Indian J. Pharmacol. 2021, 53, 270–277. [Google Scholar] [CrossRef]
- Sheu, S.Y.; Chen, W.S.; Sun, J.S.; Lin, F.H.; Wu, T. Biological characterization of oxidized hyaluronic acid/resveratrol hydrogel for cartilage tissue engineering. J. Biomed. Mater. Res.. Part A 2013, 101, 3457–3466. [Google Scholar] [CrossRef]
- Zhao, J.; Liang, G.; Zhou, G.; Hong, K.; Yang, W.; Liu, J.; Zeng, L. Efficacy and safety of curcumin therapy for knee osteoarthritis: A Bayesian network meta-analysis. J. Ethnopharmacol. 2024, 321, 117493. [Google Scholar] [CrossRef]
- Zeng, L.; Yu, G.; Hao, W.; Yang, K.; Chen, H. The efficacy and safety of Curcuma longa extract and curcumin supplements on osteoarthritis: a systematic review and meta-analysis. Biosci. Rep. 2021, 41. [Google Scholar] [CrossRef]
- Basak, S.; Hridayanka, K.S.N.; Duttaroy, A.K. Bioactives and their roles in bone metabolism of osteoarthritis: evidence and mechanisms on gut-bone axis. Front. Immunol. 2024, 14. [Google Scholar] [CrossRef]
- Heidari-Beni, M.; Moravejolahkami, A.R.; Gorgian, P.; Askari, G.; Tarrahi, M.J.; Bahreini-Esfahani, N. Herbal formulation “turmeric extract, black pepper, and ginger” versus Naproxen for chronic knee osteoarthritis: A randomized, double-blind, controlled clinical trial. Phytother. Res. PTR 2020, 34, 2067–2073. [Google Scholar] [CrossRef] [PubMed]
- Yuan, T.; Cai, D.; Hu, B.; Zhu, Y.; Qin, J. Therapeutic Effects of Curcumin on Osteoarthritis and Its Protection of Chondrocytes Through the Wnt/Β-Catenin Signaling Pathway. Altern. Ther. Health Med. 2022, 28, 28–37. [Google Scholar] [PubMed]
- Atabaki, M.; Shariati-Sarabi, Z.; Tavakkol-Afshari, J.; Mohammadi, M. Significant immunomodulatory properties of curcumin in patients with osteoarthritis; a successful clinical trial in Iran. Int Immunopharmacol 2020, 85, 106607. [Google Scholar] [CrossRef] [PubMed]
- Panahi, Y.; Alishiri, G.H.; Parvin, S.; Sahebkar, A. Mitigation of Systemic Oxidative Stress by Curcuminoids in Osteoarthritis: Results of a Randomized Controlled Trial. J. Diet. Suppl. 2016, 13, 209–220. [Google Scholar] [CrossRef]
- Dai, W.; Jin, P.; Li, X.; Zhao, J.; Lan, Y.; Li, H.; Zheng, L. A carrier-free nano-drug assembled via π-π stacking interaction for the treatment of osteoarthritis. Biomed Pharmacother 2023, 164, 114881. [Google Scholar] [CrossRef]
- Yeh, C.C.; Su, Y.H.; Lin, Y.J.; Chen, P.J.; Shi, C.S.; Chen, C.N.; Chang, H.I. Evaluation of the protective effects of curcuminoid (curcumin and bisdemethoxycurcumin)-loaded liposomes against bone turnover in a cell-based model of osteoarthritis. Drug Des. Dev. Ther. 2015, 9, 2285–2300. [Google Scholar] [CrossRef]
- Ghumman, S.A.; Ijaz, A.; Noreen, S.; Aslam, A.; Kausar, R.; Irfan, A.; Latif, S.; Shazly, G.A.; Shah, P.A.; Rana, M. , et al. Formulation and Characterization of Curcumin Niosomes: Antioxidant and Cytotoxicity Studies. Pharm. (Basel Switz. ) 2023, 16. [Google Scholar] [CrossRef]
- Tang, S.; Gao, Y.; Wang, W.; Wang, Y.; Liu, P.; Shou, Z.; Yang, R.; Jin, C.; Zan, X.; Wang, C. , et al. Self-Report Amphiphilic Polymer-Based Drug Delivery System with ROS-Triggered Drug Release for Osteoarthritis Therapy. ACS Macro Lett. 2024, 13, 58–64. [Google Scholar] [CrossRef] [PubMed]
- Kang, C.; Jung, E.; Hyeon, H.; Seon, S.; Lee, D. Acid-activatable polymeric curcumin nanoparticles as therapeutic agents for osteoarthritis. Nanomedicine 2020, 23, 102104. [Google Scholar] [CrossRef]
- Lombardi, A.F.; Ma, Y.; Jang, H.; Jerban, S.; Tang, Q.; Searleman, A.C.; Meyer, R.S.; Du, J.; Chang, E.Y. AcidoCEST-UTE MRI Reveals an Acidic Microenvironment in Knee Osteoarthritis. Int J Mol Sci 2022, 23. [Google Scholar] [CrossRef]
- Zhang, M.; Peng, X.; Ding, Y.; Ke, X.; Ren, K.; Xin, Q.; Qin, M.; Xie, J.; Li, J. A cyclic brush zwitterionic polymer based pH-responsive nanocarrier-mediated dual drug delivery system with lubrication maintenance for osteoarthritis treatment. Mater. Horiz. 2023, 10, 2554–2567. [Google Scholar] [CrossRef] [PubMed]
- Niazvand, F.; Khorsandi, L.; Abbaspour, M.; Orazizadeh, M.; Varaa, N.; Maghzi, M.; Ahmadi, K. Curcumin-loaded poly lactic-co-glycolic acid nanoparticles effects on mono-iodoacetate -induced osteoarthritis in rats. Vet Res Forum 2017, 8, 155–161. [Google Scholar]
- Hamdalla, H.M.; Ahmed, R.R.; Galaly, S.R.; Naguib, I.A.; Alghamdi, B.S.; Ahmed, O.M.; Farghali, A.; Abdul-Hamid, M. Ameliorative Effect of Curcumin Nanoparticles against Monosodium Iodoacetate-Induced Knee Osteoarthritis in Rats. Mediat. Inflamm. 2022, 2022, 8353472. [Google Scholar] [CrossRef]
- Wang, J.; Wang, X.; Cao, Y.; Huang, T.; Song, D.X.; Tao, H.R. Therapeutic potential of hyaluronic acid/chitosan nanoparticles for the delivery of curcuminoid in knee osteoarthritis and an in vitro evaluation in chondrocytes. Int J Mol Med 2018, 42, 2604–2614. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Zhang, L.; Zhu, J.; Gu, J.; Wang, X.; Tao, H. Hyaluronic Acid Modified Curcumin-Loaded Chitosan Nanoparticles Inhibit Chondrocyte Apoptosis to Attenuate Osteoarthritis via Upregulation of Activator Protein 1 and RUNX Family Transcription Factor 2. J. Biomed. Nanotechnol. 2022, 18, 144–157. [Google Scholar] [CrossRef] [PubMed]
- Sun, Q.; Yin, W.; Ru, X.; Liu, C.; Song, B.; Qian, Z. Dual role of injectable curcumin-loaded microgels for efficient repair of osteoarthritic cartilage injury. Front Bioeng Biotechnol 2022, 10, 994816–994816. [Google Scholar] [CrossRef]
- Ratanavaraporn, J.; Soontornvipart, K.; Shuangshoti, S.; Shuangshoti, S.; Damrongsakkul, S. Localized delivery of curcumin from injectable gelatin/Thai silk fibroin microspheres for anti-inflammatory treatment of osteoarthritis in a rat model. 2017.
- Crivelli, B.; Bari, E.; Perteghella, S.; Catenacci, L.; Sorrenti, M.; Mocchi, M.; Faragò, S.; Tripodo, G.; Prina-Mello, A.; Torre, M.L. Silk fibroin nanoparticles for celecoxib and curcumin delivery: ROS-scavenging and anti-inflammatory activities in an in vitro model of osteoarthritis. Eur. J. Pharm. Biopharm. : Off. J. Arbeitsgemeinschaft Fur Pharm. Verfahrenstechnik E.V 2019, 137, 37–45. [Google Scholar] [CrossRef] [PubMed]
- Yabas, M.; Orhan, C.; Er, B.; Tuzcu, M.; Durmus, A.S.; Ozercan, I.H.; Sahin, N.; Bhanuse, P.; Morde, A.A.; Padigaru, M.; et al. A Next Generation Formulation of Curcumin Ameliorates Experimentally Induced Osteoarthritis in Rats via Regulation of Inflammatory Mediators. Front Immunol 2021, 12, 609629. [Google Scholar] [CrossRef] [PubMed]
- Gugliandolo, E.A.-O.; Peritore, A.A.-O.X.; Impellizzeri, D.; Cordaro, M.A.-O.; Siracusa, R.A.-O.; Fusco, R.A.-O.; D’Amico, R.A.-O.; Paola, R.A.-O.; Schievano, C.; Cuzzocrea, S.A.-O.; et al. Dietary Supplementation with Palmitoyl-Glucosamine Co-Micronized with Curcumin Relieves Osteoarthritis Pain and Benefits Joint Mobility. LID - 10.3390/ani10101827 [doi] LID - 1827. 2020.
- Della Rocca, G.; Schievano, C.; Di Salvo, A.; Conti, M.B.; Della Valle, M.F. Palmitoyl-glucosamine co-micronized with curcumin for maintenance of meloxicam-induced pain relief in dogs with osteoarthritis pain. BMC Vet. Res. 2023, 19, 37. [Google Scholar] [CrossRef] [PubMed]
- Park, H.J.; Lee, C.K.; Song, S.H.; Yun, J.H.; Lee, A.; Park, H.J. Highly bioavailable curcumin powder suppresses articular cartilage damage in rats with mono-iodoacetate (MIA)-induced osteoarthritis. Food Sci Biotechnol 2020, 29, 251–263. [Google Scholar] [CrossRef]
- Nakagawa, Y.; Mukai, S.; Yamada, S.; Matsuoka, M.; Tarumi, E.; Hashimoto, T.; Tamura, C.; Imaizumi, A.; Nishihira, J.; Nakamura, T. Short-term effects of highly-bioavailable curcumin for treating knee osteoarthritis: a randomized, double-blind, placebo-controlled prospective study. J. Orthop. Sci. : Off. J. Jpn. Orthop. Assoc. 2014, 19, 933–939. [Google Scholar] [CrossRef] [PubMed]
- Nakagawa, Y.; Mukai, S.; Yamada, S.; Murata, S.; Yabumoto, H.; Maeda, T.; Akamatsu, S. The Efficacy and Safety of Highly-Bioavailable Curcumin for Treating Knee Osteoarthritis: A 6-Month Open-Labeled Prospective Study. Clin. Med. Insights. Arthritis Musculoskelet. Disord. 2020, 13, 1179544120948471. [Google Scholar] [CrossRef] [PubMed]
- Nakagawa, Y.; Mori, K.; Yamada, S.; Mukai, S.; Hirose, A.; Nakamura, R. The Oral Administration of Highly-Bioavailable Curcumin for One Year Has Clinical and Chondro-Protective Effects: A Randomized, Double-Blinded, Placebo-Controlled Prospective Study. Arthrosc. Sports Med. Rehabil. 2022, 4, e393–e402. [Google Scholar] [CrossRef]
- Hashemzadeh, K.; Davoudian, N.; Jaafari, R.M.; Mirfeizi, Z. The Effect of Nanocurcumin in Improvement of Knee Osteoarthritis: A Randomized Clinical Trial. Curr. Rheumatol. Rev. 2020, 16, 158–164. [Google Scholar] [CrossRef] [PubMed]
- Atabaki, M.; Shariati-Sarabi, Z.; Tavakkol-Afshari, J.; Taghipour, A.; Jafari, M.R.; Nikpoor, A.R.; Mohammadi, M. Curcumin as an effective suppressor of miRNA expression in patients with knee osteoarthritis. Avicenna J. Phytomedicine 2022, 12, 346–356. [Google Scholar] [CrossRef]
- Gupte, P.A.; Giramkar, S.A.; Harke, S.M.; Kulkarni, S.K.; Deshmukh, A.P.; Hingorani, L.L.; Mahajan, M.P.; Bhalerao, S.S. Evaluation of the efficacy and safety of Capsule Longvida(®) Optimized Curcumin (solid lipid curcumin particles) in knee osteoarthritis: a pilot clinical study. J Inflamm Res 2019, 12, 145–152. [Google Scholar] [CrossRef] [PubMed]
- Baharizade, M.; Ghetmiri, S.I.; Mohammady, M.; Mohammadi-Samani, S.; Yousefi, G. Revolutionizing Knee Osteoarthritis Treatment: Innovative Self-Nano-Emulsifying Polyethylene Glycol Organogel of Curcumin for Effective Topical Delivery. AAPS PharmSciTech 2024, 25, 80. [Google Scholar] [CrossRef]
- Deng, W.; He, Q.; Zhang, W. Analysis of the mechanism of curcumin against osteoarthritis using metabolomics and transcriptomics. Naunyn-Schmiedeberg’s Arch. Pharmacol. 2024, 397, 3313–3329. [Google Scholar] [CrossRef]
- Swallow, J.; Seidler, K.; Barrow, M. The mechanistic role of curcumin on matrix metalloproteinases in osteoarthritis. Fitoterapia 2024, 174, 105870. [Google Scholar] [CrossRef]
- Rao, C.V. Regulation of COX and LOX by curcumin. Adv Exp Med Biol 2007, 595, 213–226. [Google Scholar] [CrossRef]
- Shep, D.; Khanwelkar, C.; Gade, P.; Karad, S. Safety and efficacy of curcumin versus diclofenac in knee osteoarthritis: a randomized open-label parallel-arm study. Trials 2019, 20, 214. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Jung, J.; Lee, M.; Kim, M.; Kang, N.; Kim, O.K.; Lee, J. Curcuma longa L. extract exhibits anti-inflammatory and cytoprotective functions in the articular cartilage of monoiodoacetate-injected rats. Food Nutr. Res. 2024, 68. [Google Scholar] [CrossRef] [PubMed]
- Mathy-Hartert, M.; Jacquemond-Collet, I.; Priem, F.; Sanchez, C.; Lambert, C.; Henrotin, Y. Curcumin inhibits pro-inflammatory mediators and metalloproteinase-3 production by chondrocytes. Inflammation research : official journal of the European Histamine Research Society... [et al. 2009, 58, 899–908. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Egan, B.; Wang, J. Epigenetic mechanisms underlying the aberrant catabolic and anabolic activities of osteoarthritic chondrocytes. Int. J. Biochem. Cell Biol. 2015, 67, 101–109. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Theleman, J.L.; Lygrisse, K.A.; Wang, J. Epigenetic Mechanisms Underlying the Aging of Articular Cartilage and Osteoarthritis. Gerontology 2019, 65, 387–396. [Google Scholar] [CrossRef]
- Huber, L.C.; Brock, M.; Hemmatazad, H.; Giger, O.T.; Moritz, F.; Trenkmann, M.; Distler, J.H.; Gay, R.E.; Kolling, C.; Moch, H.; et al. Histone deacetylase/acetylase activity in total synovial tissue derived from rheumatoid arthritis and osteoarthritis patients. Arthritis Rheum 2007, 56, 1087–1093. [Google Scholar] [CrossRef] [PubMed]
- Furumatsu, T.; Tsuda, M.; Yoshida, K.; Taniguchi, N.; Ito, T.; Hashimoto, M.; Ito, T.; Asahara, H. Sox9 and p300 cooperatively regulate chromatin-mediated transcription. J. Biol. Chem. 2005, 280, 35203–35208. [Google Scholar] [CrossRef]
- Liu, C.J.; Zhang, Y.; Xu, K.; Parsons, D.; Alfonso, D.; Di Cesare, P.E. Transcriptional activation of cartilage oligomeric matrix protein by Sox9, Sox5, and Sox6 transcription factors and CBP/p300 coactivators. Front. Biosci. : A J. Virtual Libr. 2007, 12, 3899–3910. [Google Scholar] [CrossRef]
- Wan, C.; Zhang, F.; Yao, H.; Li, H.; Tuan, R.S. Histone Modifications and Chondrocyte Fate: Regulation and Therapeutic Implications. Front. Cell Dev. Biol. 2021, 9, 626708. [Google Scholar] [CrossRef]
- Huang, L.; Li, P.; Guo, L.; Li, L.; Yuan, J.; Zhao, R.; Li, H.; Wei, X. Zinc finger protein 521 attenuates osteoarthritis via the histone deacetylases 4 in the nucleus. Bioengineered 2022, 13, 14489–14502. [Google Scholar] [CrossRef]
- Hong, S.; Derfoul, A.; Pereira-Mouries, L.; Hall, D.J. A novel domain in histone deacetylase 1 and 2 mediates repression of cartilage-specific genes in human chondrocytes. Faseb J 2009, 23, 3539–3552. [Google Scholar] [CrossRef]
- Meng, F.; Li, Z.; Zhang, Z.; Yang, Z.; Kang, Y.; Zhao, X.; Long, D.; Hu, S.; Gu, M.; He, S.; et al. MicroRNA-193b-3p regulates chondrogenesis and chondrocyte metabolism by targeting HDAC3. Theranostics 2018, 8, 2862–2883. [Google Scholar] [CrossRef]
- Higashiyama, R.; Miyaki, S.; Yamashita, S.; Yoshitaka, T.; Lindman, G.; Ito, Y.; Sasho, T.; Takahashi, K.; Lotz, M.; Asahara, H. Correlation between MMP-13 and HDAC7 expression in human knee osteoarthritis. Mod. Rheumatol. 2010, 20, 11–17. [Google Scholar] [CrossRef] [PubMed]
- Bradley, E.W.; Carpio, L.R.; Olson, E.N.; Westendorf, J.J. Histone deacetylase 7 (Hdac7) suppresses chondrocyte proliferation and β-catenin activity during endochondral ossification. J. Biol. Chem. 2015, 290, 118–126. [Google Scholar] [CrossRef] [PubMed]
- Khan, N.M.; Haqqi, T.M. Epigenetics in osteoarthritis: Potential of HDAC inhibitors as therapeutics. Pharmacol Res 2018, 128, 73–79. [Google Scholar] [CrossRef]
- Wang, J.; Wang, X.; Ding, X.; Huang, T.; Song, D.; Tao, H. EZH2 is associated with cartilage degeneration in osteoarthritis by promoting SDC1 expression via histone methylation of the microRNA-138 promoter. Lab. Investig. ; A J. Tech. Methods Pathol. 2021, 101, 600–611. [Google Scholar] [CrossRef]
- Chen, L.; Wu, Y.; Wu, Y.; Wang, Y.; Sun, L.; Li, F. The inhibition of EZH2 ameliorates osteoarthritis development through the Wnt/β-catenin pathway. Sci Rep 2016, 6, 29176. [Google Scholar] [CrossRef] [PubMed]
- Camilleri, E.T.; Dudakovic, A.; Riester, S.M.; Galeano-Garces, C.; Paradise, C.R.; Bradley, E.W.; McGee-Lawrence, M.E.; Im, H.J.; Karperien, M.; Krych, A.J. , et al. Loss of histone methyltransferase Ezh2 stimulates an osteogenic transcriptional program in chondrocytes but does not affect cartilage development. J. Biol. Chem. 2018, 293, 19001–19011. [Google Scholar] [CrossRef]
- Allas, L.; Brochard, S.; Rochoux, Q.; Ribet, J.; Dujarrier, C.; Veyssiere, A.; Aury-Landas, J.; Grard, O.; Leclercq, S.; Vivien, D. , et al. EZH2 inhibition reduces cartilage loss and functional impairment related to osteoarthritis. Sci Rep 2020, 10, 19577. [Google Scholar] [CrossRef]
- Shao, R.; Suo, J.; Zhang, Z.; Kong, M.; Ma, Y.; Wen, Y.; Liu, M.; Zhuang, L.; Ge, K.; Bi, Q. , et al. H3K36 methyltransferase NSD1 protects against osteoarthritis through regulating chondrocyte differentiation and cartilage homeostasis. Cell Death Differ. 2024, 31, 106–118. [Google Scholar] [CrossRef]
- Thulson, E.; Davis, E.S.; D’Costa, S.; Coryell, P.R.; Kramer, N.E.; Mohlke, K.L.; Loeser, R.F.; Diekman, B.O.; Phanstiel, D.H. 3D chromatin structure in chondrocytes identifies putative osteoarthritis risk genes. Genetics 2022, 222. [Google Scholar] [CrossRef] [PubMed]
- Bittner, N.; Shi, C.; Zhao, D.; Ding, J.; Southam, L.; Swift, D.; Kreitmaier, P.; Tutino, M.; Stergiou, O.; Cheung, J.T.S. , et al. Primary osteoarthritis chondrocyte map of chromatin conformation reveals novel candidate effector genes. Ann. Rheum. Dis. 2024. [Google Scholar] [CrossRef]
- Alvarez-Garcia, O.; Fisch, K.M.; Wineinger, N.E.; Akagi, R.; Saito, M.; Sasho, T.; Su, A.I.; Lotz, M.K. Increased DNA Methylation and Reduced Expression of Transcription Factors in Human Osteoarthritis Cartilage. Arthritis Rheumatol. (Hoboken N.J.) 2016, 68, 1876–1886. [Google Scholar] [CrossRef] [PubMed]
- Rushton, M.D.; Reynard, L.N.; Barter, M.J.; Refaie, R.; Rankin, K.S.; Young, D.A.; Loughlin, J. Characterization of the cartilage DNA methylome in knee and hip osteoarthritis. Arthritis Rheumatol. (Hoboken N.J.) 2014, 66, 2450–2460. [Google Scholar] [CrossRef] [PubMed]
- Lafont, J.E.; Moustaghfir, S.; Durand, A.-L.; Mallein-Gerin, F. The epigenetic players and the chromatin marks involved in the articular cartilage during osteoarthritis. Front. Physiol. 2023, 14. [Google Scholar] [CrossRef]
- Imagawa, K.; de Andrés, M.C.; Hashimoto, K.; Itoi, E.; Otero, M.; Roach, H.I.; Goldring, M.B.; Oreffo, R.O. Association of reduced type IX collagen gene expression in human osteoarthritic chondrocytes with epigenetic silencing by DNA hypermethylation. Arthritis Rheumatol. (Hoboken N.J.) 2014, 66, 3040–3051. [Google Scholar] [CrossRef]
- Kim, K.I.; Park, Y.S.; Im, G.I. Changes in the epigenetic status of the SOX-9 promoter in human osteoarthritic cartilage. J. Bone Miner. Res. : Off. J. Am. Soc. Bone Miner. Res. 2013, 28, 1050–1060. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Xiao, L.; Chen, J.; Chen, X.; Chen, X.; Yao, S.; Li, H.; Zhao, G.; Ma, J. DNA methylation is involved in the pathogenesis of osteoarthritis by regulating CtBP expression and CtBP-mediated signaling. Int. J. Biol. Sci. 2020, 16, 994–1009. [Google Scholar] [CrossRef]
- Iliopoulos, D.; Malizos, K.N.; Tsezou, A. Epigenetic regulation of leptin affects MMP-13 expression in osteoarthritic chondrocytes: possible molecular target for osteoarthritis therapeutic intervention. Ann. Rheum. Dis. 2007, 66, 1616–1621. [Google Scholar] [CrossRef]
- Jeffries, M.A.; Donica, M.; Baker, L.W.; Stevenson, M.E.; Annan, A.C.; Beth Humphrey, M.; James, J.A.; Sawalha, A.H. Genome-Wide DNA Methylation Study Identifies Significant Epigenomic Changes in Osteoarthritic Subchondral Bone and Similarity to Overlying Cartilage. Arthritis Rheumatol. (Hoboken N.J.) 2016, 68, 1403–1414. [Google Scholar] [CrossRef]
- Yang, J.; Wang, N. Genome-wide expression and methylation profiles reveal candidate genes and biological processes underlying synovial inflammatory tissue of patients with osteoarthritis. Int. J. Rheum. Dis. 2015, 18, 783–790. [Google Scholar] [CrossRef]
- Fernández-Tajes, J.; Soto-Hermida, A.; Vázquez-Mosquera, M.E.; Cortés-Pereira, E.; Mosquera, A.; Fernández-Moreno, M.; Oreiro, N.; Fernández-López, C.; Fernández, J.L.; Rego-Pérez, I. , et al. Genome-wide DNA methylation analysis of articular chondrocytes reveals a cluster of osteoarthritic patients. Ann. Rheum. Dis. 2014, 73, 668–677. [Google Scholar] [CrossRef] [PubMed]
- Lin, X.; Li, L.; Liu, X.; Tian, J.; Zheng, W.; Li, J.; Wang, L. Genome-wide analysis of aberrant methylation of enhancer DNA in human osteoarthritis. BMC Med. Genom. 2020, 13, 1. [Google Scholar] [CrossRef] [PubMed]
- Papathanasiou, I.; Trachana, V.; Mourmoura, E.; Tsezou, A. DNA methylation regulates miR-140-5p and miR-146a expression in osteoarthritis. Life Sci 2019, 228, 274–284. [Google Scholar] [CrossRef]
- Fisch, K.M.; Gamini, R.; Alvarez-Garcia, O.; Akagi, R.; Saito, M.; Muramatsu, Y.; Sasho, T.; Koziol, J.A.; Su, A.I.; Lotz, M.K. Identification of transcription factors responsible for dysregulated networks in human osteoarthritis cartilage by global gene expression analysis. Osteoarthr. Cartil. 2018, 26, 1531–1538. [Google Scholar] [CrossRef]
- Yue, J.; Aobulikasimu, A.; Sun, W.; Liu, S.; Xie, W.; Sun, W. Targeted regulation of FoxO1 in chondrocytes prevents age-related osteoarthritis via autophagy mechanism. J Cell Mol Med 2022, 26, 3075–3082. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Zhang, S.; Previn, R.; Chen, D.; Jin, Y.; Zhou, G. Role of Forkhead Box O Transcription Factors in Oxidative Stress-Induced Chondrocyte Dysfunction: Possible Therapeutic Target for Osteoarthritis? Int J Mol Sci 2018, 19. [Google Scholar] [CrossRef]
- Matsuzaki, T.; Alvarez-Garcia, O.; Mokuda, S.; Nagira, K.; Olmer, M.; Gamini, R.; Miyata, K.; Akasaki, Y.; Su, A.I.; Asahara, H. , et al. FoxO transcription factors modulate autophagy and proteoglycan 4 in cartilage homeostasis and osteoarthritis. Sci. Transl. Med. 2018, 10. [Google Scholar] [CrossRef]
- Ohzono, H.; Hu, Y.; Nagira, K.; Kanaya, H.; Okubo, N.; Olmer, M.; Gotoh, M.; Kurakazu, I.; Akasaki, Y.; Kawata, M. , et al. Targeting FoxO transcription factors with HDAC inhibitors for the treatment of osteoarthritis. Ann. Rheum. Dis. 2023, 82, 262–271. [Google Scholar] [CrossRef]
- Ahmed, E.A.; Alzahrani, A.M. SOXC Transcription Factors as Diagnostic Biomarkers and Therapeutic Targets for Arthritis. Int J Mol Sci 2023, 24. [Google Scholar] [CrossRef]
- Wang, Y.P.; Di, W.J.; Yang, S.; Qin, S.L.; Xu, Y.F.; Han, P.F.; Hou, K.D. The association of growth differentiation factor 5 rs143383 gene polymorphism with osteoarthritis: a systematic review and meta-analysis. J. Orthop. Surg. Res. 2023, 18, 763. [Google Scholar] [CrossRef]
- Zhang, R.; Yao, J.; Xu, P.; Ji, B.; Luck, J.V.; Chin, B.; Lu, S.; Kelsoe, J.R.; Ma, J. A comprehensive meta-analysis of association between genetic variants of GDF5 and osteoarthritis of the knee, hip and hand. Inflammation research : official journal of the European Histamine Research Society... [et al.] 2015, 64, 405–414. [Google Scholar] [CrossRef] [PubMed]
- Murakami, T.; Ruengsinpinya, L.; Takahata, Y.; Nakaminami, Y.; Hata, K.; Nishimura, R. HOXA10 promotes Gdf5 expression in articular chondrocytes. Sci Rep 2023, 13, 22778. [Google Scholar] [CrossRef] [PubMed]
- Razmara, E.; Bitaraf, A.; Yousefi, H.; Nguyen, T.H.; Garshasbi, M.; Cho, W.C.; Babashah, S. Non-Coding RNAs in Cartilage Development: An Updated Review. Int J Mol Sci 2019, 20. [Google Scholar] [CrossRef] [PubMed]
- Kong, H.; Sun, M.L.; Zhang, X.A.; Wang, X.Q. Crosstalk Among circRNA/lncRNA, miRNA, and mRNA in Osteoarthritis. Front. Cell Dev. Biol. 2021, 9, 774370. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Zhang, H.; Xie, Q.; Feng, H.; Li, H.; Li, Z.; Yang, K.; Ding, J.; Gao, G. LncRNA-mediated cartilage homeostasis in osteoarthritis: a narrative review. Front Med (Lausanne) 2024, 11, 1326843. [Google Scholar] [CrossRef]
- Wang, R.; Shiu, H.T.; Lee, W.Y.W. Emerging role of lncRNAs in osteoarthritis: An updated review. Front Immunol 2022, 13, 982773. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, F.; Chen, G.; He, R.; Yang, L. LncRNA MALAT1 promotes osteoarthritis by modulating miR-150-5p/AKT3 axis. Cell Biosci. 2019, 9, 54. [Google Scholar] [CrossRef]
- Dang, X.; Lian, L.; Wu, D. The diagnostic value and pathogenetic role of lncRNA-ATB in patients with osteoarthritis. Cell. Mol. Biol. Lett. 2018, 23, 55. [Google Scholar] [CrossRef]
- Wang, Q.; Yang, J.; Pan, R.; Zha, Z. LncRNA SNHG1 overexpression alleviates osteoarthritis via activating PI3K/Akt signal pathway and suppressing autophagy. Immunobiology 2024, 229, 152799. [Google Scholar] [CrossRef]
- Gu, X.; Xie, T. LncRNA AC005165.1 Alleviates IL-1β-Induced Osteoarthritis via miR-199a-3p/TXNIP Axis. Biochem. Genet. 2024. [Google Scholar] [CrossRef]
- Ma, X.; Cai, D.; Zhu, Y.; Zhao, Y.; Shang, X.; Wang, C.; Zhang, H.; Bian, A.; Yu, H.; Cheng, W. L-Glutamine alleviates osteoarthritis by regulating lncRNA-NKILA expression through the TGF-β1/SMAD2/3 signalling pathway. Clinical science (London, England : 1979) 2022, 136, 1053–1069. [Google Scholar] [CrossRef]
- Losko, M.; Kotlinowski, J.; Jura, J. Long Noncoding RNAs in Metabolic Syndrome Related Disorders. Mediat. Inflamm. 2016, 2016, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Cao, Y.; Chang, C.; Huang, W.; Su, S.; Peng, Z.; Zhang, J. Knockdown of circSOD2 ameliorates osteoarthritis progression via the miR-224-5p/PRDX3 axis. J. Orthop. Surg. Res. 2023, 18, 432. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Wang, Y.; Wu, X. Knockdown of Circ_0037658 Alleviates IL-1β-Induced Osteoarthritis Progression by Serving as a Sponge of miR-665 to Regulate ADAMTS5. Front. Genet. 2022, 13, 886898. [Google Scholar] [CrossRef] [PubMed]
- Qian, W.; Mei, K.; Zhu, L.; Chu, Y.; Lv, J.; Yun, C. Circ_0044235 regulates the development of osteoarthritis by the modulation of miR-375/PIK3R3 axis. J. Orthop. Surg. Res. 2024, 19, 241. [Google Scholar] [CrossRef] [PubMed]
- Li, H.Z.; Lin, Z.; Xu, X.H.; Lin, N.; Lu, H.D. The potential roles of circRNAs in osteoarthritis: a coming journey to find a treasure. Biosci. Rep. 2018, 38. [Google Scholar] [CrossRef]
- Lian, W.S.; Ko, J.Y.; Wu, R.W.; Sun, Y.C.; Chen, Y.S.; Wu, S.L.; Weng, L.H.; Jahr, H.; Wang, F.S. MicroRNA-128a represses chondrocyte autophagy and exacerbates knee osteoarthritis by disrupting Atg12. Cell Death Dis 2018, 9, 919. [Google Scholar] [CrossRef]
- Cao, Y.; Tang, S.; Nie, X.; Zhou, Z.; Ruan, G.; Han, W.; Zhu, Z.; Ding, C. Decreased miR-214-3p activates NF-κB pathway and aggravates osteoarthritis progression. EBioMedicine 2021, 65, 103283. [Google Scholar] [CrossRef]
- Li, F.; Yao, J.; Hao, Q.; Duan, Z. miRNA-103 promotes chondrocyte apoptosis by down-regulation of Sphingosine kinase-1 and ameliorates PI3K/AKT pathway in osteoarthritis. Biosci. Rep. 2019, 39. [Google Scholar] [CrossRef]
- Kopańska, M.; Szala, D.; Czech, J.; Gabło, N.; Gargasz, K.; Trzeciak, M.; Zawlik, I.; Snela, S. MiRNA expression in the cartilage of patients with osteoarthritis. J. Orthop. Surg. Res. 2017, 12, 51. [Google Scholar] [CrossRef]
- Zheng, W.D.; Zhou, F.L.; Lin, N.; Liu, J. Investigation for the role of CTX-III and microRNA-98 in diagnosis and treatment of osteoarthritis. Eur Rev Med Pharmacol Sci 2018, 22, 5424–5428. [Google Scholar] [CrossRef]
- Song, J.; Jin, E.H.; Kim, D.; Kim, K.Y.; Chun, C.H.; Jin, E.J. MicroRNA-222 regulates MMP-13 via targeting HDAC-4 during osteoarthritis pathogenesis. BBA Clin. 2015, 3, 79–89. [Google Scholar] [CrossRef] [PubMed]
- Xu, B.; Li, Y.Y.; Ma, J.; Pei, F.X. Roles of microRNA and signaling pathway in osteoarthritis pathogenesis. J. Zhejiang University. Science. B 2016, 17, 200–208. [Google Scholar] [CrossRef] [PubMed]
- Shorter, E.; Avelar, R.; Zachariou, M.; Spyrou, G.M.; Raina, P.; Smagul, A.; Ashraf Kharaz, Y.; Peffers, M.; Goljanek-Whysall, K.; de Magalhães, J.P. , et al. Identifying Novel Osteoarthritis-Associated Genes in Human Cartilage Using a Systematic Meta-Analysis and a Multi-Source Integrated Network. Int J Mol Sci 2022, 23. [Google Scholar] [CrossRef]
- Jiang, Y.; Shen, Y.; Ding, L.; Xia, S.; Jiang, L. Identification of transcription factors and construction of a novel miRNA regulatory network in primary osteoarthritis by integrated analysis. BMC Musculoskelet. Disord. 2021, 22, 1008. [Google Scholar] [CrossRef] [PubMed]
- Reuter, S.; Gupta, S.C.; Park, B.; Goel, A.; Aggarwal, B.B. Epigenetic changes induced by curcumin and other natural compounds. Genes Nutr. 2011, 6, 93–108. [Google Scholar] [CrossRef] [PubMed]
- Arias, C.; Salazar, L.A. Autophagy and Polyphenols in Osteoarthritis: A Focus on Epigenetic Regulation. Int J Mol Sci 2021, 23. [Google Scholar] [CrossRef]
- Joven, J.; Micol, V.; Segura-Carretero, A.; Alonso-Villaverde, C.; Menéndez, J.A. Polyphenols and the modulation of gene expression pathways: can we eat our way out of the danger of chronic disease? Crit. Rev. Food Sci. Nutr. 2014, 54, 985–1001. [Google Scholar] [CrossRef]
- Boyanapalli, S.S.S.; Huang, Y.; Su, Z.; Cheng, D.; Zhang, C.; Guo, Y.; Rao, R.; Androulakis, I.P.; Kong, A.N. Pharmacokinetics and Pharmacodynamics of Curcumin in regulating anti-inflammatory and epigenetic gene expression. Biopharm. Drug Dispos. 2018, 39, 289–297. [Google Scholar] [CrossRef]
- Cheng, D.; Li, W.; Wang, L.; Lin, T.; Poiani, G.; Wassef, A.; Hudlikar, R.; Ondar, P.; Brunetti, L.; Kong, A.N. Pharmacokinetics, Pharmacodynamics, and PKPD Modeling of Curcumin in Regulating Antioxidant and Epigenetic Gene Expression in Healthy Human Volunteers. Mol Pharm 2019, 16, 1881–1889. [Google Scholar] [CrossRef]
- Marcu, M.G.; Jung, Y.J.; Lee, S.; Chung, E.J.; Lee, M.J.; Trepel, J.; Neckers, L. Curcumin is an inhibitor of p300 histone acetylatransferase. Med. Chem. (Shariqah (United Arab Emir. )) 2006, 2, 169–174. [Google Scholar] [CrossRef]
- Yuan, Z.; Syed, M.A.; Panchal, D.; Rogers, D.; Joo, M.; Sadikot, R.T. Curcumin mediated epigenetic modulation inhibits TREM-1 expression in response to lipopolysaccharide. Int. J. Biochem. Cell Biol. 2012, 44, 2032–2043. [Google Scholar] [CrossRef]
- Hassan, F.U.; Rehman, M.S.; Khan, M.S.; Ali, M.A.; Javed, A.; Nawaz, A.; Yang, C. Curcumin as an Alternative Epigenetic Modulator: Mechanism of Action and Potential Effects. Front. Genet. 2019, 10, 514. [Google Scholar] [CrossRef] [PubMed]
- Sanchez, C.; Zappia, J.; Lambert, C.; Foguenne, J.; Dierckxsens, Y.; Dubuc, J.-E.; Delcour, J.-P.; Gothot, A.; Henrotin, Y. Curcuma longa and Boswellia serrata Extracts Modulate Different and Complementary Pathways on Human Chondrocytes In Vitro: Deciphering of a Transcriptomic Study. Front. Pharmacol. 2022, 13. [Google Scholar] [CrossRef] [PubMed]
- Mishra, S.; Verma, S.S.; Rai, V.; Awasthee, N.; Chava, S.; Hui, K.M.; Kumar, A.P.; Challagundla, K.B.; Sethi, G.; Gupta, S.C. Long non-coding RNAs are emerging targets of phytochemicals for cancer and other chronic diseases. Cell Mol Life Sci 2019, 76, 1947–1966. [Google Scholar] [CrossRef] [PubMed]
- Shishodia, S.; Singh, T.; Chaturvedi, M.M. Modulation of transcription factors by curcumin. Adv Exp Med Biol 2007, 595, 127–148. [Google Scholar] [CrossRef]
- Nguyen, H.D.; Kim, M.S. The protective effects of curcumin on metabolic syndrome and its components: In-silico analysis for genes, transcription factors, and microRNAs involved. Arch. Biochem. Biophys. 2022, 727, 109326. [Google Scholar] [CrossRef]
- JZ, A.L.; AlFaris, N.A.; Al-Farga, A.M.; Alshammari, G.M.; BinMowyna, M.N.; Yahya, M.A. Curcumin reverses diabetic nephropathy in streptozotocin-induced diabetes in rats by inhibition of PKCβ/p(66)Shc axis and activation of FOXO-3a. J Nutr Biochem 2021, 87, 108515. [Google Scholar] [CrossRef]
- Li, X.; Zhu, R.; Jiang, H.; Yin, Q.; Gu, J.; Chen, J.; Ji, X.; Wu, X.; Fu, H.; Wang, H. , et al. Autophagy enhanced by curcumin ameliorates inflammation in atherogenesis via the TFEB-P300-BRD4 axis. Acta Pharm. Sinica. B 2022, 12, 2280–2299. [Google Scholar] [CrossRef]
- Xiang, L.; Nakamura, Y.; Lim, Y.M.; Yamasaki, Y.; Kurokawa-Nose, Y.; Maruyama, W.; Osawa, T.; Matsuura, A.; Motoyama, N.; Tsuda, L. Tetrahydrocurcumin extends life span and inhibits the oxidative stress response by regulating the FOXO forkhead transcription factor. Aging 2011, 3, 1098–1109. [Google Scholar] [CrossRef]
- Arora, I.; Sharma, M.; Tollefsbol, T.O. Combinatorial Epigenetics Impact of Polyphenols and Phytochemicals in Cancer Prevention and Therapy. Int J Mol Sci 2019, 20. [Google Scholar] [CrossRef]
- Yao, J.; Liu, X.; Sun, Y.; Dong, X.; Liu, L.; Gu, H. Curcumin-Alleviated Osteoarthritic Progression in Rats Fed a High-Fat Diet by Inhibiting Apoptosis and Activating Autophagy via Modulation of MicroRNA-34a. J Inflamm Res 2021, 14, 2317–2331. [Google Scholar] [CrossRef] [PubMed]
- Saber, M.M.; Mahmoud, M.M.; Amin, H.M.; Essam, R.M. Therapeutic effects of combining curcumin and swimming in osteoarthritis using a rat model. Biomed Pharmacother 2023, 166, 115309. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Stöckl, S.; Lukas, C.; Herrmann, M.; Brochhausen, C.; König, M.A.; Johnstone, B.; Grässel, S. Curcumin-primed human BMSC-derived extracellular vesicles reverse IL-1β-induced catabolic responses of OA chondrocytes by upregulating miR-126-3p. Stem Cell Res. Ther. 2021, 12, 252. [Google Scholar] [CrossRef] [PubMed]
- Qiu, B.; Xu, X.; Yi, P.; Hao, Y. Curcumin reinforces MSC-derived exosomes in attenuating osteoarthritis via modulating the miR-124/NF-kB and miR-143/ROCK1/TLR9 signalling pathways. J Cell Mol Med 2020, 24, 10855–10865. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.J.; Hou, Y.K.; Chen, M.W.; Yu, X.Z.; Chen, S.Y.; Yue, Y.R.; Guo, X.T.; Chen, J.X.; Zhou, Q. A pH-responsive metal-organic framework for the co-delivery of HIF-2α siRNA and curcumin for enhanced therapy of osteoarthritis. J Nanobiotechnology 2023, 21, 18. [Google Scholar] [CrossRef]
- Tong, K.M.; Shieh, D.C.; Chen, C.P.; Tzeng, C.Y.; Wang, S.P.; Huang, K.C.; Chiu, Y.C.; Fong, Y.C.; Tang, C.H. Leptin induces IL-8 expression via leptin receptor, IRS-1, PI3K, Akt cascade and promotion of NF-kappaB/p300 binding in human synovial fibroblasts. Cell. Signal. 2008, 20, 1478–1488. [Google Scholar] [CrossRef]
- Cai, Z.; Long, T.; Zhao, Y.; Lin, R.; Wang, Y. Epigenetic Regulation in Knee Osteoarthritis. Front. Genet. 2022, 13. [Google Scholar] [CrossRef]
- Caldo, D.; Massarini, E.; Rucci, M.; Deaglio, S.; Ferracini, R. Epigenetics in Knee Osteoarthritis: A 2020–2023 Update Systematic Review. Life 2024, 14, 269. [Google Scholar] [CrossRef]
- Che, X.; Chen, T.; Wei, L.; Gu, X.; Gao, Y.; Liang, S.; Li, P.; Shi, D.; Liang, B.; Wang, C. , et al. MicroRNA-1 regulates the development of osteoarthritis in a Col2a1-Cre-ERT2/GFPfl/fl-RFP-miR-1 mouse model of osteoarthritis through the downregulation of Indian hedgehog expression. Int J Mol Med 2020, 46, 360–370. [Google Scholar] [CrossRef]
- Zhang, C.; Zhang, Z.; Chang, Z.; Mao, G.; Hu, S.; Zeng, A.; Fu, M. miR-193b-5p regulates chondrocytes metabolism by directly targeting histone deacetylase 7 in interleukin-1β-induced osteoarthritis. J Cell Biochem 2019, 120, 12775–12784. [Google Scholar] [CrossRef]
- Mao, G.; Zhang, Z.; Huang, Z.; Chen, W.; Huang, G.; Meng, F.; Zhang, Z.; Kang, Y. MicroRNA-92a-3p regulates the expression of cartilage-specific genes by directly targeting histone deacetylase 2 in chondrogenesis and degradation. Osteoarthr. Cartil. 2017, 25, 521–532. [Google Scholar] [CrossRef] [PubMed]
- Xu, P.; Zhang, X.; Li, Q. Modeling Osteoarthritis: MiR-16-5p Attenuates IL-1β Induced Chondrocyte Dysfunction by Targeting MAP2K1 through the MAPK Pathway. Ann. Clin. Lab. Sci. 2023, 53, 248–258. [Google Scholar] [PubMed]
- Ding, Y.; Wang, L.; Zhao, Q.; Wu, Z.; Kong, L. MicroRNA-93 inhibits chondrocyte apoptosis and inflammation in osteoarthritis by targeting the TLR4/NF-κB signaling pathway. Int J Mol Med 2019, 43, 779–790. [Google Scholar] [CrossRef]
- Dai, Y.; Liu, S.; Xie, X.; Ding, M.; Zhou, Q.; Zhou, X. MicroRNA-31 promotes chondrocyte proliferation by targeting C-X-C motif chemokine ligand 12. Mol Med Rep 2019, 19, 2231–2237. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.D.; Miao, W.H.; Zhang, Y.Y.; Zou, M.J.; Yan, X.F. Inhibition of miR-126 protects chondrocytes from IL-1β induced inflammation via upregulation of Bcl-2. Bone Jt. Res. 2018, 7, 414–421. [Google Scholar] [CrossRef]
- Shi, J.; Guo, K.; Su, S.; Li, J.; Li, C. miR-486-5p is upregulated in osteoarthritis and inhibits chondrocyte proliferation and migration by suppressing SMAD2. Mol Med Rep 2018, 18, 502–508. [Google Scholar] [CrossRef]
- Ji, M.L.; Jiang, H.; Wu, F.; Geng, R.; Ya, L.K.; Lin, Y.C.; Xu, J.H.; Wu, X.T.; Lu, J. Precise targeting of miR-141/200c cluster in chondrocytes attenuates osteoarthritis development. Ann. Rheum. Dis. 2021, 80, 356–366. [Google Scholar] [CrossRef]
- Zhang, L.; Peng, H.; Zhang, W.; Li, Y.; Liu, L.; Leng, T. Yeast Cell wall Particle mediated Nanotube-RNA delivery system loaded with miR365 Antagomir for Post-traumatic Osteoarthritis Therapy via Oral Route. Theranostics 2020, 10, 8479–8493. [Google Scholar] [CrossRef]
- Yan, H.; Duan, X.; Pan, H.; Holguin, N.; Rai, M.F.; Akk, A.; Springer, L.E.; Wickline, S.A.; Sandell, L.J.; Pham, C.T. Suppression of NF-κB activity via nanoparticle-based siRNA delivery alters early cartilage responses to injury. Proc Natl Acad Sci U S A 2016, 113, E6199–e6208. [Google Scholar] [CrossRef]
- Tang, X.; Muhammad, H.; McLean, C.; Miotla-Zarebska, J.; Fleming, J.; Didangelos, A.; Önnerfjord, P.; Leask, A.; Saklatvala, J.; Vincent, T.L. Connective tissue growth factor contributes to joint homeostasis and osteoarthritis severity by controlling the matrix sequestration and activation of latent TGFβ. Ann. Rheum. Dis. 2018, 77, 1372–1380. [Google Scholar] [CrossRef]
- Rai, M.F.; Pan, H.; Yan, H.; Sandell, L.J.; Pham, C.T.N.; Wickline, S.A. Applications of RNA interference in the treatment of arthritis. Transl. Res. : J. Lab. Clin. Med. 2019, 214, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Conte, R.; Finicelli, M.; Borrone, A.; Margarucci, S.; Peluso, G.; Calarco, A.; Bosetti, M. MMP-2 Silencing through siRNA Loaded Positively-Charged Nanoparticles (AcPEI-NPs) Counteracts Chondrocyte De-Differentiation. Polymers 2023, 15. [Google Scholar] [CrossRef]
- Bedingfield, S.K.; Colazo, J.M.; Yu, F.; Liu, D.D.; Jackson, M.A.; Himmel, L.E.; Cho, H.; Crofford, L.J.; Hasty, K.A.; Duvall, C.L. Amelioration of post-traumatic osteoarthritis via nanoparticle depots delivering small interfering RNA to damaged cartilage. Nat. Biomed. Eng. 2021, 5, 1069–1083. [Google Scholar] [CrossRef] [PubMed]
- Bedingfield, S.K.; Colazo, J.M.; Di Francesco, M.; Yu, F.; Liu, D.D.; Di Francesco, V.; Himmel, L.E.; Gupta, M.K.; Cho, H.; Hasty, K.A. , et al. Top-Down Fabricated microPlates for Prolonged, Intra-articular Matrix Metalloproteinase 13 siRNA Nanocarrier Delivery to Reduce Post-traumatic Osteoarthritis. ACS Nano 2021, 15, 14475–14491. [Google Scholar] [CrossRef]
- Yan, Y.; Lu, A.; Dou, Y.; Zhang, Z.; Wang, X.Y.; Zhai, L.; Ai, L.Y.; Du, M.Z.; Jiang, L.X.; Zhu, Y.J. , et al. Nanomedicines Reprogram Synovial Macrophages by Scavenging Nitric Oxide and Silencing CA9 in Progressive Osteoarthritis. Adv. Sci. (Weinh. Baden-Wurtt. Ger. ) 2023, 10, e2207490. [Google Scholar] [CrossRef]
- Kondreddy, V.; Banerjee, R.; Devi, B.; Muralidharan, K.; Piramanayagam, S. Inhibition of the MALT1-LPCAT3 axis protects cartilage degeneration and osteoarthritis. Cell Commun. Signal. : CCS 2024, 22, 189. [Google Scholar] [CrossRef]
- Gabbianelli, R.; Bordoni, L.; Morano, S.; Calleja-Agius, J.; Lalor, J.G. Nutri-Epigenetics and Gut Microbiota: How Birth Care, Bonding and Breastfeeding Can Influence and Be Influenced? Int J Mol Sci 2020, 21. [Google Scholar] [CrossRef] [PubMed]
- Hao, X.; Shang, X.; Liu, J.; Chi, R.; Zhang, J.; Xu, T. The gut microbiota in osteoarthritis: where do we stand and what can we do? Arthritis Res Ther 2021, 23, 42. [Google Scholar] [CrossRef]
- Jeyaraman, M.; Ram, P.R.; Jeyaraman, N.; Yadav, S. The Gut-Joint Axis in Osteoarthritis. Cureus 2023, 15, e48951. [Google Scholar] [CrossRef]
- Favazzo, L.J.; Hendesi, H.; Villani, D.A.; Soniwala, S.; Dar, Q.A.; Schott, E.M.; Gill, S.R.; Zuscik, M.J. The gut microbiome-joint connection: implications in osteoarthritis. Curr. Opin. Rheumatol. 2020, 32, 92–101. [Google Scholar] [CrossRef]
- Wei, Z.; Li, F.; Pi, G. Association Between Gut Microbiota and Osteoarthritis: A Review of Evidence for Potential Mechanisms and Therapeutics. Front. Cell. Infect. Microbiol. 2022, 12, 812596. [Google Scholar] [CrossRef]
- Yu, X.H.; Yang, Y.Q.; Cao, R.R.; Bo, L.; Lei, S.F. The causal role of gut microbiota in development of osteoarthritis. Osteoarthr. Cartil. 2021, 29, 1741–1750. [Google Scholar] [CrossRef] [PubMed]
- Gleason, B.; Chisari, E.; Parvizi, J. Osteoarthritis Can Also Start in the Gut: The Gut-Joint Axis. Indian J. Orthop. 2022, 56, 1150–1155. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Ding, W.; Wang, H.L.; Dai, L.L.; Zong, W.H.; Wang, Y.Z.; Bi, J.; Han, W.; Dong, G.J. Gut microbiota and obesity-associated osteoarthritis. Osteoarthr. Cartil. 2019, 27, 1257–1265. [Google Scholar] [CrossRef]
- Amabebe, E.; Robert, F.O.; Agbalalah, T.; Orubu, E.S.F. Microbial dysbiosis-induced obesity: role of gut microbiota in homoeostasis of energy metabolism. Br. J. Nutr. 2020, 123, 1127–1137. [Google Scholar] [CrossRef]
- Berthelot, J.M.; Sellam, J.; Maugars, Y.; Berenbaum, F. Cartilage-gut-microbiome axis: a new paradigm for novel therapeutic opportunities in osteoarthritis. RMD Open 2019, 5, e001037. [Google Scholar] [CrossRef]
- Arora, V.; Singh, G.; O-Sullivan, I.; Ma, K.; Natarajan Anbazhagan, A.; Votta-Velis, E.G.; Bruce, B.; Richard, R.; van Wijnen, A.J.; Im, H.-J. Gut-microbiota modulation: The impact of thegut-microbiotaon osteoarthritis. Gene 2021, 785, 145619. [Google Scholar] [CrossRef]
- Biver, E.; Berenbaum, F.; Valdes, A.M.; Araujo de Carvalho, I.; Bindels, L.B.; Brandi, M.L.; Calder, P.C.; Castronovo, V.; Cavalier, E.; Cherubini, A. , et al. Gut microbiota and osteoarthritis management: An expert consensus of the European society for clinical and economic aspects of osteoporosis, osteoarthritis and musculoskeletal diseases (ESCEO). Ageing Res. Rev. 2019, 55, 100946. [Google Scholar] [CrossRef]
- Li, D.; Li, Y.; Yang, S.; Lu, J.; Jin, X.; Wu, M. Diet-gut microbiota-epigenetics in metabolic diseases: From mechanisms to therapeutics. Biomed Pharmacother 2022, 153, 113290. [Google Scholar] [CrossRef]
- Cuevas-Sierra, A.; Ramos-Lopez, O.; Riezu-Boj, J.I.; Milagro, F.I.; Martinez, J.A. Diet, Gut Microbiota, and Obesity: Links with Host Genetics and Epigenetics and Potential Applications. Adv. Nutr. (Bethesda Md.) 2019, 10, S17–s30. [Google Scholar] [CrossRef]
- Remely, M.; Aumueller, E.; Merold, C.; Dworzak, S.; Hippe, B.; Zanner, J.; Pointner, A.; Brath, H.; Haslberger, A.G. Effects of short chain fatty acid producing bacteria on epigenetic regulation of FFAR3 in type 2 diabetes and obesity. Gene 2014, 537, 85–92. [Google Scholar] [CrossRef] [PubMed]
- Bhat, M.I.; Kapila, R. Dietary metabolites derived from gut microbiota: critical modulators of epigenetic changes in mammals. Nutr Rev 2017, 75, 374–389. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, S.; Singh, A.; Sandeep, K.; Yadav, D. Epigenetic Regulation of Gut Microbial Dysbiosis. Indian J. Microbiol. 2021, 61, 125–129. [Google Scholar] [CrossRef] [PubMed]
- Woo, V.; Alenghat, T. Epigenetic regulation by gut microbiota. Gut Microbes 2022, 14, 2022407. [Google Scholar] [CrossRef] [PubMed]
- Marín-Tello, C.; Jintaridth, P.; Sanchez, F.; González, C.; Zelada-Castillo, L.; Vásquez-Arqueros, A.; Guevara-Vásquez, A.; Vieira, A. Epigenetic regulation by metabolites from the gut microbiome. Benef. Microbes 2022, 13, 437–444. [Google Scholar] [CrossRef]
- Berni Canani, R.; Di Costanzo, M.; Leone, L. The epigenetic effects of butyrate: potential therapeutic implications for clinical practice. Clin. Epigenetics 2012, 4, 4. [Google Scholar] [CrossRef]
- Fellows, R.; Denizot, J.; Stellato, C.; Cuomo, A.; Jain, P.; Stoyanova, E.; Balázsi, S.; Hajnády, Z.; Liebert, A.; Kazakevych, J. , et al. Microbiota derived short chain fatty acids promote histone crotonylation in the colon through histone deacetylases. Nat. Commun. 2018, 9, 105. [Google Scholar] [CrossRef]
- Alenghat, T.; Osborne, L.C.; Saenz, S.A.; Kobuley, D.; Ziegler, C.G.; Mullican, S.E.; Choi, I.; Grunberg, S.; Sinha, R.; Wynosky-Dolfi, M. , et al. Histone deacetylase 3 coordinates commensal-bacteria-dependent intestinal homeostasis. Nature 2013, 504, 153–157. [Google Scholar] [CrossRef]
- Liu, K.; He, X.; Huang, J.; Yu, S.; Cui, M.; Gao, M.; Liu, L.; Qian, Y.; Xie, Y.; Hui, M. , et al. Short-chain fatty acid-butyric acid ameliorates granulosa cells inflammation through regulating METTL3-mediated N6-methyladenosine modification of FOSL2 in polycystic ovarian syndrome. Clin. Epigenetics 2023, 15, 86. [Google Scholar] [CrossRef]
- Noureldein, M.H.; Bitar, S.; Youssef, N.; Azar, S.; Eid, A.A. Butyrate modulates diabetes-linked gut dysbiosis: epigenetic and mechanistic modifications. J. Mol. Endocrinol. 2020, 64, 29–42. [Google Scholar] [CrossRef]
- Kumar, H.; Lund, R.; Laiho, A.; Lundelin, K.; Ley, R.E.; Isolauri, E.; Salminen, S. Gut microbiota as an epigenetic regulator: pilot study based on whole-genome methylation analysis. mBio 2014, 5. [Google Scholar] [CrossRef] [PubMed]
- Ramos-Molina, B.; Sánchez-Alcoholado, L.; Cabrera-Mulero, A.; Lopez-Dominguez, R.; Carmona-Saez, P.; Garcia-Fuentes, E.; Moreno-Indias, I.; Tinahones, F.J. Gut Microbiota Composition Is Associated With the Global DNA Methylation Pattern in Obesity. Front. Genet. 2019, 10, 613. [Google Scholar] [CrossRef] [PubMed]
- Tahara, T.; Hirata, I.; Nakano, N.; Tahara, S.; Horiguchi, N.; Kawamura, T.; Okubo, M.; Ishizuka, T.; Yamada, H.; Yoshida, D. , et al. Potential link between Fusobacterium enrichment and DNA methylation accumulation in the inflammatory colonic mucosa in ulcerative colitis. Oncotarget 2017, 8, 61917–61926. [Google Scholar] [CrossRef] [PubMed]
- Ansari, I.; Raddatz, G.; Gutekunst, J.; Ridnik, M.; Cohen, D.; Abu-Remaileh, M.; Tuganbaev, T.; Shapiro, H.; Pikarsky, E.; Elinav, E. , et al. The microbiota programs DNA methylation to control intestinal homeostasis and inflammation. Nat. Microbiol. 2020, 5, 610–619. [Google Scholar] [CrossRef]
- Shi, W.; Cassmann, T.J.; Bhagwate, A.V.; Hitosugi, T.; Ip, W.K.E. Lactic acid induces transcriptional repression of macrophage inflammatory response via histone acetylation. Cell Rep 2024, 43, 113746. [Google Scholar] [CrossRef]
- Remely, M.; Aumueller, E.; Jahn, D.; Hippe, B.; Brath, H.; Haslberger, A.G. Microbiota and epigenetic regulation of inflammatory mediators in type 2 diabetes and obesity. Benef. Microbes 2014, 5, 33–43. [Google Scholar] [CrossRef]
- Fitzgerald, K.N.; Hodges, R.; Hanes, D.; Stack, E.; Cheishvili, D.; Szyf, M.; Henkel, J.; Twedt, M.W.; Giannopoulou, D.; Herdell, J. , et al. Potential reversal of epigenetic age using a diet and lifestyle intervention: a pilot randomized clinical trial. Aging 2021, 13, 9419–9432. [Google Scholar] [CrossRef]
- Zhang, Q.; Zhao, Q.; Li, T.; Lu, L.; Wang, F.; Zhang, H.; Liu, Z.; Ma, H.; Zhu, Q.; Wang, J. , et al. Lactobacillus plantarum-derived indole-3-lactic acid ameliorates colorectal tumorigenesis via epigenetic regulation of CD8(+) T cell immunity. Cell Metab 2023, 35, 943–960.e949. [Google Scholar] [CrossRef]
- Novakovic, B.; Habibi, E.; Wang, S.Y.; Arts, R.J.W.; Davar, R.; Megchelenbrink, W.; Kim, B.; Kuznetsova, T.; Kox, M.; Zwaag, J. , et al. β-Glucan Reverses the Epigenetic State of LPS-Induced Immunological Tolerance. Cell 2016, 167, 1354–1368.e1314. [Google Scholar] [CrossRef]
- Wu, S.E.; Hashimoto-Hill, S.; Woo, V.; Eshleman, E.M.; Whitt, J.; Engleman, L.; Karns, R.; Denson, L.A.; Haslam, D.B.; Alenghat, T. Microbiota-derived metabolite promotes HDAC3 activity in the gut. Nature 2020, 586, 108–112. [Google Scholar] [CrossRef]
- Guan, Z.; Jin, X.; Guan, Z.; Liu, S.; Tao, K.; Luo, L. The gut microbiota metabolite capsiate regulate SLC2A1 expression by targeting HIF-1α to inhibit knee osteoarthritis-induced ferroptosis. Aging Cell 2023, 22, e13807. [Google Scholar] [CrossRef] [PubMed]
- Rice, S.J.; Beier, F.; Young, D.A.; Loughlin, J. Interplay between genetics and epigenetics in osteoarthritis. Nat. Rev.. Rheumatol. 2020, 16, 268–281. [Google Scholar] [CrossRef] [PubMed]
- Coutinho de Almeida, R.; Ramos, Y.F.M.; Meulenbelt, I. Involvement of epigenetics in osteoarthritis. Best Pract. Res.. Clin. Rheumatol. 2017, 31, 634–648. [Google Scholar] [CrossRef] [PubMed]
- Sun, D.; Zhuang, X.; Xiang, X.; Liu, Y.; Zhang, S.; Liu, C.; Barnes, S.; Grizzle, W.; Miller, D.; Zhang, H.G. A novel nanoparticle drug delivery system: the anti-inflammatory activity of curcumin is enhanced when encapsulated in exosomes. Mol. Ther. : J. Am. Soc. Gene Ther. 2010, 18, 1606–1614. [Google Scholar] [CrossRef]
- Trivedi, M.K.; Mondal, S.C.; Gangwar, M.; Jana, S. Immunomodulatory potential of nanocurcumin-based formulation. Inflammopharmacology 2017, 25, 609–619. [Google Scholar] [CrossRef]
- Ghosh, S.S.; He, H.; Wang, J.; Gehr, T.W.; Ghosh, S. Curcumin-mediated regulation of intestinal barrier function: The mechanism underlying its beneficial effects. Tissue Barriers 2018, 6, e1425085. [Google Scholar] [CrossRef] [PubMed]
- Bertoncini-Silva, C.; Fassini, P.G.; Carlos, D.; de Paula, N.A.; Ramalho, L.N.Z.; Rodrigues Giuliani, M.; Pereira Í, S.; Guimarães, J.B.; Suen, V.M.M. The Dose-Dependent Effect of Curcumin Supplementation on Inflammatory Response and Gut Microbiota Profile in High-Fat Fed C57BL/6 Mice. Mol Nutr Food Res 2023, 67, e2300378. [Google Scholar] [CrossRef]
- Pluta, R.; Januszewski, S.; Ułamek-Kozioł, M. Mutual Two-Way Interactions of Curcumin and Gut Microbiota. Int J Mol Sci 2020, 21. [Google Scholar] [CrossRef]
- Scazzocchio, B.; Minghetti, L.; D’Archivio, M. Interaction between Gut Microbiota and Curcumin: A New Key of Understanding for the Health Effects of Curcumin. Nutrients 2020, 12. [Google Scholar] [CrossRef]
| Polyphenol nanoformulation |
Osteoarthritis model | Effects in vitro | Effects in vivo | Refs. |
|---|---|---|---|---|
| Tannic acid/Sr2+ coated Silk/graphene oxide-based meniscus scaffold | -In vitro – LPS-induced rabbit-synovial MSC -In vivo – Papain-induced OA rat model |
Increased extracellular matrix secretion and promoted cell migration | Reduced cartilage degeneration and OA damage by downregulating MMP, IL6, IL8 | [47] |
| pH responsive - metal organic framework of Hyaluronic acid loaded with protocatechuic acid |
- In vitro - IL1β-induced rat primary chondrocytes -In vivo - ALCT induced OA rat model |
-Downregulated IL6, COX2, MMP1, MMP3, MMP13, ADAMTS5 and iNOS; -Reduced synovial inflammation |
Promoted cartilage regeneration | [48] |
| Liposome-anchored teriparatide incorporated into Gallic acid-grafted gelatin hydrogel | -In vitro - IL1β-induced mouse chondrocyte cell line, ATDC5 -In vivo - DMM- induced OA mice model |
- Activated expression of p-PI3K and p-AKT - Promoted anti-apoptotic effect by upregulating Bcl-2 - Downregulated ADAMTS5 |
- Upregulated expression of ACAN, SOX9; -Promoted glycosaminoglycan secretion and ROS scavenging |
[49] |
| Gallic acid encapsulated polymeric nanoliposome | -In vitro - H2O2-induced human chondrocyte cell line, C28/I2 -In vivo - MIA- induced OA rat model |
-Promoted ROS scavenging; -Lowered cartilage damage via upregulation of aggrecan and collagen II |
-Mitigated joint wear by improving cartilage lubrication; -Lowered cartilage erosion; -Promoted ROS scavenging |
[50] |
| Gallic acid loaded liposome with hyaluronan grafted poly (2-acrylamide-2-methylpropanesulfonic acid sodium salt) | -In vitro - H2O2-induced human chondrocyte cell line, C28/I2 -In vivo - MIA- induced OA rat model |
-Upregulated expression of Col II and ACAN; -Reduced chondrocyte degeneration; -Promoted antioxidant effect |
-Lowered cartilage erosion and chondrocyte degeneration; -Promoted glycosaminoglycan deposition |
[51] |
| Polydopamine- coated Hesperetin-loaded Gd2 (CO3)3 nanoparticles |
-In vitro – IL1β – induced chondrocytes -In vivo – ACLT- induced OA mice model |
Downregulated TLR2, decreased inflammation, cellular apoptosis and promoted chondrocyte maturation by inactivating NFκB/Akt pathway | Displayed cartilage binding ability, mitigated cartilage degeneration | [52] |
| pH-responsive Polycaprolactone/polyethylene glycol naringenin nanofiber |
-In vitro – IL1β-induced primary rat chondrocytes -In vivo – ACLT-induced OA rat model |
Inhibited expression of IL6, IL1β, MMP3 and MMP13; Promoted COL2A1 | Reduced cartilage damage; increased proteoglycan retention and glycosaminoglycan content | [53] |
| Scaffold of Berberine oleanolic acid complex grafted onto Hyaluronic Acid/Silk fibroin composite |
In vitro – IL1β-induced OA model in primary rabbit articular chondrocytes |
Upregulated COL1, COL2 and SOX9; restored chondrocyte morphology |
Promoted cartilage tissue regeneration in nude mice post subcutaneous implantation | [54] |
| NIR-responsive Epigallocatechin gallate decorated Au-Ag nano-jars |
-In vitro – H2O2-induced primary rat chondrocytes -In vivo – ACLT-induced OA rat model |
Lowered chondrocyte apoptosis; downregulated p-NFκB, iNOS and COX2; promoted cell migration |
Reduced cartilage erosion; improved cartilage thickness; lowered chondrocyte apoptosis | [55] |
| Hyaluronic acid- coated gelatin nanoparticles loaded with kaempferol | -In vitro – IL1β- induced primary rat chondrocytes -In vivo – ACLT-induced OA rat model |
Downregulated expression of inflammatory cytokines – COX2, MMP9, MMP13, TNFα, IL1β; | Attenuated inflammation and matrix degradation; restored cartilage thickness | [56] |
| Poly p-Coumaric Acid Nanoparticles | In vivo – Temporomandibular joint OA rat model | -- | Inhibited chondrocyte ferroptosis; reduced ECM degradation; exhibited long-term efficacy and alleviated cartilage repair | [57] |
| Nano-naringenin |
In vivo – MIA-induced OA rat model | -- | Upregulated GSH and TIMP-3; downregulated MDA, ADAMTS5 and MMP3 | [58] |
| Composition | Dosage & Delivery | Duration/number of subjects (N) | Key outcomes | Refs. |
|---|---|---|---|---|
| Curcumin in water-dispersible form | 180mg/day, Oral | 8 weeks, N=50 | Reduced dependence on celecoxib vs placebo group; lower VAS scores of knee pain |
[104] |
| Curcumin in water-dispersible form | 180mg/day, Oral | 6 months, N=50 | Improved JOA, VAS and JKOM scores, observed no major side effects, 75.6% effective compared to placebo | [105] |
| Curcumin in water-dispersible form | 180mg/day, Oral | 12 months, N=50 | Reduced cartilage stiffness and time-dependent decrease in scores of JOA, VAS and JKOM | [106] |
| Curcumin nanomicelle | 80mg/day, Oral | 6 weeks, N=71 | Significant decrease in WOMAC score | [107] |
| Curcumin nanomicelle |
80mg/day, Oral | 3 months, N=30 | Decrease in VAS score, lower CRP levels, Immunomodulatory effects on T cells and B cells | [84] |
| Curcumin nanomicelle | 80mg/day, Oral | 3 months, N=30 | Suppressed expression of key miRNAs | [108] |
| Solid lipid curcumin particles | 160mg/day, Oral | 3 months, N=50 | Improved WOMAC and VAS scores comparable to ibuprofen. No significant change was observed in inflammatory markers | [109] |
| Curcumin self-nano-emulsifying-PEG organogel | 1.5g/twice per day, Topical | 8 weeks, N=75 | Significantly reduced WOMAC scores | [110] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





