1. Introduction
Rapid and accurate diagnosis of highly infectious viral diseases is a key to controlling disease outbreaks in susceptible animals. Devastating infectious animal diseases have huge negative impacts on agriculture and the economy in affected countries. As global trade and transportation become an integral part of the economy and growth of countries, they also risk accidental or intentional (bioterrorism) introduction of diseases into areas with naive livestock that can lead to disease outbreaks. Timely and accurate diagnosis of transboundary animal diseases (TADs) is an important first step to detect then prevent, control, and stop the spread of diseases. Peste des petits ruminants (PPR), goatpox (GP), sheeppox (SP), lumpy skin disease (LSD), and African swine fever (ASF) are TADs, caused by peste des petits ruminants virus (PPRV), goatpox virus (GTPV), sheeppox virus (SPPV), lumpy skin disease virus (LSDV), and African swine fever virus (ASFV), respectively. The disease caused by these agents are reportable to the World Organization for Animal Health (WOAH, previously OIE).
PPR is a disease of small ruminants, such as goats and sheep, that is currently considered as one of the major TADs with outbreaks in many parts of the world, including Europe, Asia, and Africa [
1,
2,
3,
4]. PPRV belongs to the genus
Morbillivirus in the family
Paramyxoviridae. At the genomic level, PPRV contains a single-stranded, non-segmented, negative-sense RNA genome of approximately 16 kb [
1].
GP, SP, and LSD are the diseases of goats, sheep, and cattle, respectively, caused by GTPV, SPPV and LSDV, respectively. These viruses belong to the genus
Capripoxvirus (CaPV) in the family
Poxviridae. Capripoxviruses (CaPVs) are endemic to many countries in the Asia subcontinent and Africa [
5] with recent outbreaks (LSD) reported in many Asian countries [
6]. All CaPVs have a double-stranded DNA genome of approximately 150 kb and share 147 putative genes that are highly conserved (96–97%) between the species [
7].
ASF is a highly infectious and lethal hemorrhagic viral disease of domestic swine and related wild reservoir hosts, including wild boar, warthogs, and feral swine. The disease is currently present in many countries in Asia and Europe and, more recently, in the Caribbean [
8,
9]. The causative agent, ASFV, is a DNA virus belonging to the genus
Asfivirus and the family
Asfarviridae [
10]. The ASFV genome consists of double-stranded DNA of 170-193 kb that contains between 151 and 167 open reading frames (ORFs), depending on the virus strain [
10].
Whether routine surveillance or disease outbreak, rapid and sensitive detection of the causative agent(s) is an important first step towards the prevention and control of animal diseases. Molecular detection of viral DNA/RNA by PCR or loop-mediated isothermal amplification (LAMP) is rapid, specific, and sensitive; and the turnaround time is much shorter (~1-2 hours) than traditional methods such as VI and antigen ELISA. Genomic identification based on the amplification of viral DNA/RNA by PCR or LAMP has been routinely used for rapid detection of PPRV [
11,
12], CaPVs [
13,
14,
15], and ASFV [
16,
17,
18].
Despite of high sensitivity and specificity, the performance of PCR or LAMP can be compromised by multiple factors such as, PCR inhibition caused by naturally occurring PCR inhibitors or failure to detect the causative agents (virus) in specimens during the initial (asymptomatic) phase of infection when the viral load remains low. In both cases, a true positive sample can be tested as false negative that may lead to failure of disease control and prevention efforts.
The Nanotrap
® Microbiome A Particles (NMAPs) are highly porous, thermostable hydrogel particles coupled with chemical affinity baits that can capture and concentrate a broad range of analytes including virions [
19,
20]. Recently, we have shown that NMAPs can capture and concentrate PPRV from diluted suspensions, and the captured viruses remained infectious and recoverable by virus isolation (VI) using susceptible cell lines (21). In this study, NMAPs were used to capture, concentrate, and recover several highly infectious animal disease viruses, including GTPV, SPPV, LSDV, PPRV, and ASFV. The virus capture and recovery were analyzed by VI and virus-specific qPCR/RT-qPCR.
2. Materials and methods
2.1. Virus Strains, Cell Culture, and Virus Isolation
All viruses used in this study were obtained from the biorepository of the Reagents and Vaccine Services Section (RVSS) of the Foreign Animal Disease Diagnostic Laboratory (FADDL) at the Plum Island Animal Disease Center (PIADC). The viruses include GTPV strain Pendik (GTPV-Pendik), SPPV strain HELD (SPPV-HELD), LSDV strain Cameroon (LSDV-Cameroon), PPRV strain Egypt (PPRV-Egypt), PPRV strain Turkey (PPRV- Türkiye) and a Vero adapted ASFV Lisbon-60 vaccine strain BA71V. A virulent ASFV strain Georgia (ASFV-Georgia) was also used and was provided by the Proficiency and Validation Services Section (PVSS) of FADDL. The TCID50/mL titers of the original supernatants of the virus cultures used in virus capture experiments were GTPV, SPPV, LSDV, and PPRV (Egypt) at 106.6, 106.6, and 106.8, and 105.5, respectively. The titers of the ASFV strains were not available.
For VI, the viruses were inoculated onto susceptible cells grown on 6-well microtiter plates. Vero E6 cells were used for PPRV and ASFV (BA17V), while primary LK cells were used for GTPV, SPPV, and LSDV. The Vero and LK cells were grown in Eagle’s Minimum Essential Medium (EMEM) supplemented with FBS (10% v/v for LK and 7% v/v for Vero) plus 20% (v/v) of an antibiotic/antimycotic cocktail containing penicillin, streptomycin, and amphotericin B (Gibco/Thermo Fisher Scientific). The plates were incubated in a CO2 incubator (5% v/v) at 37 oC until the cell density reached 70-75% confluency (2-3 days post-incubation). Freshly prepared cells at the desired confluency (70-75%) were inoculated with the virus for VI. After inoculation, the plates were incubated in a CO2 incubator for 1 hour for adsorption. Next, the plates were overlaid with EMEM supplemented with 4% FBS plus antibiotics (above) and further incubated in a CO2 incubator for an extended period, and the cells were examined for cytopathic effect (CPE) by microscopy (Evos XL Core, Invitrogen/Thermo Fisher Scientific, Waltham MA).
2.2. Animal Experiments and Collection of Diagnostic Samples
Diagnostic samples used in this study were collected either from in-house (PIADC) animal experiments (SPPV, PPRV or ASFV) or from natural infections (ASFV). Animal experiments were carried out in BSL-3 Ag isolation rooms at PIADC. For SP and PPR, sheep and goats 6–8-month-old, weight 40-60Ibs, mixed breed, were used, respectively. For ASF, female swine of approximately 60-90Ibs, Yorkshire breed, were used. Sheep (n=6) were inoculated intravenously with SPPV-HELD while goats (n=8) were inoculated intranasally with PPRV-Türkiye. Swine (n=20) were inoculated intramuscularly (IM) with ASFV-Georgia. All animal procedures were performed following Protocol 225-10-R approved by the Plum Island Animal Disease Center Institutional Animal Care and Use Committee (IACUC), which ensured ethical and humane treatment of experimental animals. After collection of diagnostic samples, the animals (sheep, goats and swine) were given xylazine intramuscularly to sedate and then Fatal Plus IV to euthanize.
Experimental samples used in the study include swabs (nasal, oral and conjunctival) immersed in 1 mL of EMEM plus 5% antibiotic/antimycotic in cryovials and EDTA whole blood (EWB) in EDTA tube (Becton, Dickinson and co., Franklin Lakes, NJ). All samples were collected between 7- and 10-days post inoculation when the animals exhibited mild to severe clinical signs typical for the inoculating viruses including fever, conjunctivitis and swelling of skin (papules) for sheep (SPPV); fever, diarrhea and nasal discharges for goats (PPRV); and fever, anorexia, depression, diarrhea staggering gait, and purple skin discoloration for swine (ASFV). The ASFV samples (EWB) were kindly provided by Agriculture Research Services of USDA (USDA-ARS) at PIADC. After collection, all samples were transferred to the laboratory (BSL3) and stored at −70 °C until they were analyzed. The ASFV samples (clinical specimens) were also obtained from the recent ASF outbreaks in Dominical Republic (DR) which included EWB from naturally infected swine (n=20) collected in 2023 by the Laboratorio Veterinario Central (LAVECEN) in the Dominican Republic (DR) and kindly provided to us by PVSS (Proficiency and Validation Services) at FADDL. The EWB samples were hemolyzed by freeze-thaw (freeze at -70 oC for 15 min followed by thawing at RT for 15 min) to prevent blood clot prior to use in downstream applications, including virus capture and nucleic acid extractions.
2.3. Optimization of Virus Capture Using NAMPs
Previously, we showed that 100 μl of NMAPs (Microbiome A Particles; SKU 44202; CERES Nanosciences Inc., Manassas, VA) was optimum to capture PPRV from diluted suspensions (21). We repeated the virus capture protocol on other viruses used in this study. A fixed amount of each virus (GTPV, SPPV, LSDV or ASFV) was diluted in PBS or EMEM in multiple volumes (2-, 5-, 10-, 20-, 50-μl) and then subjected to virus capture using different amounts of NMAPs (50-, 75-, 100-, 150- and 200-μl). The NMAPs and the captured viruses were clarified on a magnetic stand (DynaMag-2, -5, -15, or -50; Thermo Fisher Scientific), reconstituted in PBS (200 μl), extracted (viral DNA/RNA), and analyzed by virus-specific qPCR/RT-qPCR. Based on the Ct values, it was found that the optimum recovery (capture) of the viruses was achieved using 100-, 150- or 200-μl of NMAPs (not shown). Therefore, unless or otherwise stated, 100 ml of NMAPs were used in all virus capture experiments reported onwards.
2.4. Assessment of Virus Capture by PCR and Virus Titration
This experiment was carried out using PPRV as the target. Two hundred μl of PPRV-Egypt (original stock; TCID50/ml 105.5) was used as the starting material; the virus was diluted 1:50 in EMEM (final volume 10 ml) and then subjected to virus capture using 100 μl of NMAPs. The NMAPs and the captured viruses were clarified on a magnetic stand, washed (1 ml of EMEM), and reconstituted in 200 μl of EMEM. Reconstituted NMAPs and the captured viruses, referred to as “treated” afterwards, were serially diluted in EMEM and analyzed separately by PPRV RT-qPCR and virus titration (described below). For positive control (PC), referred to as “untreated” afterwards, 200 μl of PPRV (TCID50/ml 105.5) was serially diluted in EMEM and analyzed separately by RT-qPCR and virus titration.
Virus (PPRV) titrations were performed on Vero E6 cells grown in 96-well plates. The plates were inoculated with 100 μl of Vero E6 at 10
4 cells/well and then incubated at 37
oC in a CO
2 incubator. Once the cells reached 70-75% confluency (~ 2-3 days post-incubation), they were inoculated with serially diluted PPRV (untreated) or reconstituted NMAPs and the captured viruses (treated) at a rate of 50 μl/well in duplicate. After absorption of the viruses (1 hour incubation at 37
oC in CO
2 incubator), the plates were overlaid with 50 μl of EMEM (4% FBS plus antibiotics) and re-incubated in CO
2 incubator for an extended period (up to 10 days). Monolayers were examined by microscopy (Evos XL Core) for the development of CPE. The virus titers were expressed as Log
10 TCID
50/ml based on a calculation described by Cottral [
22].
2.5. Virus Capture from Suspensions Containing Multiple Viruses
In this experiment, we examined whether NMAPs can capture and concentrate multiple viruses from suspensions. Five viruses used in the study were mixed in three different combinations in PBS in a final volume of 5 ml as follows: GTPV + PPRV + ASFV; SPPV + PPRV + ASFV; and LSDV + PPRV + ASFV. Each virus alone (GTPV, SPPV, LSDV, PPRV, or ASFV) was also diluted in PBS (5 ml final volume) and used as a positive control (PC). The virus suspensions containing either single virus (homogeneous) or multiple viruses (heterogeneous) were then subjected to virus capture using 100 ml of NMAPs. The NMAPs and the captured viruses were clarified on a magnetic stand, reconstituted (200 μl of PBS), extracted (viral DNA/RNA), and analyzed by virus-specific qPCR/RT-qPCR (section 2.8 below).
2.6. Virus Capture from Whole Blood
In preliminary studies, we found that the viruses captured from whole blood (EWB) was partially blocked, which was most likely due to hemoglobin (HMB) that also bound to NMAPs and outcompeted the viruses (not shown). To improve virus-capture from EWB (spiked), two different protocols were tested. In one protocol, EWB (spiked with virus) was diluted 1:10 in PBS (pH 7.2; filtered through a 0.2 μ membrane filter) and then subjected to virus capture using NMAPs. In the other protocol, EWB (spiked) was treated with HemogloBind™ (HGB; Biotech Support Group, Monmouth Junction, NJ), which specifically binds and precipitates HMB without interfering with virus capture. EMEM could not be used as a diluent since it caused the formation of blood clots that significantly inhibited virus-capture and recovery. EWB from healthy goats, sheep, cattle, or swine (Innovative Research Inc.; Novi, MI) was used in this experiment.
In the dilution protocol, 2 ml of EWB samples (undiluted or diluted 1:10 in PBS) were spiked with the virus (20 μl; original stock) and then subjected to virus capture using 100 μl of NMAPs. For positive control (PC), the same amount of virus (20 μl) was spiked into PBS (2 ml) and then subjected to virus capture using NMAPs. The NMAPs and the captured viruses were clarified on a magnetic stand, washed (1 ml PBS), reconstituted (200 μl PBS), extracted (viral DNA/RNA), and analyzed separately by VI and virus-specific qPCR/RT-qPCR.
In the HGB protocol, EWB spiked with the virus was treated with HGB to remove/precipitate HMB according to the manufacturer’s instructions. Accordingly, EWB from goat, sheep, cattle and swine was spiked with GTPV or PPRV, SPPV, LSDV, and AFV, respectively. Briefly, 200 μl of EWB was mixed with the virus (20 μl) in a 1.5 ml Eppendorf tube and incubated at RT on a shaker for 5 min for equilibration. Next, 200 μl of HGB was added, and the contents were further incubated at RT on shaker at RT for 10 min to facilitate binding of HMB to HGB. The contents were centrifuged at 10,000 x g for 4 min at RT. The precipitate (HMB + HGB) was discarded, and the clear supernatant containing the residual viruses was diluted to 2 ml in PBS and then subjected to virus capture using NMAPs (100 μl). The NMAPs and the captured viruses were clarified on a magnetic stand, washed in PBS (1 ml), reconstituted in PBS or EMEM (for VI), and analyzed separately by qPCR/RT-qPCR and VI.
2.7. Virus Capture from Experimental and Clinical Samples and Diagnostic Sensitivity
The NMAPs were used to capture and concentrate viruses from experimental and clinical samples to determine diagnostic sensitivity (DSe). Briefly, 200 μl of swabs or 100 μl of EWB was diluted in PBS (2 ml final volume) and then subjected to virus capture using 100 μl of NMAPs. The NMAPs and the captured viruses were clarified on a magnetic stand, reconstituted (200 μl PBS), extracted (viral DNA/RNA), and analyzed by virus-specific qPCR/RT-qPCR as described below (section 2.8). For validation/comparison purposes, nucleic acids (viral DNA/RNA) were also extracted from untreated diagnostic samples as well as negative extraction control (NEC; 200 μl of PBS or NMAPs) and analyzed by virus-specific qPCR/RT-qPCR.
2.8. Nucleic Acid (DNA/RNA) Extractions and qPCR/RT-qPCR
The QIAmp® Viral RNA Mini Kit (Germantown, MD) was used for the purification of viral RNA (PPRV), and the Cyclone DNA/RNA Purification Kit (DTPM; Fort Payne, AL) was used for the purification of viral DNA (GTPV, SPPV, LSDV, and ASFV). The latter kit (DTPM) was also used for the purification of viral DNA/RNA from EWB samples. All blood samples (EWB) were hemolyzed by freeze-thaw (freezing at -80 oC and thawing at RT; above) prior to extractions. Briefly, 200 μl of sample was used for each extraction and the extracted nucleic acids (DNA/RNA) was eluted with 100 μl of elution buffer.
Virus-specific qPCR/RT-qPCR assays were carried out on an Applied Biosystems 7500 Fast thermocycler (Thermo Fisher Scientific). The PPRV RT-qPCR was carried out according to Batten et al. [
15] using the Path-ID™ Multiplex One-Step RT-PCR Kit (Thermo Fisher Scientific) as described [
24]. The ASFV qPCR was carried out according to Zsak et al. [
19] using TaqMan™ Fast Virus 1-Step Mastermix (Thermo Fisher Scientific). The CaPV (GTPV, SPPV, or LSDV) qPCR assays were carried out using Path-ID™ qPCR Mastermix according to Das et al. [
17,
18]. The oligonucleotide primers (forward and reverse) for the PCR assays were purchased from Integrated DNA Technology (Coralville, IA), and the TaqMan probes (labeled with FAM as reporter dye at the 5’-end and MGB as the quencher dye at the 3’-end) were purchased from Thermo Fisher Scientific. The PCR reaction master mixes were prepared as per the manufacturer’s instructions to include 1x buffer, enzymes (
Taq DNA polymerase for qPCR and a reverse transcriptase plus
Taq DNA polymerase for RT-qPCR), primers, probe, and 5 μl of template (extracted viral DNA/RNA) plus required amount of nuclease-free water in a final volume of 25 μl. The thermocycling conditions for PCR amplification were as follows:
ASFV qPCR: one cycle of 95 oC for 20s followed by 40 cycles of amplification with each cycle consisting of 95 oC for 10s and 60 oC for 30s;
CaPV qPCR: one cycle of 95 oC for 10 min (enzyme activation/template denaturation) followed by 40 cycles of amplification with each cycle consisting of 95 oC for 15 s and 60 oC for 60 s; and
PPRV RT-qPCR: one cycle of 45 oC for 10 min (reverse transcription), one cycle of 95 oC for 10 min (enzyme activation/template denaturation) and 40 cycles of amplification with each cycle consisting of 95 oC for 15 s and 60 oC for 60 s.
4. Discussion
One of the objectives of this study was to examine and evaluate the efficacy of Nanotrap
® particles to capture and concentrate highly infectious animal disease viruses from diluted suspensions or diagnostic samples to improve their recovery and sensitivity of detection. Nanotrap
® particles are known to capture low abundance targets/analytes from different types of matrices, such as gas, liquids, or biological fluids [
19]. NMAPs have been previously used for the enrichment of several infectious human and animal disease viruses, including Rift Valley Fever virus (RVFV), coronaviruses, influenza viruses, and respiratory syncytial viruses [
20,
23,
24,
25]. In this study, hydrogel Nanotrap™ particles, also referred to as Nanotrap
® microbiome A particles or NMAPs, were used to capture and concentrate several highly infectious transboundary animal disease viruses, including GTPV, SPPV, LSDV, PPRV, and ASFV. Initial optimization of virus-capture was carried out using virus suspensions in PBS or EMEM, and the efficiency of virus capture was assessed using virus-specific qPCR/RT-qPCR and VI. The optimized protocol was subsequently used to capture and concentrate viruses from diagnostic (experimental and clinical) samples to determined DSe.
Optimum recovery of the viruses (GTPV, SPPV, LSDV, PPRV, or ASFV) from diluted suspensions was obtained using 100 ml of NMAPs, also reported earlier by us [
21] and others [
24,
25,
26]. All the viruses (GTPV, SPPV, LSDV, PPRV, and ASFV) captured by NMAPs were recoverable by VI using virus-specific susceptible cell lines (
Figure 1 and
Figure 2), indicating that the infectivity of the viruses was not compromised by NMAPs.
To improve the recovery and sensitivity of detection of the viruses in diluted suspensions, a fixed amount (20 μl) of appropriately diluted virus (GTPV, SPPV, LSDV, PPRV, or ASFV) was further diluted (in PBS) from 10-fold (200 μl) to 10,000-fold (200 ml) and then subjected to virus capture (except 200 μl) using NMAPs and analyzed by virus specific qPCR/RT-qPCR. The results (
Table 2) show untreated viruses were detectable only up to a 10-fold dilution (2 ml), while they were detectable up to 1000-fold dilution (200 ml) after being captured using NMAPs (treated), a 100-fold increase in the sensitivity of detection compared to the untreated viruses. Furthermore, the NMAPs and the captured viruses from all dilutions exhibited similar Ct values (between 34 and 35), indicating a very similar efficiency (~ 100%) of virus capture irrespective of the virus or the dilution. Indeed, improved sensitivity of detection (up to 10-fold) of SARS-CoV-2 using NMAPs has also been reported by others (24).
Further assessment of virus capture was carried out by virus titration using PPRV as the target. The results (
Table 1) show comparable titers (TCID
50/ml) corresponding to the serial dilutions of the viruses either treated or untreated, further confirming efficient recovery of the virus using NMAPs.
The amplification efficiency (AE) of the viral DNA/RNA extracted from NMAPs and the captured viruses (LSDV, PPRV, or ASFV) from serial dilutions was determined and compared against that extracted from same dilutions of the untreated viruses. The linear regression standard curves (Ct vs serial dilution; Supplemental Figure 1) showed AEs were comparable and within the acceptable range (between 90 and 100%) for the viral DNA/RNA extracted from either treated or untreated viruses. The combined results of VI, virus titration, and qPCR/RT-qPCR indicate there were no adverse effects of NMAPs on the cells (Vero or LK) or the infectivity of the viruses and there is minimal or no interference of the NMAPs on nucleic acid extraction or amplification.
The virus-capture (NMAPs) from EWB (spiked) and their analysis by qPCR/RT-qPCR (
Table 3) showed partial recovery of the viruses (detectable at higher Cts) from undiluted EWB compared to that from diluted (1:10) EWB. Further analysis of the NMAPs and the captured viruses by VI showed cells inoculated with the viruses captured from diluted (1:10) EWB developed CPE after the 1
st passage, while that captured from undiluted EWB developed CPE after the 2
nd passage (
Table 4). The delayed recovery (2
nd passage) of the viruses captured from undiluted EWB was most likely due to partial blocking of virus capture by HMB since it also binds NMAPs and outcompeted the viruses. This was corroborated by the treatment of the EWB with HGB to remove HMB that resulted in a faster recovery (1
st passage) (
Table 4;
Figure 3).
The NAMPs were also used to capture and concentrate viruses (PPRV, SPPV, or ASFV) from experimental and clinical samples to determine DSe. The DSe of the virus-specific qPCR/RT-qPCR on the viruses determined using NMAPs was shown to be comparable (100%) to that determined using untreated samples (
Table 5 and Supplemental tables S1, S2, S3 and S4). The PCR results (Supplemental tables S1, S2, S3 and S4) also show Ct values were slightly but consistently lower for the viruses captured from EWB compared to that from untreated EWB. The higher Ct values of the viruses from untreated EWB could be due to PCR inhibition by HMB, which was partially separated and removed in the treated EWB after virus capture.
It should be noted that all viruses (GTPV, SPPV, LSDV, PPRV, and ASFV) were captured/recovered from diluted suspensions at very similar efficiencies (~ 100%), which could have been due to all being enveloped viruses with a strong affinity for binding to the NMAPs as previously reported [
20,
21,
24,
25]. NMAPs were also shown to capture and concentrate multiple viruses form diluted suspensions (heterogeneous) at similar efficiencies as that from suspensions (homogeneous) containing a single virus (
Table 6). Capture of and enrichment of multiple respiratory viruses (influenza virus, respiratory syncytial virus, and coronavirus) from specimens using NMAPs has also been reported by others (20). These findings show NMAPs can be used to detect multiple viral pathogens in specimens from animals during a co-infection scenario.
One of the major challenges of diagnostic PCR is the false negatives that can occur either due to PCR inhibition by naturally occurring PCR inhibitors or low level of virus in the samples. Naturally occurring PCR inhibitors present in body fluids or environmental samples include hemoglobin (blood), bile salts and complex polysaccharides (feces), urea (urine), calcium ions (bone, milk), or environmental polysaccharides and humic acid (soil, water) [
26,
27]. These inhibitors often co-extract and co-purified with the template (DNA/RNA) during extractions and interfere (i.e., PCR inhibition) in the downstream applications (e.g., amplification). There are methodologies/protocols available to neutralize the effect of PCR inhibition, including: 1) use of inhibitor-resistant recombinant
Taq DNA polymerases [
28,
29] or native thermostable DNA polymerases [
30]; 2) use of enhancers or facilitators such as dimethyl sulfoxide (DMSO), betaine, cattle serum albumin (BSA), or glycerol [
29]; 3) modification of nucleic acid extraction protocols by adding extra washing steps [
31]; or 4) decreasing the inhibitor concentration by sample dilution [
32]. Alternatively, the interference of PCR inhibitors can be neutralized by sample dilution followed by virus capture using NMAPs (this study). Therefore, NMAPs can be used to enhance the detection of viruses in biological fluids containing PCR inhibitors or in diluted samples where the virus concentrations remain below the limit of detection (LOD) of PCR as in the specimens from animals at the early stages of infection.
Viruses can be concentrated from diluted suspensions/samples by other methods including membrane filtration, skim milk flocculation, polyethylene glycol (PEG) precipitation, or adsorption-extraction [
33,
34]. These methods can be expensive (equipment) and/or labor-intensive; therefore, they are not applicable in low-resource settings. One of the prime examples of concentrating viruses/pathogens is the wastewater-based surveillance (WBS) of communicable diseases, which has now become one of the most powerful tools used in monitoring public health since the outbreak of SARS-CoV-2 [
35,
36,
37,
38,
39]. This study shows NMAPs efficiently captured viruses from a wide range of dilutions (1:10 or 1:1000), and therefore concentrating viruses using NMAPs can be more effective than other methods. The NMAPs have been successfully used in WBS of several human pathogens, including SARS-CoV-2, monkeypox, enterovirus, human norovirus, human adenovirus, bocavirus, Epstein Barr virus, influenza A virus, and respiratory syncytial virus B [
35,
36,
37,
38,
39]. Likewise, WBS can be a powerful tool in disease surveillance to monitor highly infectious animal disease viruses in wastewater/discharge from livestock farms, wet markets, or slaughterhouses that are reservoirs and the sites of amplification of infectious agents [
40].
NMAPs have been successfully used to monitor (by PCR) virus loads in biological fluids for detection of several zoonotic and animal disease viruses, including Rift valley fever virus, Venezuelan equine encephalitis virus, and influenza viruses [
41]. One of the potential advantages of the NMAPs is to counter false negatives (PCR) that occurs due to either PCR inhibition or low level of virus (below the LOD of PCR) in the sample. NMAPs can be used to capture, concentrate and separate viruses from PCR inhibitors in diluted suspensions to enhance recovery and improve their detection. These applications of NMAPs can aid in disease surveillance and accurate diagnosis of animal disease viruses by diagnostic PCR.
Supplemental Figure 1.
Linear regression standard curves and amplification efficiencies (AE) of the viral DNA/RNA extracted from serial dilutions of the viruses (LSDV, PPRV or ASFV) with (+NMAPS) or without (-NMAPs) virus capture. Viruses were serially (10-fold) diluted in PBS and used as the starting material. For untreated samples, 200 ml of virus from each dilution was extracted directly and analyzed by qPCR/RT-qPCR. For treated (+NMAPs) samples, the same amount of virus (200 μl) from each dilution was further diluted 1:50 (10 ml) in PBS and then subjected to virus capture using NMAPs. The NMAPs and the captured viruses were clarified on a magnetic stand, reconstituted in PBS (200 ml), extracted (viral DNA/RNA), and analyzed by virus-specific qPCR/RT-qPCR. A, standard curve of ASFV DNA extracted from serial dilutions; B, standard curve of PPRV RNA extracted from serial dilutions; and C, standard curve of LSDV DNA extracted from serial dilutions. The inset shows amplification efficiency (AE) calculated from the standard curves. Each data point (Ct values) represents an average of 3 replicates with a standard deviation of “mean ± 0.155”.
Supplemental Figure 1.
Linear regression standard curves and amplification efficiencies (AE) of the viral DNA/RNA extracted from serial dilutions of the viruses (LSDV, PPRV or ASFV) with (+NMAPS) or without (-NMAPs) virus capture. Viruses were serially (10-fold) diluted in PBS and used as the starting material. For untreated samples, 200 ml of virus from each dilution was extracted directly and analyzed by qPCR/RT-qPCR. For treated (+NMAPs) samples, the same amount of virus (200 μl) from each dilution was further diluted 1:50 (10 ml) in PBS and then subjected to virus capture using NMAPs. The NMAPs and the captured viruses were clarified on a magnetic stand, reconstituted in PBS (200 ml), extracted (viral DNA/RNA), and analyzed by virus-specific qPCR/RT-qPCR. A, standard curve of ASFV DNA extracted from serial dilutions; B, standard curve of PPRV RNA extracted from serial dilutions; and C, standard curve of LSDV DNA extracted from serial dilutions. The inset shows amplification efficiency (AE) calculated from the standard curves. Each data point (Ct values) represents an average of 3 replicates with a standard deviation of “mean ± 0.155”.
Table S1.
Diagnostic sensitivity of ASFV qPCR with DNA extracted from specimens of experimentally infected animals after virus capture using NMAPs.
Table S1.
Diagnostic sensitivity of ASFV qPCR with DNA extracted from specimens of experimentally infected animals after virus capture using NMAPs.
| Virus |
Host |
# Animals |
Dpi |
Specimen |
Animal ID |
Cycle threshold (qPCR) |
Untreated*
(-NMAPs) |
Treated#
(+NMAPs) |
| ASFV |
Swine |
20 |
7 |
EWB |
52468 |
17.939 |
17.111 |
| 52469 |
17.481 |
17.084 |
| 52470 |
16.115 |
16.124 |
| 52471 |
16.855 |
16.243 |
| 52.792 |
18.479 |
18.120 |
| 52793 |
20.388 |
20.072 |
| 52795 |
19.375 |
19.165 |
| 52796 |
17.905 |
17.649 |
| 53465 |
28.951 |
28.541 |
| 53466 |
20.162 |
19.326 |
| 52467 |
22.593 |
21.554 |
| 53468 |
20.900 |
20.495 |
| 53469 |
19.637 |
19.100 |
| 53470 |
16.111 |
15.224 |
| 53471 |
16.855 |
16.243 |
| 53838 |
30.569 |
30.211 |
| 53839 |
21.758 |
21.123 |
| 53841 |
17.283 |
17.032 |
| 53842 |
18.500 |
17.632 |
| 53843 |
16.752 |
16.068 |
Table S2.
Diagnostic sensitivity of PPRV RT-qPCR with RNA extracted from specimens of experimentally infected animals with (treated) or without (untreated) virus capture using NMAPs.
Table S2.
Diagnostic sensitivity of PPRV RT-qPCR with RNA extracted from specimens of experimentally infected animals with (treated) or without (untreated) virus capture using NMAPs.
| Virus |
Host |
# Animals |
Dpi |
Specimen |
Animal ID |
Cycle threshold (qPCR) |
Untreated*
(-NMAPs) |
Treated#
(+NMAPs) |
| PPRV |
Goat |
8 |
8 |
EWB |
23-1 |
29.814 |
29.156 |
| 23-2 |
28.097 |
28.898 |
| 23-3 |
30.107 |
27.365 |
| 23-4 |
28.025 |
27.430 |
| 23-13 |
32.076 |
30.060 |
| 23-14 |
28.198 |
27.535 |
| 23-15 |
33.507 |
33.197 |
| 23-16 |
31.424 |
31.160 |
| Nasal swab |
23-1 |
20.993 |
19.201 |
| 23-2 |
28.477 |
26.065 |
| 23-3 |
29.109 |
27.197 |
| 23-4 |
20.364 |
18.708 |
| 23-13 |
24.098 |
25.993 |
| 23-14 |
26.279 |
27.479 |
| 23-15 |
28.097 |
30.306 |
| 23-16 |
26.814 |
24.128 |
| Oral |
23-13 |
25.407 |
27.432 |
| 23-14 |
27.801 |
29.069 |
| 23-15 |
31.847 |
30.905 |
| 23-16 |
28.884 |
27.814 |
| Conjunctival swab |
23-1 |
21.767 |
19.994 |
| 23-2 |
25.942 |
25.302 |
| 23-3 |
29.991 |
26.679 |
| 23-4 |
26.068 |
25.326 |
| 23-13 |
23.608 |
27.526 |
| 23-14 |
26.845 |
26.492 |
| 23-15 |
22.841 |
23.954 |
| 23-16 |
22.502 |
23.016 |
Table S3.
Diagnostic sensitivity of SPPV qPCR with DNA extracted from specimens of experimentally infected animals after virus capture using NMAPs.
Table S3.
Diagnostic sensitivity of SPPV qPCR with DNA extracted from specimens of experimentally infected animals after virus capture using NMAPs.
| Virus |
Host |
# Animals |
Dpi |
Specimen |
Animal ID |
Cycle threshold (qPCR) |
Untreated*
(-NMAPs) |
Treated# (+NMAPs) |
| SPPV |
Sheep |
6 |
10 |
Oral swab |
23-20 |
26.344 |
31.046 |
| 23-21 |
28.602 |
29.857 |
| 23-22 |
33.764 |
32.054 |
| 23-28 |
30.836 |
32.176 |
| 23-29 |
33.162 |
32.513 |
| 23-30 |
32.458 |
32.871 |
| Nasal swab |
23-20 |
29.088 |
22.960 |
| 23.21 |
25.898 |
26.889 |
| 23-22 |
37.022 |
37.407 |
| 23-28 |
24.412 |
25.454 |
| 23-29 |
29.240 |
31.502 |
| 23-30 |
29.284 |
31.072 |
| Conjunctival; swab |
23-20 |
27.779 |
29.219 |
| 23-21 |
27.600 |
29.739 |
| 23-22 |
35.546 |
35.869 |
| 23-28 |
18.085 |
19.427 |
| 23-29 |
26.118 |
28.356 |
| 23-30 |
28.167 |
28.871 |
| EWB |
23-20 |
24.550 |
23.989 |
| 23-21 |
27.415 |
27.479 |
| 23-22 |
23.393 |
21.898 |
| 23-28 |
27.8097 |
27.066 |
| 23-29 |
23.406 |
23.050 |
| 23-30 |
21.470 |
20.604 |
Table S4.
Diagnostic sensitivity of ASFV qPCR with DNA extracted from EWB of naturally infected swine after virus capture using NMAPs.
Table S4.
Diagnostic sensitivity of ASFV qPCR with DNA extracted from EWB of naturally infected swine after virus capture using NMAPs.
| Virus |
Host |
# Animals |
Specimen |
Animal ID |
Cycle threshold (qPCR) |
Untreated*
(-NMAPs) |
Treated#
(+NMAPs) |
| ASFV |
Swine |
20 |
EWB |
14 |
18.06 |
17.470 |
| 27 |
19.526 |
19.056 |
| 29 |
19.423 |
19.154 |
| 30 |
18.722 |
18.372 |
| 32 |
22.583 |
20,540 |
| 36 |
22.432 |
21.468 |
| 37 |
20.704 |
19.779 |
| 39 |
18.155 |
17.823 |
| 41 |
18.618 |
18.214 |
| 42 |
22.822 |
22.890 |
| 43 |
36.399 |
35.919 |
| 44 |
22.325 |
22.221 |
| 45 |
18.859 |
17.599 |
| 46 |
20.383 |
20.061 |
| 52 |
19.443 |
19.137 |
| 53 |
19.309 |
18.443 |
| 54 |
18.489 |
18.786 |
| 58 |
17.624 |
17.362 |
| 59 |
22.920 |
22.330 |
| 60 |
20.967 |
20.260 |
Figure 1.
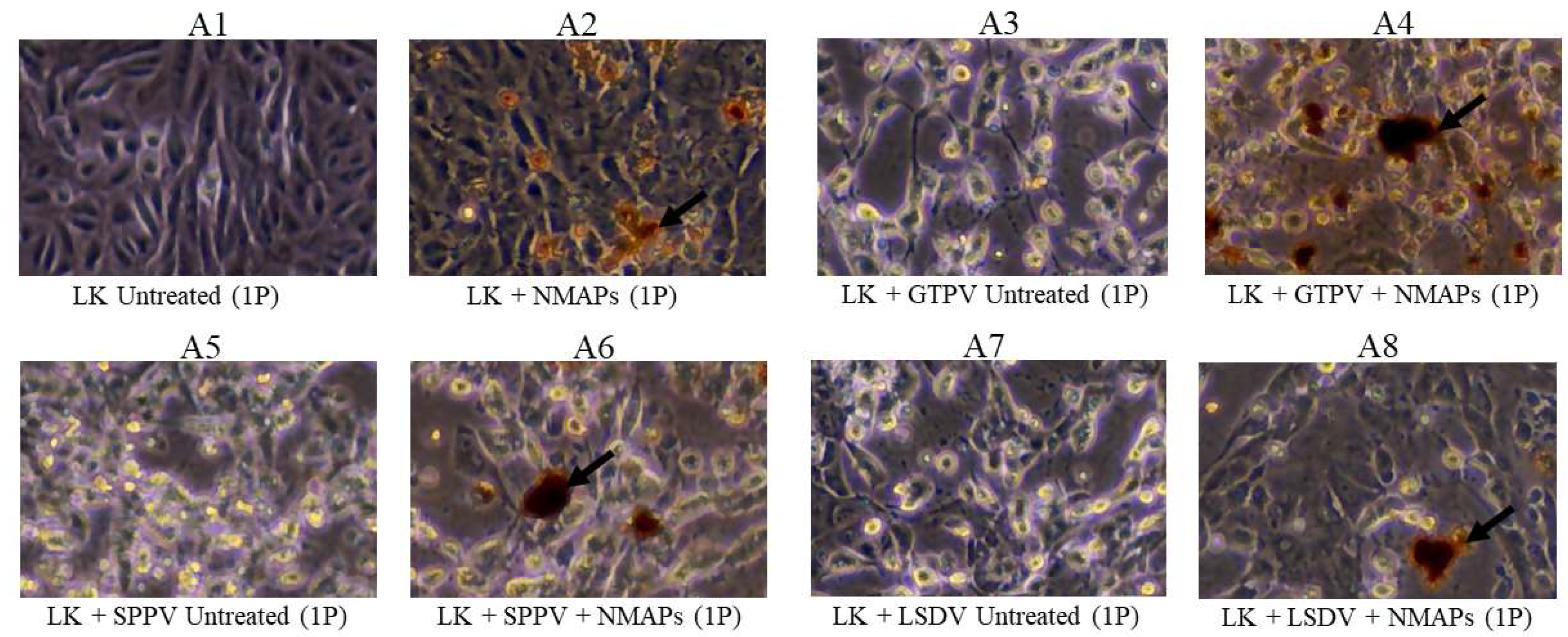
Capture and recovery of GTPV, SPPV, and LSDV from diluted suspensions using NMAPs. Viruses were captured and concentrated using NMAPs as described in the materials and methods. The NMAPs and the captured viruses were washed, reconstituted in EMEM, and inoculated onto LK cells for VI and examined by microscopy for the development of CPE for 144 h (1P). A3, cells inoculated with untreated GTPV; A4, cells inoculated with treated (+NMAPs) GTPV; A5, cells inoculated with untreated SPPV; A6, cells inoculated with treated (+NMAPs) SPPV; A7, cells inoculated with untreated (-NMAPs) LSDV; and A8, cells inoculated with treated (+NMAPs) LSDV. For negative controls, cells were inoculated with either EMEM (A1) or NMAPs (A2). NMAPs are shown by arrows.
Figure 1.
Capture and recovery of GTPV, SPPV, and LSDV from diluted suspensions using NMAPs. Viruses were captured and concentrated using NMAPs as described in the materials and methods. The NMAPs and the captured viruses were washed, reconstituted in EMEM, and inoculated onto LK cells for VI and examined by microscopy for the development of CPE for 144 h (1P). A3, cells inoculated with untreated GTPV; A4, cells inoculated with treated (+NMAPs) GTPV; A5, cells inoculated with untreated SPPV; A6, cells inoculated with treated (+NMAPs) SPPV; A7, cells inoculated with untreated (-NMAPs) LSDV; and A8, cells inoculated with treated (+NMAPs) LSDV. For negative controls, cells were inoculated with either EMEM (A1) or NMAPs (A2). NMAPs are shown by arrows.
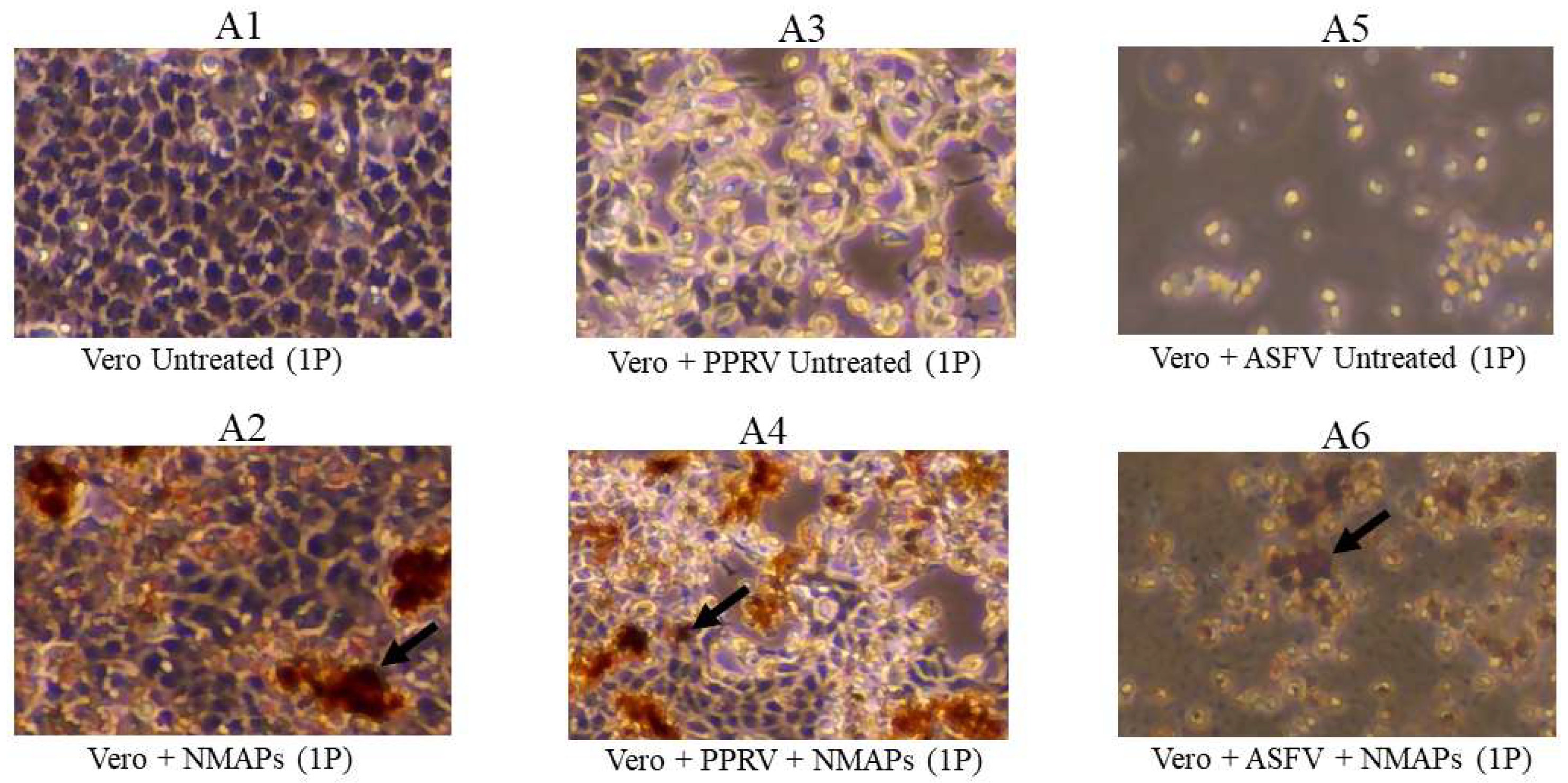
Figure 2.
Capture and recovery of PPRV and ASFV from diluted suspensions using NMAPs. Viruses were captured and concentrated using NMAPs as described in the materials and methods. The NMAPs and the captured viruses were washed, reconstituted in EMEM, and inoculated onto Vero cells for VI and monitored for the development of CPE by microscopy for up to 168-hour (1P). A3, cells inoculated with untreated PPRV; A4, cells inoculated with treated (+NMAPs) PPRV; A5, cells inoculated with untreated ASFV; A6, cells inoculated with treated (+NMAPs) ASFV. For negative controls, cells were inoculated with either EMEM (A1) or NMAPs (A2). NMAPs are shown by arrows.
Figure 2.
Capture and recovery of PPRV and ASFV from diluted suspensions using NMAPs. Viruses were captured and concentrated using NMAPs as described in the materials and methods. The NMAPs and the captured viruses were washed, reconstituted in EMEM, and inoculated onto Vero cells for VI and monitored for the development of CPE by microscopy for up to 168-hour (1P). A3, cells inoculated with untreated PPRV; A4, cells inoculated with treated (+NMAPs) PPRV; A5, cells inoculated with untreated ASFV; A6, cells inoculated with treated (+NMAPs) ASFV. For negative controls, cells were inoculated with either EMEM (A1) or NMAPs (A2). NMAPs are shown by arrows.
Figure 3.
Capture and recovery of residual viruses (spiked) from the supernatants of EWB after treatment with HemogloBind™ (HGB) using NMAPs. Viruses were spiked into EWB of the sensitive host animals and then treated with HGB to precipitate HMB as described in the materials and methods (section 2.6). After clarification (centrifugation), the supernatants containing the residual viruses were diluted in PBS (2 ml) and then subjected to virus capture using NMAPs. The NMAPs and the captured viruses were washed, reconstituted in EMEM, and inoculated onto virus-specific cells for VI (LK for GPV, SPV, and LSDV; Vero for PPRV or ASFV) and were monitored for the development of CPE for 144 h (1P). A1, LK cells inoculated with NMAPs and the captured GTPV; A2, LK cells inoculated with NMAPs and the captured SPPV; A3, LK inoculated with NMAPs and the captured LSDV; A4, Vero cells inoculated with NMAPs and the captured PPRV; and A5, Vero cells inoculated with NMAPs and the captured ASFV. NMAPs are shown by arrows.
Figure 3.
Capture and recovery of residual viruses (spiked) from the supernatants of EWB after treatment with HemogloBind™ (HGB) using NMAPs. Viruses were spiked into EWB of the sensitive host animals and then treated with HGB to precipitate HMB as described in the materials and methods (section 2.6). After clarification (centrifugation), the supernatants containing the residual viruses were diluted in PBS (2 ml) and then subjected to virus capture using NMAPs. The NMAPs and the captured viruses were washed, reconstituted in EMEM, and inoculated onto virus-specific cells for VI (LK for GPV, SPV, and LSDV; Vero for PPRV or ASFV) and were monitored for the development of CPE for 144 h (1P). A1, LK cells inoculated with NMAPs and the captured GTPV; A2, LK cells inoculated with NMAPs and the captured SPPV; A3, LK inoculated with NMAPs and the captured LSDV; A4, Vero cells inoculated with NMAPs and the captured PPRV; and A5, Vero cells inoculated with NMAPs and the captured ASFV. NMAPs are shown by arrows.
Table 1.
Assessment of virus (PPRV) capture analyzed by RT-qPCR and virus titration.
Table 1.
Assessment of virus (PPRV) capture analyzed by RT-qPCR and virus titration.
| PPRV-Egypt |
PPRV RT-qPCR
(Cycle threshold) |
Virus titer
(TCID50/ml) |
Serial dilution
Untreated*
|
Undiluted |
17.333 |
5.5 |
| 10-1 Dilution |
20.374 |
4.3 |
| 10-2 Dilution |
22.914 |
3.2 |
| 10-3 Dilution |
26.582 |
2.4 |
| 10-4 Dilution |
30.440 |
Below LOD |
| 10-5 Dilution |
32.318 |
Below LOD |
| 10-6 - 10-10 Dilution |
Undetectable |
Below LOD |
Serial dilution
Treated#
|
Undiluted |
18.547 |
5.1 |
| 10-1 Dilution |
20.991 |
4.5 |
| 10-2 Dilution |
24.846 |
3.5 |
| 10-3 Dilution |
27.147 |
2.4 |
| 10-4 Dilution |
29.810 |
1.9 |
| 10-5 Dilution |
32.772 |
Below LOD |
| 10-6 - 10-10 Dilution |
Undetectable |
Below LOD |
Table 2.
Virus capture from serial dilutions using NMAPs analyzed by qPCR/RT-qPCR.
Table 2.
Virus capture from serial dilutions using NMAPs analyzed by qPCR/RT-qPCR.
Working stock/
Dilution |
Cycle Threshold (qPCR/RT-qPCR) |
| PPRV |
GTPV |
SPPV |
LSDV |
ASFV |
| UT#
|
Treated¶
|
UT |
Treated |
UT |
Treated |
UT |
Treated |
UT |
Treated |
| WS* (0:0) |
32.279 |
ND§
|
31.754 |
ND |
31.141 |
ND |
32.700 |
ND |
32.289 |
ND |
| WS 1:10 |
35.285 |
35.543 |
34.281 |
34.455 |
34.295 |
34.298 |
35.180 |
35.756 |
35.958 |
35.289 |
| WS 1:25 |
UD†
|
35.869 |
UD |
34.722 |
UD |
34.208 |
UD |
35.980 |
UD |
35.448 |
| WS 1:50 |
UD |
35.577 |
UD |
34.929 |
UD |
34.627 |
UD |
35.237 |
UD |
35.238 |
| WS 1:100 |
UD |
35.587 |
UD |
35.993 |
UD |
34.124 |
UD |
35.970 |
UD |
35.316 |
| WS1:250 |
UD |
35.554 |
UD |
34.889 |
UD |
34.688 |
UD |
35.605 |
UD |
35.636 |
| WS 1:500 |
UD |
35.931 |
UD |
34.559 |
UD |
35.943 |
UD |
35.642 |
UD |
35.920 |
| WS 1:1000 |
UD |
35.339 |
UD |
34.986 |
UD |
34.995 |
UD |
35.192 |
UD |
35.509 |
Table 3.
Virus-capture from EDTA whole blood (spiked) after treatment with HemogloBind™ (HGB) analyzed by qPCR/RT-qPCR.
Table 3.
Virus-capture from EDTA whole blood (spiked) after treatment with HemogloBind™ (HGB) analyzed by qPCR/RT-qPCR.
| Virus |
Cycle threshold (spiked virus) of qPCR/RT-qPCR
with or without virus capture or HGB treatment |
| PBS |
EWB
(0:0) |
EWB (1:10) |
EWB (0:0) + HGB¶
(Supernatant) |
| Untreated*
|
Treated#
|
Untreated |
Treated |
Untreated |
Treated |
Untreated |
Treated |
| GTPV |
16.907 |
17.928 |
21.033 |
21.375 |
19.235 |
18.174 |
21.439 |
22.177 |
| SPPV |
17.956 |
18.350 |
20.231 |
20.669 |
20.056 |
19.523 |
21.617 |
22.221 |
| LSDV |
17.776 |
18.711 |
20.623 |
19.864 |
19.742 |
18.995 |
21.731 |
22.108 |
| PPRV |
18.561 |
19.552 |
20.615 |
20.718 |
20.145 |
19.630 |
21.846 |
22.604 |
| ASFV |
19.915 |
20.155 |
22.033 |
22.298 |
19.444 |
20.603 |
22.525 |
23.138 |
Table 4.
Virus capture and recovery from spiked EWB using NMAPs after dilution or treatment with HGB analyzed by VI and qPCR/RT-qPCR*.
Table 4.
Virus capture and recovery from spiked EWB using NMAPs after dilution or treatment with HGB analyzed by VI and qPCR/RT-qPCR*.
Virus
(spiked)
|
EWB
(host)
|
EWB
(dilution/treatment)
|
NMVPs Added
(Y/N) |
CPE (VI) |
Cycle threshold
(qPCR/RT-qPCR) |
| 1P |
2P |
1P |
2P |
| GTPV |
Goat |
0:0 |
Y |
N |
Y |
23.342 |
18.045 |
| 1:10 |
Y |
Y |
Y |
17.631 |
16.495 |
| HGB (Sup) |
Y |
Y |
Y |
18.569 |
17.663 |
| SPPV |
Sheep |
0:0 |
Y |
N |
Y |
23.657 |
18.322 |
| 1:10 |
Y |
Y |
Y |
17.913 |
16.724 |
| HGB (Sup) |
Y |
Y |
Y |
17.812 |
16.583 |
| LSDV |
Cattle |
0:0 |
Y |
N |
Y |
23.644 |
18.509 |
| 1:10 |
Y |
Y |
Y |
19.766 |
18.609 |
| HGB (Sup) |
Y |
Y |
Y |
18.578 |
17.455 |
| PPRV |
Goat |
0:0 |
Y |
N |
Y |
25.257 |
20.311 |
| 1:10 |
Y |
Y |
Y |
19.984 |
18.766 |
| HGB (Sup) |
Y |
Y |
Y |
20.572 |
19.427 |
| ASFV |
Swine |
0:0 |
Y |
N |
Y |
23.712 |
18.116 |
| 1:10 |
Y |
Y |
Y |
17.913 |
16.893 |
| HGB (Sup) |
Y |
Y |
Y |
18.579 |
17.398 |
Table 5.
Diagnostic sensitivity of virus specific qPCR/RT-qPCR after virus capture using NMAPs from specimens of experimentally or naturally infected animals*. .
Table 5.
Diagnostic sensitivity of virus specific qPCR/RT-qPCR after virus capture using NMAPs from specimens of experimentally or naturally infected animals*. .
| Virus |
Host animal |
# of Animals |
# of Specimens |
Infection
route
|
Ct (qPCR/RT-qPCR) |
Diagnostic
sensitivity
(%) |
Untreated
(-NMAPs) |
Treated
(+NMAPs) |
| # Pos |
# Neg |
# Pos |
# Neg |
| PPRV |
Goat |
8 |
28 |
EI# |
28 |
0 |
28 |
0 |
100 |
| SPPV |
Sheep |
6 |
24 |
24 |
0 |
24 |
0 |
100 |
| ASFV |
Swine |
20 |
20 |
20 |
0 |
20 |
0 |
100 |
| ASFV |
Swine |
20 |
20 |
NI¶
|
20 |
0 |
20 |
0 |
100 |
Table 6.
Virus capture from suspensions containing multiple viruses using NMAPs analyzed by qPCR/RT-qPCR*.
Table 6.
Virus capture from suspensions containing multiple viruses using NMAPs analyzed by qPCR/RT-qPCR*.
| Virus |
Virus suspension (PBS)
Single (S)/Mixed (M) |
Cycle threshold (Ct) (qPCR/RT-qPCR) |
| CaPV |
GTPV (S) |
27.420 (GTPV) |
| GTPV + PPRV + ASFV (M) |
27.078 (GTPV) |
| SPPV (S) |
27.182 (SPPV) |
| SPPV + PPRV + ASFV (M) |
27.470 (SPPV) |
| LSDV (S) |
29.754 (LSDV) |
| LSDV + PPRV + ASFV (M) |
29.116 (LSDV) |
| PPRV |
PPRV (S) |
29.001 (PPRV) |
| PPRV + GTPV + ASFV (M) |
29.157 (PPRV) |
| PPRV + SPPV + ASFV (M) |
29.893 (PPRV) |
| PPRV + LSDV + ASFV (M) |
29.846 (PPRV) |
| ASFV |
ASFV (S) |
31.659 (ASFV) |
| ASFV + GTPV + PPRV (M) |
31.397 (ASFV) |
| ASFV + SPPV + PPRV (M) |
31.785 (ASFV) |
| ASFV + LSDV + PPRV (M) |
31.307 (ASFV) |