Submitted:
23 September 2024
Posted:
24 September 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Aptamers: Discovery and First Applications
3. Aptamer-Guided Approaches for Delivering Cancer Gene Therapy
3.1. Aptamer-Based Conjugates for miRNA and AntimiR Delivery
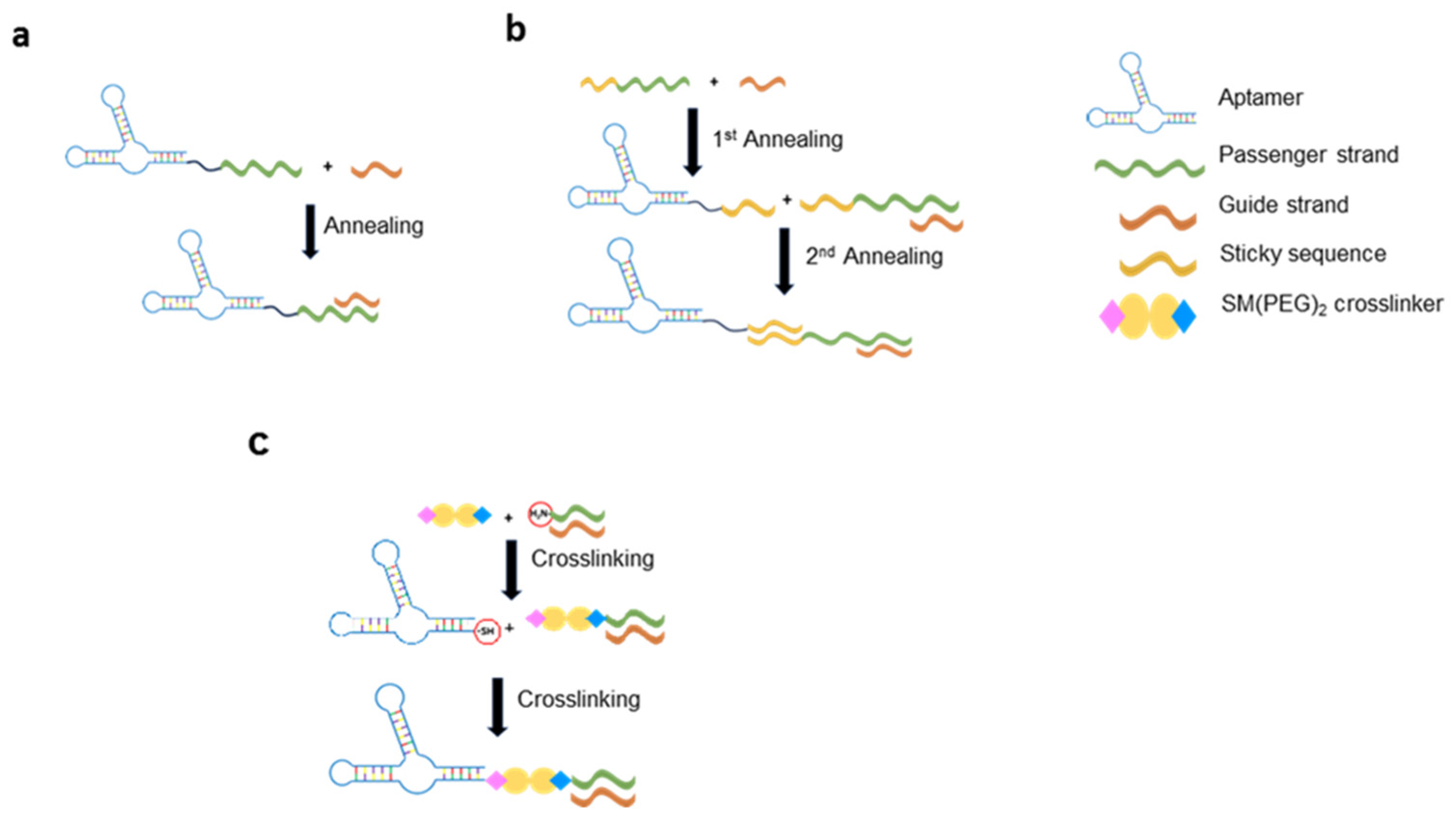
3.2. Aptamer-Based Conjugates for siRNA Delivery
3.3. Aptamer-Based Conjugated for DNAzyme Delivery
3.4. Aptamer-Coated Viral Vectors
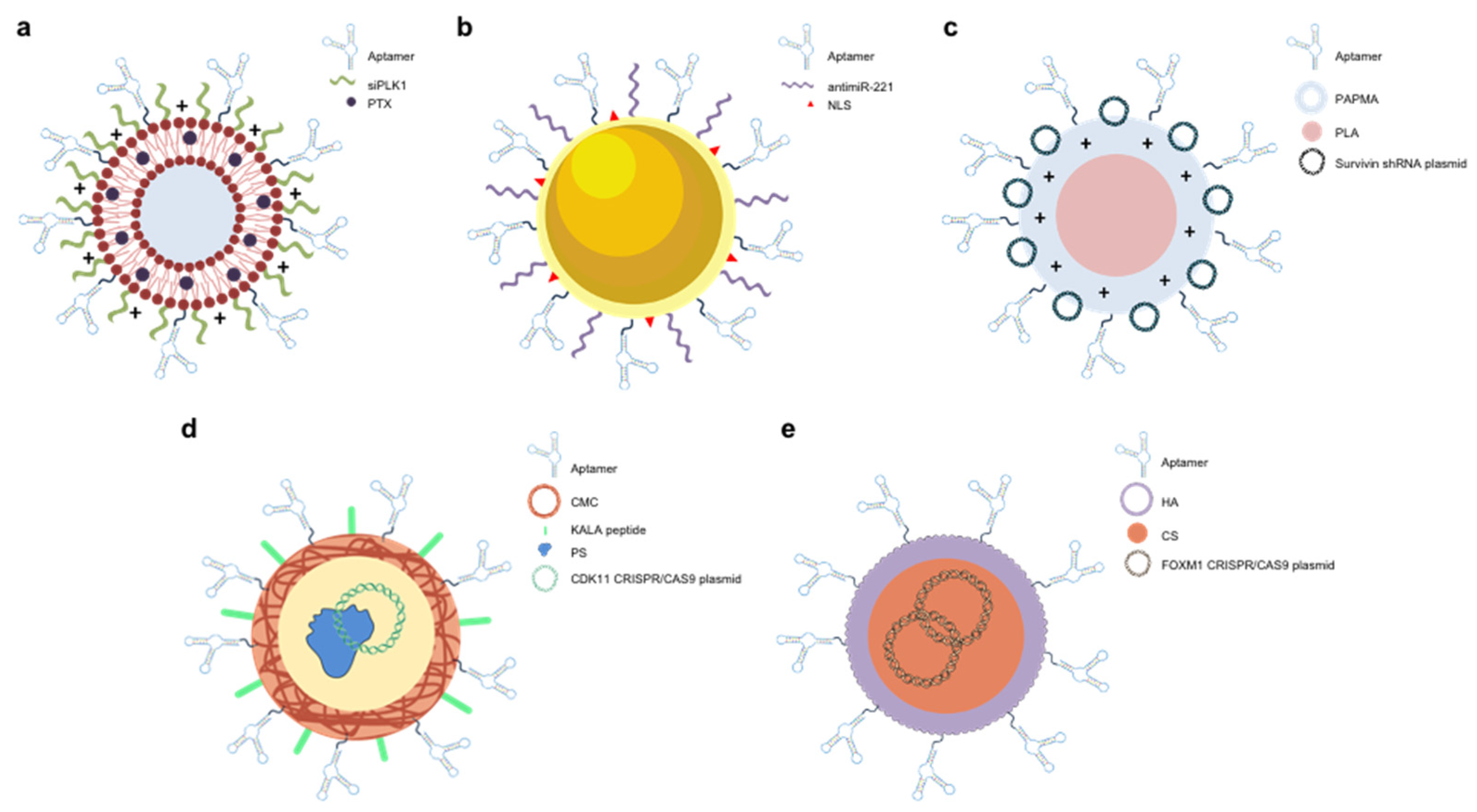
3.5. Aptamer-Functionalized NPs
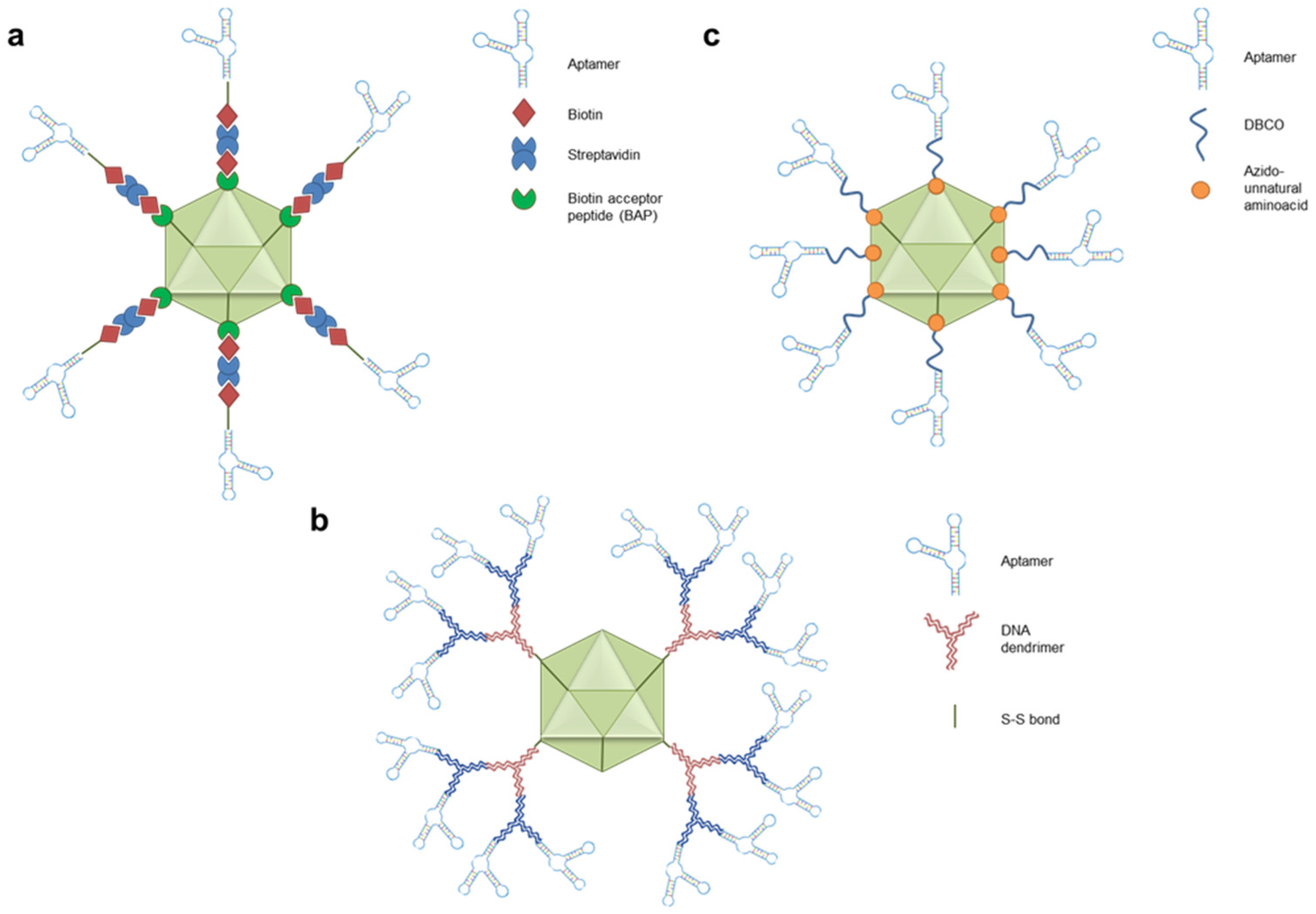
4. Aptamers as CRISPR/Cas9 Regulators and Epigenetic Modifiers
5. Discussion
6. Conclusions
Supplementary Materials
Author Contributions
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Chakravarthi, B.V.S.K.; Nepal, S.; Varambally, S. Genomic and Epigenomic Alterations in Cancer. Am J Pathol 2016, 186, 1724–1735. [Google Scholar] [CrossRef] [PubMed]
- The Lancet 20 Years of Precision Medicine in Oncology. The Lancet 2021, 397, 1781. [CrossRef] [PubMed]
- The Global Challenge of Cancer. Nat Cancer 2020, 1, 1–2. [CrossRef] [PubMed]
- Haier, J.; Schaefers, J. Economic Perspective of Cancer Care and Its Consequences for Vulnerable Groups. Cancers (Basel) 2022, 14, 3158. [Google Scholar] [CrossRef]
- Bauer, G.; Anderson, J.S. History of Gene Therapy. In; 2014; pp. 9–15.
- Landhuis, E. The Definition of Gene Therapy Has Changed. Nature 2021. [Google Scholar] [CrossRef]
- Dowdy, S.F. Overcoming Cellular Barriers for RNA Therapeutics. Nat Biotechnol 2017, 35, 222–229. [Google Scholar] [CrossRef]
- Roberts, T.C.; Langer, R.; Wood, M.J.A. Advances in Oligonucleotide Drug Delivery. Nat Rev Drug Discov 2020, 19, 673–694. [Google Scholar] [CrossRef]
- Zhao, Z.; Anselmo, A.C.; Mitragotri, S. Viral Vector-based Gene Therapies in the Clinic. Bioeng Transl Med 2022, 7. [Google Scholar] [CrossRef]
- Qi, L.; Li, G.; Li, P.; Wang, H.; Fang, X.; He, T.; Li, J. Twenty Years of Gendicine® RAd-P53 Cancer Gene Therapy: The First-in-Class Human Cancer Gene Therapy in the Era of Personalized Oncology. Genes Dis 2024, 11, 101155. [Google Scholar] [CrossRef]
- Wang, J.-H.; Gessler, D.J.; Zhan, W.; Gallagher, T.L.; Gao, G. Adeno-Associated Virus as a Delivery Vector for Gene Therapy of Human Diseases. Signal Transduct Target Ther 2024, 9, 78. [Google Scholar] [CrossRef]
- Anguela, X.M.; High, K.A. Entering the Modern Era of Gene Therapy. Annu Rev Med 2019, 70, 273–288. [Google Scholar] [CrossRef] [PubMed]
- Sayour, E.J.; Boczkowski, D.; Mitchell, D.A.; Nair, S.K. Cancer MRNA Vaccines: Clinical Advances and Future Opportunities. Nat Rev Clin Oncol 2024. [Google Scholar] [CrossRef] [PubMed]
- Chaudhary, N.; Weissman, D.; Whitehead, K.A. MRNA Vaccines for Infectious Diseases: Principles, Delivery and Clinical Translation. Nat Rev Drug Discov 2021, 20, 817–838. [Google Scholar] [CrossRef] [PubMed]
- Mirón-Barroso, S.; Domènech, E.B.; Trigueros, S. Nanotechnology-Based Strategies to Overcome Current Barriers in Gene Delivery. Int J Mol Sci 2021, 22, 8537. [Google Scholar] [CrossRef] [PubMed]
- Guan, B.; Zhang, X. Aptamers as Versatile Ligands for Biomedical and Pharmaceutical Applications. Int J Nanomedicine 2020, Volume 15, 1059–1071. [Google Scholar] [CrossRef]
- Bauer, M.; Strom, M.; Hammond, D.S.; Shigdar, S. Anything You Can Do, I Can Do Better: Can Aptamers Replace Antibodies in Clinical Diagnostic Applications? Molecules 2019, 24, 4377. [Google Scholar] [CrossRef]
- Porciani, D.; Cardwell, L.N.; Tawiah, K.D.; Alam, K.K.; Lange, M.J.; Daniels, M.A.; Burke, D.H. Modular Cell-Internalizing Aptamer Nanostructure Enables Targeted Delivery of Large Functional RNAs in Cancer Cell Lines. Nat Commun 2018, 9, 2283. [Google Scholar] [CrossRef]
- Esposito, C.L.; Catuogno, S.; Condorelli, G.; Ungaro, P.; De Franciscis, V. Aptamer Chimeras for Therapeutic Delivery: The Challenging Perspectives. Genes (Basel) 2018, 9, 529. [Google Scholar] [CrossRef]
- Tuerk, C.; Gold, L. Systematic Evolution of Ligands by Exponential Enrichment: RNA Ligands to Bacteriophage T4 DNA Polymerase. Science (1979) 1990, 249, 505–510. [Google Scholar] [CrossRef]
- Ellington, A.D.; Szostak, J.W. In Vitro Selection of RNA Molecules That Bind Specific Ligands. Nature 1990, 346, 818–822. [Google Scholar] [CrossRef]
- Zhu, C.; Feng, Z.; Qin, H.; Chen, L.; Yan, M.; Li, L.; Qu, F. Recent Progress of SELEX Methods for Screening Nucleic Acid Aptamers. Talanta 2024, 266, 124998. [Google Scholar] [CrossRef] [PubMed]
- Catuogno, S.; Esposito, C.L. Aptamer Cell-Based Selection: Overview and Advances. Biomedicines 2017, 5, 49. [Google Scholar] [CrossRef] [PubMed]
- Mayer, G.; Wulffen, B. The Chemical Biology of Aptamers: Synthesis and Applications. In The Chemical Biology of Nucleic Acids; Wiley, 2010; pp. 377–400. [Google Scholar]
- Li, N.; Nguyen, H.H.; Byrom, M.; Ellington, A.D. Inhibition of Cell Proliferation by an Anti-EGFR Aptamer. PLoS One 2011, 6, e20299. [Google Scholar] [CrossRef]
- McNamara, J.O.; Kolonias, D.; Pastor, F.; Mittler, R.S.; Chen, L.; Giangrande, P.H.; Sullenger, B.; Gilboa, E. Multivalent 4-1BB Binding Aptamers Costimulate CD8+ T Cells and Inhibit Tumor Growth in Mice. Journal of Clinical Investigation 2008, 118, 376–386. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Rossi, J. Aptamers as Targeted Therapeutics: Current Potential and Challenges. Nat Rev Drug Discov 2017, 16, 181–202. [Google Scholar] [CrossRef] [PubMed]
- Ali, M.H.; Elsherbiny, M.E.; Emara, M. Updates on Aptamer Research. Int J Mol Sci 2019, 20, 2511. [Google Scholar] [CrossRef]
- Esawi, E.; Nsairat, H.; Mahmoud, I.S.; Lafi, Z.; Al-Kadash, A.; Al-Ragheb, B.A.; Ismail, S.I.; Alhaer, W. Clinical Use and Future Perspective of Aptamers. In Aptamers Engineered Nanocarriers for Cancer Therapy; Elsevier, 2023; pp. 481–520. [Google Scholar]
- Gragoudas, E.S.; Adamis, A.P.; Cunningham, E.T.; Feinsod, M.; Guyer, D.R. Pegaptanib for Neovascular Age-Related Macular Degeneration. New England Journal of Medicine 2004, 351, 2805–2816. [Google Scholar] [CrossRef]
- Mullard, A. FDA Approves Second RNA Aptamer. Nat Rev Drug Discov 2023, 22, 774–774. [Google Scholar] [CrossRef]
- Xiao, X.; Li, H.; Zhao, L.; Zhang, Y.; Liu, Z. Oligonucleotide Aptamers: Recent Advances in Their Screening, Molecular Conformation and Therapeutic Applications. Biomedicine & Pharmacotherapy 2021, 143, 112232. [Google Scholar] [CrossRef]
- Wan, L.-Y.; Yuan, W.-F.; Ai, W.-B.; Ai, Y.-W.; Wang, J.-J.; Chu, L.-Y.; Zhang, Y.-Q.; Wu, J.-F. An Exploration of Aptamer Internalization Mechanisms and Their Applications in Drug Delivery. Expert Opin Drug Deliv 2019, 16, 207–218. [Google Scholar] [CrossRef]
- Yoon, S.; Rossi, J.J. Aptamers: Uptake Mechanisms and Intracellular Applications. Adv Drug Deliv Rev 2018, 134, 22–35. [Google Scholar] [CrossRef] [PubMed]
- Thiel, W.H.; Thiel, K.W.; Flenker, K.S.; Bair, T.; Dupuy, A.J.; McNamara, J.O.; Miller, F.J.; Giangrande, P.H. Cell-Internalization SELEX: Method for Identifying Cell-Internalizing RNA Aptamers for Delivering SiRNAs to Target Cells. In; 2015; pp. 187–199.
- Thiel, K.W.; Hernandez, L.I.; Dassie, J.P.; Thiel, W.H.; Liu, X.; Stockdale, K.R.; Rothman, A.M.; Hernandez, F.J.; McNamara, J.O.; Giangrande, P.H. Delivery of Chemo-Sensitizing SiRNAs to HER2+-Breast Cancer Cells Using RNA Aptamers. Nucleic Acids Res 2012, 40, 6319–6337. [Google Scholar] [CrossRef] [PubMed]
- Gamboa, J.; Lourenço, P.; Cruz, C.; Gallardo, E. Aptamers for the Delivery of Plant-Based Compounds: A Review. Pharmaceutics 2024, 16, 541. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef] [PubMed]
- Shang, R.; Lee, S.; Senavirathne, G.; Lai, E.C. MicroRNAs in Action: Biogenesis, Function and Regulation. Nat Rev Genet 2023, 24, 816–833. [Google Scholar] [CrossRef]
- Iwakawa, H.; Tomari, Y. Life of RISC: Formation, Action, and Degradation of RNA-Induced Silencing Complex. Mol Cell 2022, 82, 30–43. [Google Scholar] [CrossRef]
- Peng, Y.; Croce, C.M. The Role of MicroRNAs in Human Cancer. Signal Transduct Target Ther 2016, 1, 15004. [Google Scholar] [CrossRef]
- Menon, A.; Abd-Aziz, N.; Khalid, K.; Poh, C.L.; Naidu, R. MiRNA: A Promising Therapeutic Target in Cancer. Int J Mol Sci 2022, 23, 11502. [Google Scholar] [CrossRef]
- Chen, Y.; Gao, D.-Y.; Huang, L. In Vivo Delivery of MiRNAs for Cancer Therapy: Challenges and Strategies. Adv Drug Deliv Rev 2015, 81, 128–141. [Google Scholar] [CrossRef]
- Odeh, F.; Nsairat, H.; Alshaer, W.; Ismail, M.A.; Esawi, E.; Qaqish, B.; Bawab, A. Al; Ismail, S.I. Aptamers Chemistry: Chemical Modifications and Conjugation Strategies. Molecules 2019, 25, 3. [Google Scholar] [CrossRef]
- Esposito, C.L.; Catuogno, S.; de Franciscis, V. Aptamer-MiRNA Conjugates for Cancer Cell-Targeted Delivery. In; 2016; pp. 197–208.
- Esposito, C.L.; Cerchia, L.; Catuogno, S.; De Vita, G.; Dassie, J.P.; Santamaria, G.; Swiderski, P.; Condorelli, G.; Giangrande, P.H.; de Franciscis, V. Multifunctional Aptamer-MiRNA Conjugates for Targeted Cancer Therapy. Mol Ther 2014, 22, 1151–1163. [Google Scholar] [CrossRef] [PubMed]
- Cerchia, L.; Esposito, C.L.; Camorani, S.; Rienzo, A.; Stasio, L.; Insabato, L.; Affuso, A.; de Franciscis, V. Targeting Axl With an High-Affinity Inhibitory Aptamer. Molecular Therapy 2012, 20, 2291–2303. [Google Scholar] [CrossRef] [PubMed]
- Kumar, M.S.; Erkeland, S.J.; Pester, R.E.; Chen, C.Y.; Ebert, M.S.; Sharp, P.A.; Jacks, T. Suppression of Non-Small Cell Lung Tumor Development by the Let-7 MicroRNA Family. Proceedings of the National Academy of Sciences 2008, 105, 3903–3908. [Google Scholar] [CrossRef] [PubMed]
- Iaboni, M.; Russo, V.; Fontanella, R.; Roscigno, G.; Fiore, D.; Donnarumma, E.; Esposito, C.L.; Quintavalle, C.; Giangrande, P.H.; de Franciscis, V.; et al. Aptamer-MiRNA-212 Conjugate Sensitizes NSCLC Cells to TRAIL. Mol Ther Nucleic Acids 2016, 5, e289. [Google Scholar] [CrossRef]
- Chen, W.; Song, J.; Bian, H.; Yang, X.; Xie, X.; Zhu, Q.; Qin, C.; Qi, J. The Functions and Targets of MiR-212 as a Potential Biomarker of Cancer Diagnosis and Therapy. J Cell Mol Med 2020, 24, 2392–2401. [Google Scholar] [CrossRef]
- Russo, V.; Paciocco, A.; Affinito, A.; Roscigno, G.; Fiore, D.; Palma, F.; Galasso, M.; Volinia, S.; Fiorelli, A.; Esposito, C.L.; et al. Aptamer-MiR-34c Conjugate Affects Cell Proliferation of Non-Small-Cell Lung Cancer Cells. Mol Ther Nucleic Acids 2018, 13, 334–346. [Google Scholar] [CrossRef]
- Nuzzo, S.; Catuogno, S.; Capuozzo, M.; Fiorelli, A.; Swiderski, P.; Boccella, S.; de Nigris, F.; Esposito, C.L. Axl-Targeted Delivery of the Oncosuppressor MiR-137 in Non-Small-Cell Lung Cancer. Mol Ther Nucleic Acids 2019, 17, 256–263. [Google Scholar] [CrossRef]
- Quirico, L.; Orso, F.; Esposito, C.L.; Bertone, S.; Coppo, R.; Conti, L.; Catuogno, S.; Cavallo, F.; de Franciscis, V.; Taverna, D. Axl-148b Chimeric Aptamers Inhibit Breast Cancer and Melanoma Progression. Int J Biol Sci 2020, 16, 1238–1251. [Google Scholar] [CrossRef]
- Zhao, N.; Pei, S.-N.; Qi, J.; Zeng, Z.; Iyer, S.P.; Lin, P.; Tung, C.-H.; Zu, Y. Oligonucleotide Aptamer-Drug Conjugates for Targeted Therapy of Acute Myeloid Leukemia. Biomaterials 2015, 67, 42–51. [Google Scholar] [CrossRef]
- Tanno, T.; Zhang, P.; Lazarski, C.A.; Liu, Y.; Zheng, P. An Aptamer-Based Targeted Delivery of MiR-26a Protects Mice against Chemotherapy Toxicity While Suppressing Tumor Growth. Blood Adv 2017, 1, 1107–1119. [Google Scholar] [CrossRef]
- Ramezanpour, M.; Daei, P.; Tabarzad, M.; Khanaki, K.; Elmi, A.; Barati, M. Preliminary Study on the Effect of Nucleolin Specific Aptamer–MiRNA Let-7d Chimera on Janus Kinase-2 Expression Level and Activity in Gastric Cancer (MKN-45) Cells. Mol Biol Rep 2019, 46, 207–215. [Google Scholar] [CrossRef] [PubMed]
- Daei, P.; Ramezanpour, M.; Khanaki, K.; Tabarzad, M.; Nikokar, I.; Hedayati Ch, M.; Elmi, A. Aptamer-Based Targeted Delivery of MiRNA Let-7d to Gastric Cancer Cells as a Novel Anti-Tumor Therapeutic Agent. Iran J Pharm Res 2018, 17, 1537–1549. [Google Scholar] [PubMed]
- Crooke, S.T.; Baker, B.F.; Crooke, R.M.; Liang, X. Antisense Technology: An Overview and Prospectus. Nat Rev Drug Discov 2021, 20, 427–453. [Google Scholar] [CrossRef] [PubMed]
- Catuogno, S.; Rienzo, A.; Di Vito, A.; Esposito, C.L.; de Franciscis, V. Selective Delivery of Therapeutic Single Strand AntimiRs by Aptamer-Based Conjugates. Journal of Controlled Release 2015, 210, 147–159. [Google Scholar] [CrossRef]
- Esposito, C.L.; Nuzzo, S.; Kumar, S.A.; Rienzo, A.; Lawrence, C.L.; Pallini, R.; Shaw, L.; Alder, J.E.; Ricci-Vitiani, L.; Catuogno, S.; et al. A Combined MicroRNA-Based Targeted Therapeutic Approach to Eradicate Glioblastoma Stem-like Cells. Journal of Controlled Release 2016, 238, 43–57. [Google Scholar] [CrossRef]
- Fang, Y.; Shu, D.; Xiao, F.; Guo, P.; Qin, P.Z. Modular Assembly of Chimeric Phi29 Packaging RNAs That Support DNA Packaging. Biochem Biophys Res Commun 2008, 372, 589–594. [Google Scholar] [CrossRef]
- Shu, D.; Li, H.; Shu, Y.; Xiong, G.; Carson, W.E.; Haque, F.; Xu, R.; Guo, P. Systemic Delivery of Anti-MiRNA for Suppression of Triple Negative Breast Cancer Utilizing RNA Nanotechnology. ACS Nano 2015, 9, 9731–9740. [Google Scholar] [CrossRef]
- Arnold, C. Theranostics Could Be Big Business in Precision Oncology. Nat Med 2022, 28, 606–608. [Google Scholar] [CrossRef]
- Dana, H.; Chalbatani, G.M.; Mahmoodzadeh, H.; Karimloo, R.; Rezaiean, O.; Moradzadeh, A.; Mehmandoost, N.; Moazzen, F.; Mazraeh, A.; Marmari, V.; et al. Molecular Mechanisms and Biological Functions of SiRNA. Int J Biomed Sci 2017, 13, 48–57. [Google Scholar] [CrossRef]
- Zhang, M.M.; Bahal, R.; Rasmussen, T.P.; Manautou, J.E.; Zhong, X. The Growth of SiRNA-Based Therapeutics: Updated Clinical Studies. Biochem Pharmacol 2021, 189, 114432. [Google Scholar] [CrossRef]
- Friedrich, M.; Aigner, A. Therapeutic SiRNA: State-of-the-Art and Future Perspectives. BioDrugs 2022, 36, 549–571. [Google Scholar] [CrossRef] [PubMed]
- Ali Zaidi, S.S.; Fatima, F.; Ali Zaidi, S.A.; Zhou, D.; Deng, W.; Liu, S. Engineering SiRNA Therapeutics: Challenges and Strategies. J Nanobiotechnology 2023, 21, 381. [Google Scholar] [CrossRef] [PubMed]
- McNamara, J.O.; Andrechek, E.R.; Wang, Y.; Viles, K.D.; Rempel, R.E.; Gilboa, E.; Sullenger, B.A.; Giangrande, P.H. Cell Type–Specific Delivery of SiRNAs with Aptamer-SiRNA Chimeras. Nat Biotechnol 2006, 24, 1005–1015. [Google Scholar] [CrossRef] [PubMed]
- Dassie, J.P.; Liu, X.; Thomas, G.S.; Whitaker, R.M.; Thiel, K.W.; Stockdale, K.R.; Meyerholz, D.K.; McCaffrey, A.P.; McNamara, J.O.; Giangrande, P.H. Systemic Administration of Optimized Aptamer-SiRNA Chimeras Promotes Regression of PSMA-Expressing Tumors. Nat Biotechnol 2009, 27, 839–846. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Tiemann, K.; Chomchan, P.; Alluin, J.; Swiderski, P.; Burnett, J.; Zhang, X.; Forman, S.; Chen, R.; Rossi, J. Dual Functional BAFF Receptor Aptamers Inhibit Ligand-Induced Proliferation and Deliver SiRNAs to NHL Cells. Nucleic Acids Res 2013, 41, 4266–4283. [Google Scholar] [CrossRef]
- Mohtar, M.; Syafruddin, S.; Nasir, S.; Low, T.Y. Revisiting the Roles of Pro-Metastatic EpCAM in Cancer. Biomolecules 2020, 10, 255. [Google Scholar] [CrossRef]
- Wang, T.; Gantier, M.P.; Xiang, D.; Bean, A.G.; Bruce, M.; Zhou, S.-F.; Khasraw, M.; Ward, A.; Wang, L.; Wei, M.Q.; et al. EpCAM Aptamer-Mediated Survivin Silencing Sensitized Cancer Stem Cells to Doxorubicin in a Breast Cancer Model. Theranostics 2015, 5, 1456–1472. [Google Scholar] [CrossRef]
- Shigdar, S.; Lin, J.; Yu, Y.; Pastuovic, M.; Wei, M.; Duan, W. RNA Aptamer against a Cancer Stem Cell Marker Epithelial Cell Adhesion Molecule. Cancer Sci 2011, 102, 991–998. [Google Scholar] [CrossRef]
- Gilboa-Geffen, A.; Hamar, P.; Le, M.T.N.; Wheeler, L.A.; Trifonova, R.; Petrocca, F.; Wittrup, A.; Lieberman, J. Gene Knockdown by EpCAM Aptamer–SiRNA Chimeras Suppresses Epithelial Breast Cancers and Their Tumor-Initiating Cells. Mol Cancer Ther 2015, 14, 2279–2291. [Google Scholar] [CrossRef]
- Esposito, C.L.; Nuzzo, S.; Catuogno, S.; Romano, S.; de Nigris, F.; de Franciscis, V. STAT3 Gene Silencing by Aptamer-SiRNA Chimera as Selective Therapeutic for Glioblastoma. Mol Ther Nucleic Acids 2018, 10, 398–411. [Google Scholar] [CrossRef]
- Esposito, C.L.; Nuzzo, S.; Ibba, M.L.; Ricci-Vitiani, L.; Pallini, R.; Condorelli, G.; Catuogno, S.; de Franciscis, V. Combined Targeting of Glioblastoma Stem-Like Cells by Neutralizing RNA-Bio-Drugs for STAT3. Cancers (Basel) 2020, 12, 1434. [Google Scholar] [CrossRef] [PubMed]
- Ibba, M.L.; Ciccone, G.; Rotoli, D.; Coppola, G.; Fiorelli, A.; Catuogno, S.; Esposito, C.L. STAT3 Silencing by an Aptamer-Based Strategy Hampers the Crosstalk between NSCLC Cells and Cancer-Associated Fibroblasts. Mol Ther Nucleic Acids 2023, 32, 111–126. [Google Scholar] [CrossRef] [PubMed]
- Oostindie, S.C.; Lazar, G.A.; Schuurman, J.; Parren, P.W.H.I. Avidity in Antibody Effector Functions and Biotherapeutic Drug Design. Nat Rev Drug Discov 2022, 21, 715–735. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.Y.; Yu, X.; Liu, H.; Wu, D.; She, J.-X. Co-Targeting EGFR and Survivin with a Bivalent Aptamer-Dual SiRNA Chimera Effectively Suppresses Prostate Cancer. Sci Rep 2016, 6, 30346. [Google Scholar] [CrossRef] [PubMed]
- Xue, L.; Maihle, N.J.; Yu, X.; Tang, S.-C.; Liu, H.Y. Synergistic Targeting HER2 and EGFR with Bivalent Aptamer-SiRNA Chimera Efficiently Inhibits HER2-Positive Tumor Growth. Mol Pharm 2018, 15, 4801–4813. [Google Scholar] [CrossRef]
- Yu, X.; Ghamande, S.; Liu, H.; Xue, L.; Zhao, S.; Tan, W.; Zhao, L.; Tang, S.-C.; Wu, D.; Korkaya, H.; et al. Targeting EGFR/HER2/HER3 with a Three-in-One Aptamer-SiRNA Chimera Confers Superior Activity against HER2+ Breast Cancer. Mol Ther Nucleic Acids 2018, 10, 317–330. [Google Scholar] [CrossRef]
- Walter, N.G.; Engelke, D.R. Ribozymes: Catalytic RNAs That Cut Things, Make Things, and Do Odd and Useful Jobs. Biologist (London) 2002, 49, 199–203. [Google Scholar]
- Goleij, P.; Babamohamadi, M.; Rezaee, A.; Sanaye, P.M.; Tabari, M.A.K.; Sadreddini, S.; Arefnezhad, R.; Motedayyen, H. Types of RNA Therapeutics. In; 2024; pp. 41–63.
- Reza, Md.S.; Mim, F.; Quader, M.R.; Khan, M.J.R.; Hossain, Md.S.; Uddin, K.R.; Akter, S.; Rahman, S.; Roy, S.; Rumman, Md.A. The Possibility of Nucleic Acids to Act as Anti-Viral Therapeutic Agents—A Review. Open J Med Microbiol 2021, 11, 198–248. [Google Scholar] [CrossRef]
- Khan, A.U. Ribozyme: A Clinical Tool. Clinica Chimica Acta 2006, 367, 20–27. [Google Scholar] [CrossRef]
- Feng, R.; Patil, S.; Zhao, X.; Miao, Z.; Qian, A. RNA Therapeutics - Research and Clinical Advancements. Front Mol Biosci 2021, 8. [Google Scholar] [CrossRef]
- Sioud, M. Ribozymes and SiRNAs: From Structure to Preclinical Applications. In RNA Towards Medicine; Springer-Verlag: Berlin/Heidelberg; pp. 223–242.
- Yan, J.; Bhadane, R.; Ran, M.; Ma, X.; Li, Y.; Zheng, D.; Salo-Ahen, O.M.H.; Zhang, H. Development of Aptamer-DNAzyme Based Metal-Nucleic Acid Frameworks for Gastric Cancer Therapy. Nat Commun 2024, 15, 3684. [Google Scholar] [CrossRef] [PubMed]
- Gu, C.; Liu, X.; Luo, L.; Chen, J.; Zhou, X.; Chen, G.; Huang, X.; Yu, L.; Chen, Q.; Yang, Y.; et al. Metal-DNA Nanocomplexes Enhance Chemo-dynamic Therapy by Inhibiting Autophagy-Mediated Resistance. Angewandte Chemie International Edition 2023, 62. [Google Scholar] [CrossRef] [PubMed]
- Nomaguchi, M.; Fujita, M.; Miyazaki, Y.; Adachi, A. Viral Tropism. Front Microbiol 2012, 3. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.; Sumerel, L.A.; Belousova, N.; Lyons, G.R.; Carey, D.E.; Krasnykh, V.; Douglas, J.T. The Coxsackievirus and Adenovirus Receptor Acts as a Tumour Suppressor in Malignant Glioma Cells. Br J Cancer 2003, 88, 1411–1416. [Google Scholar] [CrossRef]
- Tessarollo, N.G.; Domingues, A.C.M.; Antunes, F.; Luz, J.C. dos S. da; Rodrigues, O.A.; Cerqueira, O.L.D.; Strauss, B.E. Nonreplicating Adenoviral Vectors: Improving Tropism and Delivery of Cancer Gene Therapy. Cancers (Basel) 2021, 13, 1863. [Google Scholar] [CrossRef]
- Chen, H.; Zheng, X.; Di, B.; Wang, D.; Zhang, Y.; Xia, H.; Mao, Q. Aptamer Modification Improves the Adenoviral Transduction of Malignant Glioma Cells. J Biotechnol 2013, 168, 362–366. [Google Scholar] [CrossRef]
- Dhungel, B.P.; Bailey, C.G.; Rasko, J.E.J. Journey to the Center of the Cell: Tracing the Path of AAV Transduction. Trends Mol Med 2021, 27, 172–184. [Google Scholar] [CrossRef]
- Wu, Y.; Zhang, L.; Cui, C.; Cansiz, S.; Liang, H.; Wu, C.; Teng, I.-T.; Chen, W.; Liu, Y.; Hou, W.; et al. Enhanced Targeted Gene Transduction: AAV2 Vectors Conjugated to Multiple Aptamers via Reducible Disulfide Linkages. J Am Chem Soc 2018, 140, 2–5. [Google Scholar] [CrossRef]
- Puzzo, F.; Zhang, C.; Powell Gray, B.; Zhang, F.; Sullenger, B.A.; Kay, M.A. Aptamer-Programmable Adeno-Associated Viral Vectors as a Novel Platform for Cell-Specific Gene Transfer. Mol Ther Nucleic Acids 2023, 31, 383–397. [Google Scholar] [CrossRef]
- Dogbey, D.M.; Torres, V.E.S.; Fajemisin, E.; Mpondo, L.; Ngwenya, T.; Akinrinmade, O.A.; Perriman, A.W.; Barth, S. Technological Advances in the Use of Viral and Non-Viral Vectors for Delivering Genetic and Non-Genetic Cargos for Cancer Therapy. Drug Deliv Transl Res 2023, 13, 2719–2738. [Google Scholar] [CrossRef]
- Germain, M.; Caputo, F.; Metcalfe, S.; Tosi, G.; Spring, K.; Åslund, A.K.O.; Pottier, A.; Schiffelers, R.; Ceccaldi, A.; Schmid, R. Delivering the Power of Nanomedicine to Patients Today. Journal of Controlled Release 2020, 326, 164–171. [Google Scholar] [CrossRef] [PubMed]
- Rosenberg, J.E.; Bambury, R.M.; Van Allen, E.M.; Drabkin, H.A.; Lara, P.N.; Harzstark, A.L.; Wagle, N.; Figlin, R.A.; Smith, G.W.; Garraway, L.A.; et al. A Phase II Trial of AS1411 (a Novel Nucleolin-Targeted DNA Aptamer) in Metastatic Renal Cell Carcinoma. Invest New Drugs 2014, 32, 178–187. [Google Scholar] [CrossRef] [PubMed]
- Van den Avont, A.; Sharma-Walia, N. Anti-Nucleolin Aptamer AS1411: An Advancing Therapeutic. Front Mol Biosci 2023, 10. [Google Scholar] [CrossRef] [PubMed]
- Mendes, B.B.; Conniot, J.; Avital, A.; Yao, D.; Jiang, X.; Zhou, X.; Sharf-Pauker, N.; Xiao, Y.; Adir, O.; Liang, H.; et al. Nanodelivery of Nucleic Acids. Nature Reviews Methods Primers 2022, 2, 24. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez, F.; Caruana, P.; De la Fuente, N.; Español, P.; Gámez, M.; Balart, J.; Llurba, E.; Rovira, R.; Ruiz, R.; Martín-Lorente, C.; et al. Nano-Based Approved Pharmaceuticals for Cancer Treatment: Present and Future Challenges. Biomolecules 2022, 12, 784. [Google Scholar] [CrossRef] [PubMed]
- Luiz, M.T.; Dutra, J.A.P.; Tofani, L.B.; de Araújo, J.T.C.; Di Filippo, L.D.; Marchetti, J.M.; Chorilli, M. Targeted Liposomes: A Nonviral Gene Delivery System for Cancer Therapy. Pharmaceutics 2022, 14, 821. [Google Scholar] [CrossRef]
- Wong, K.-Y.; Wong, M.-S.; Liu, J. Aptamer-Functionalized Liposomes for Drug Delivery. Biomed J 2024, 47, 100685. [Google Scholar] [CrossRef]
- Yu, S.; Bi, X.; Yang, L.; Wu, S.; Yu, Y.; Jiang, B.; Zhang, A.; Lan, K.; Duan, S. Co-Delivery of Paclitaxel and PLK1-Targeted SiRNA Using Aptamer-Functionalized Cationic Liposome for Synergistic Anti-Breast Cancer Effects In Vivo. J Biomed Nanotechnol 2019, 15, 1135–1148. [Google Scholar] [CrossRef]
- Deng, R.; Shen, N.; Yang, Y.; Yu, H.; Xu, S.; Yang, Y.-W.; Liu, S.; Meguellati, K.; Yan, F. Targeting Epigenetic Pathway with Gold Nanoparticles for Acute Myeloid Leukemia Therapy. Biomaterials 2018, 167, 80–90. [Google Scholar] [CrossRef]
- Ayatollahi, S.; Salmasi, Z.; Hashemi, M.; Askarian, S.; Oskuee, R.K.; Abnous, K.; Ramezani, M. Aptamer-Targeted Delivery of Bcl-XL ShRNA Using Alkyl Modified PAMAM Dendrimers into Lung Cancer Cells. Int J Biochem Cell Biol 2017, 92, 210–217. [Google Scholar] [CrossRef]
- Aliabadi, A.; Vakili-Azghandi, M.; Abnous, K.; Taghdisi, S.M.; Ramezani, M.; Alibolandi, M. Nucleolin-Targeted Cationic Nanoparticle for Delivery of Survivin ShRNA against Colorectal Cancer in Vitro and in Vivo. Eur Polym J 2024, 208, 112872. [Google Scholar] [CrossRef]
- Fu, Z.; Xiang, J. Aptamer-Functionalized Nanoparticles in Targeted Delivery and Cancer Therapy. Int J Mol Sci 2020, 21, 9123. [Google Scholar] [CrossRef] [PubMed]
- Yin, X.; He, Z.; Ge, W.; Zhao, Z. Application of Aptamer Functionalized Nanomaterials in Targeting Therapeutics of Typical Tumors. Front Bioeng Biotechnol 2023, 11. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Yang, Y.; Qi, H.; Cui, W.; Zhang, L.; Fu, X.; He, X.; Liu, M.; Li, P.; Yu, T. CRISPR/Cas9 Therapeutics: Progress and Prospects. Signal Transduct Target Ther 2023, 8, 36. [Google Scholar] [CrossRef] [PubMed]
- Ehrke-Schulz, E.; Schiwon, M.; Leitner, T.; Dávid, S.; Bergmann, T.; Liu, J.; Ehrhardt, A. CRISPR/Cas9 Delivery with One Single Adenoviral Vector Devoid of All Viral Genes. Sci Rep 2017, 7, 17113. [Google Scholar] [CrossRef] [PubMed]
- Duan, L.; Ouyang, K.; Xu, X.; Xu, L.; Wen, C.; Zhou, X.; Qin, Z.; Xu, Z.; Sun, W.; Liang, Y. Nanoparticle Delivery of CRISPR/Cas9 for Genome Editing. Front Genet 2021, 12. [Google Scholar] [CrossRef]
- Yip, B. Recent Advances in CRISPR/Cas9 Delivery Strategies. Biomolecules 2020, 10, 839. [Google Scholar] [CrossRef]
- Liu, B.-Y.; He, X.-Y.; Zhuo, R.-X.; Cheng, S.-X. Tumor Targeted Genome Editing Mediated by a Multi-Functional Gene Vector for Regulating Cell Behaviors. Journal of Controlled Release 2018, 291, 90–98. [Google Scholar] [CrossRef]
- Khademi, Z. , Ramezani, M., Alibolandi, M., Zirak, M. R., Salmasi, Z., Abnous, K., & Taghdisi, S. M.. A novel dual-targeting delivery system for specific delivery of CRISPR/Cas9 using hyaluronic acid, chitosan and AS1411. Carbohydrate Polymers 2022, 292, 119691. [Google Scholar] [CrossRef]
- Kundert, K.; Lucas, J.E.; Watters, K.E.; Fellmann, C.; Ng, A.H.; Heineike, B.M.; Fitzsimmons, C.M.; Oakes, B.L.; Qu, J.; Prasad, N.; et al. Controlling CRISPR-Cas9 with Ligand-Activated and Ligand-Deactivated SgRNAs. Nat Commun 2019, 10, 2127. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, Y.; Lin, J.; Xu, L. Theophylline-Induced Synergic Activation of Guide RNA to Control CRISPR/Cas9 Function. Chemical Communications 2021, 57, 5418–5421. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.E.; Kim, M.-Y. Cancer Epigenetics: Past, Present and Future. Semin Cancer Biol 2022, 83, 4–14. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Lee, J.Y.; Gao, L.; Yin, J.; Duan, Y.; Jimenez, L.A.; Adkins, G.B.; Ren, W.; Li, L.; Fang, J.; et al. A DNA Aptamer for Binding and Inhibition of DNA Methyltransferase 1. Nucleic Acids Res 2019. [Google Scholar] [CrossRef] [PubMed]
- Di Ruscio, A.; Ebralidze, A.K.; Benoukraf, T.; Amabile, G.; Goff, L.A.; Terragni, J.; Figueroa, M.E.; De Figueiredo Pontes, L.L.; Alberich-Jorda, M.; Zhang, P.; et al. DNMT1-Interacting RNAs Block Gene-Specific DNA Methylation. Nature 2013, 503, 371–376. [Google Scholar] [CrossRef] [PubMed]
- Esposito, C.L.; Autiero, I.; Sandomenico, A.; Li, H.; Bassal, M.A.; Ibba, M.L.; Wang, D.; Rinaldi, L.; Ummarino, S.; Gaggi, G.; et al. Targeted Systematic Evolution of an RNA Platform Neutralizing DNMT1 Function and Controlling DNA Methylation. Nat Commun 2023, 14, 99. [Google Scholar] [CrossRef]
- Klett-Mingo, J.I.; Pinto-Díez, C.; Cambronero-Plaza, J.; Carrión-Marchante, R.; Barragán-Usero, M.; Pérez-Morgado, M.I.; Rodríguez-Martín, E.; Toledo-Lobo, M.V.; González, V.M.; Martín, M.E. Potential Therapeutic Use of Aptamers against HAT1 in Lung Cancer. Cancers (Basel) 2022, 15, 227. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




