Submitted:
27 September 2024
Posted:
30 September 2024
Read the latest preprint version here
Abstract
Keywords:
1. Introduction
2. Methods
2.1. Sample and Procedures
2.2. Features
2.2.1. Demographic and Clinical Data Including Inflammatory Markers
2.2.2. Telomere Length (TL) Measurement
2.2.3. Social Determinants of Health (SDOH)
2.3. Outcome
2.3.1. GI Health
2.4. Data Analyses
2.4.1. Initial Data Analysis
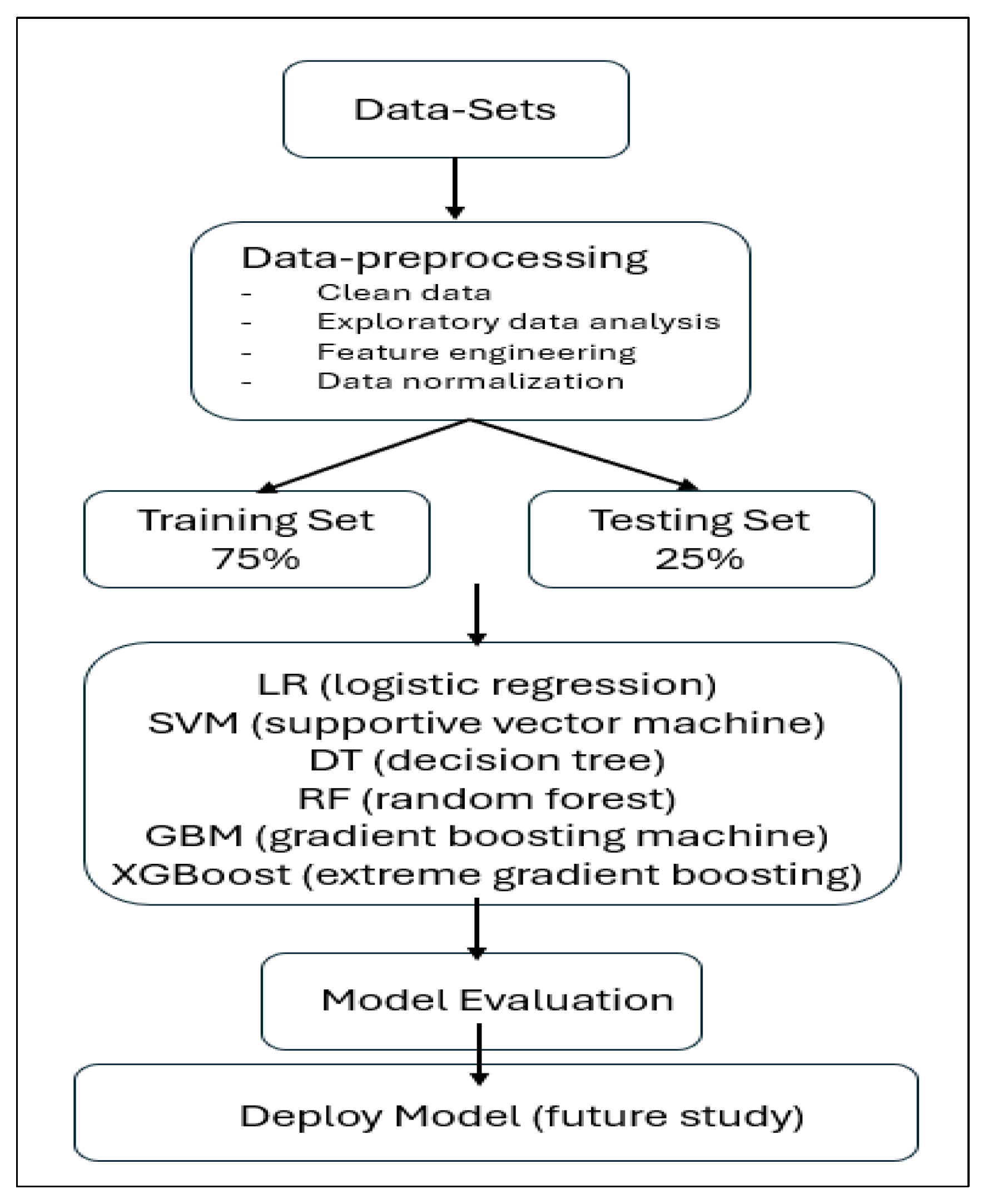
2.4.2. Machine Learning Model
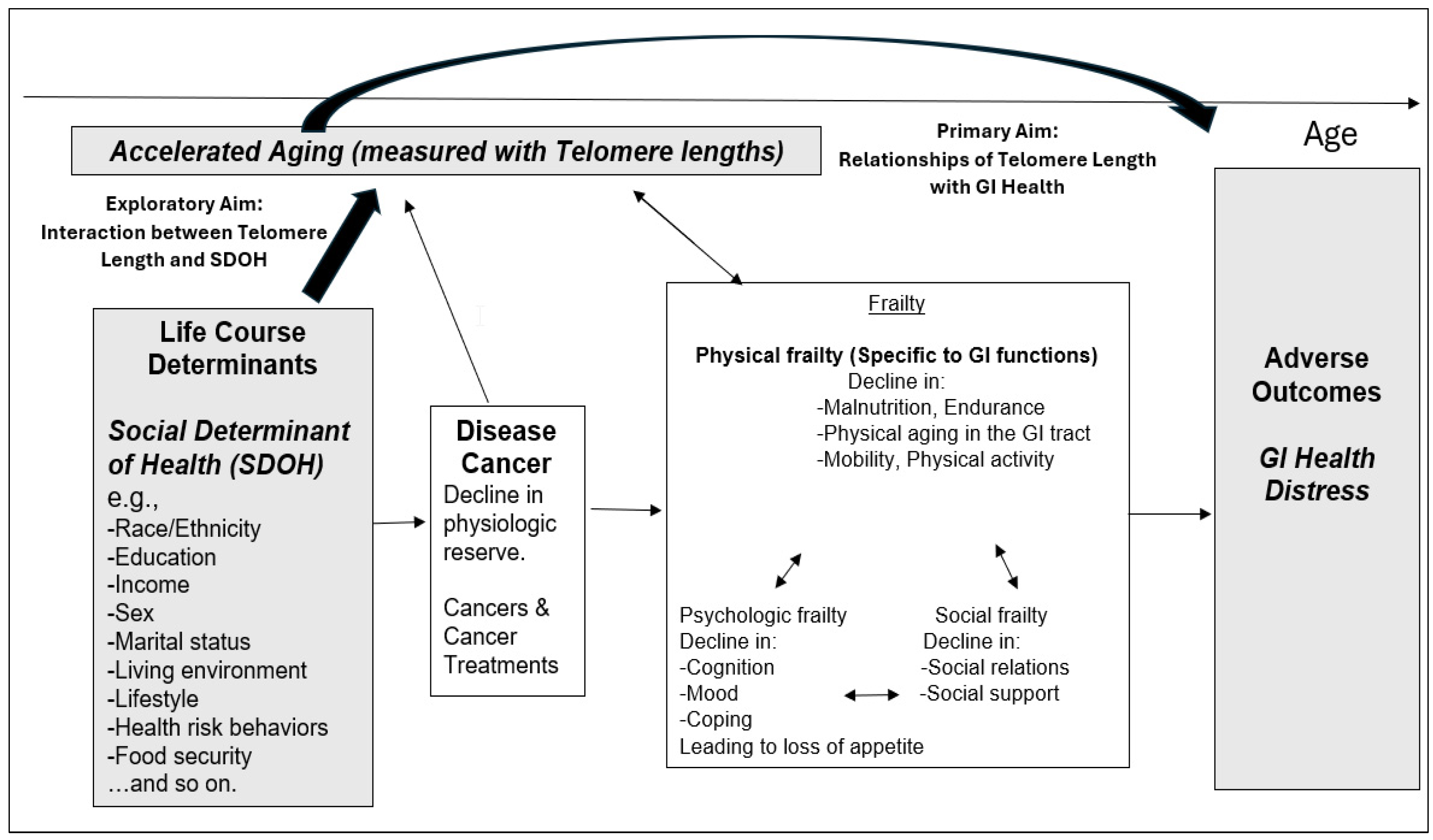
2.5. Conceptual Framework
3. Results
3.1. Initial Descriptive Analyses
3.1.1. Participant Characteristics, Clinical Data Including Inflammatory Markers, TL, and GI Health
3.1.2. SDOH Variables
3.1.3. Potential Risk Factors for GI Health within the Training Dataset
3.2. Machine Learning Models for GI Health
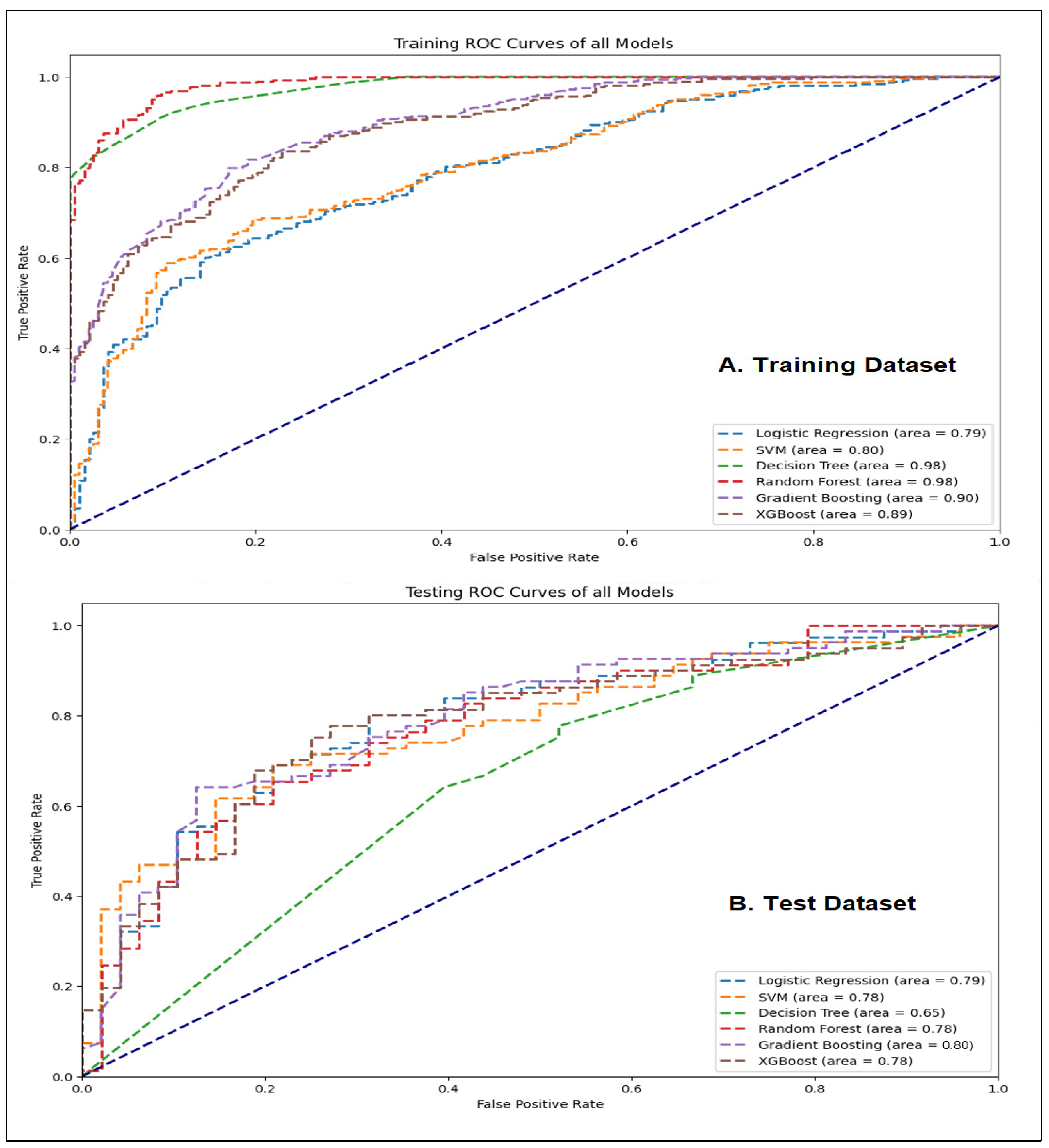
3.2.1. Performance Comparison for Classification Models
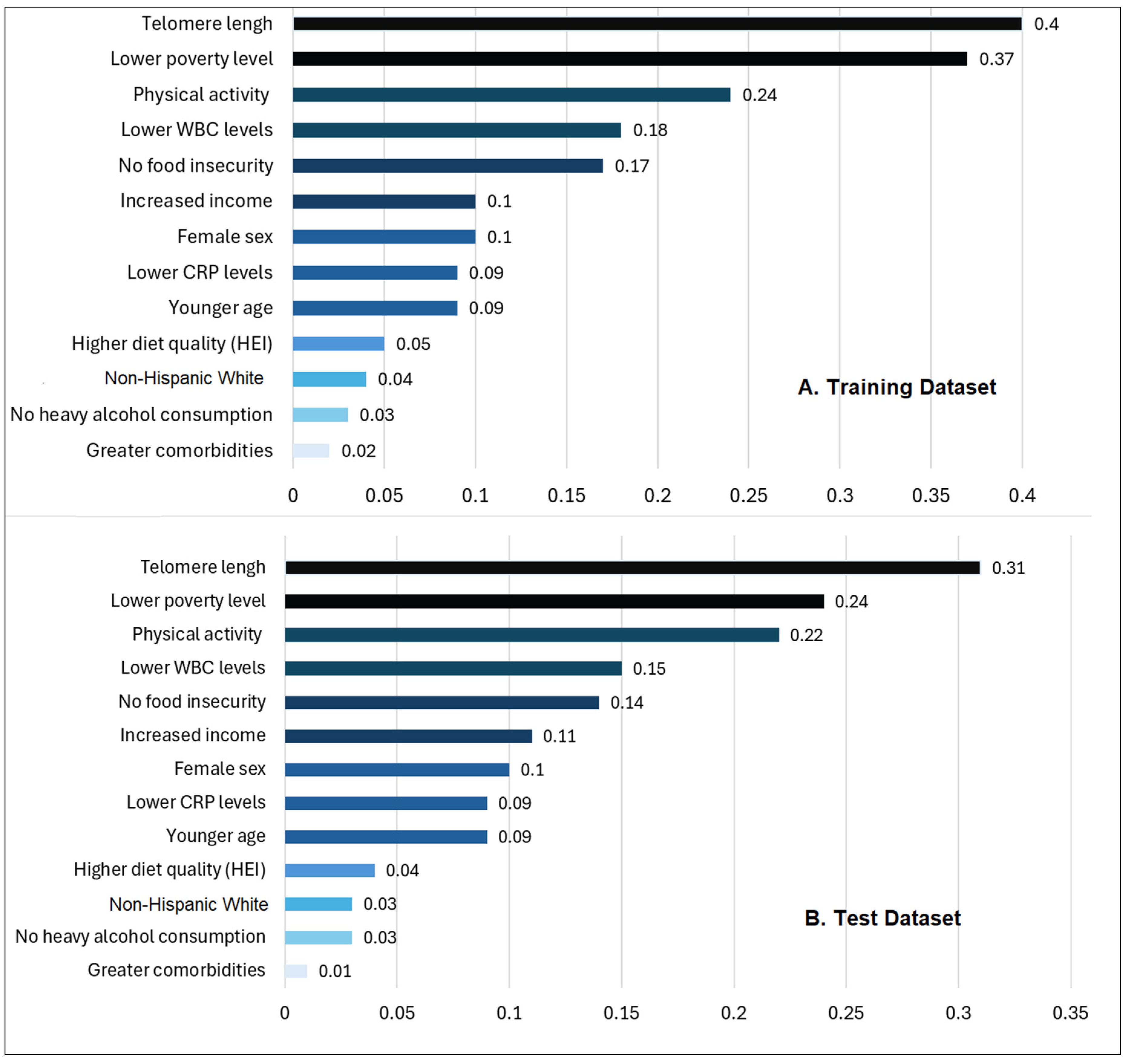
3.2.2. Feature Importance
4. Discussion
5. Conclusions
Supplementary Material
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Siegel, R.L.; Giaquinto, A.N.; Jemal, A. Cancer statistics, 2024. CA Cancer J Clin. 2024, 74(1), 12-49. [CrossRef]
- Lustberg, M.B.; Kuderer, N.M.; Desai, A.; Bergerot, C.; Lyman, G.H. Mitigating long-term and delayed adverse events associated with cancer treatment: implications for survivorship. Nat Rev Clin Oncol. 2023, 20(8), 527-542. [CrossRef]
- Firkins, J.; Hansen, L.; Driessnack, M.; Dieckmann, N. Quality of life in “chronic” cancer survivors: a meta-analysis. J Cancer Surviv. 2020, 14(4), 504-517. [CrossRef]
- Miller, K.D.; Nogueira, L.; Devasia, T.; Mariotto, A.B.; Yabroff, K.R.; Jemal, A.; Kramer, J.; Siegel, R.L. Cancer treatment and survivorship statistics, 2022. CA Cancer J Clin. 2022, 72(5), 409-436. [CrossRef]
- Song, B.C.; Bai, J. Microbiome-gut-brain axis in cancer treatment-related psychoneurological toxicities and symptoms: a systematic review. Support Care Cancer. 2021, 29(2), 605-617. [CrossRef]
- Mitra, A.; Biegert, G.W.G.; Delgado, A.Y.; Karpinets, T.V.; Solley, T.N.; Mezzari, M.P.; Yoshida-Court, K.; Petrosino, J.F.; Mikkelson, M.D.; Lin, L. Microbial diversity and composition is associated with patient-reported toxicity during chemoradiation therapy for cervical cancer. Int J Radiant Oncol Biol Phys. 2020, 107(1), 163-171. [CrossRef]
- Han, C.J.; Yang, G.S.; Syrjala, K. Symptom experiences in colorectal cancer survivors after cancer treatments: a systematic review and meta-analysis. Cancer Nurs. 2020, 43(3), E132-E158. [CrossRef]
- Han, C.J.; Reding, K.W.; Kalady, M.F.; Yung, R.; Greenlee, H.; Paskett, E.D. Factors associated with long-term gastrointestinal symptoms in colorectal cancer survivors in the women’s health initiatives (WHI study). Plos one 2023, 18(5), e0286058. [CrossRef]
- Deleemans, J.M.; Chleilat, F.; Reimer, R.A.; Baydoun, M.; Piedalue, K.-A.; Lowry, D.E.; Henning, J.-W.; Carlson, L.E. The chemo-gut pilot study: Associations between gut microbiota, gastrointestinal symptoms, and psychosocial health outcomes in a cross-sectional sample of young adult cancer survivors. Current Oncology 2022, 29(5), 2973-2994. . [CrossRef]
- Muls, A.C.; Watson, L.; Shaw, C.; Andreyev, H.J.N. Managing gastrointestinal symptoms after cancer treatment: a practical approach for gastroenterologists. Frontline Gastroenterology 2013, 4(1), 57-68. [CrossRef]
- Wiranata, J.A.; Hutajulu, S.H.; Astari, Y.K.; Leo, B.; Bintoro, B.S.; Hardianti, M.S.; Taroeno-Hariadi, K.W.; Kurnianda, J.; Purwanto, I. Patient-reported outcomes and symptom clusters pattern of chemotherapy-induced toxicity in patients with early breast cancer. PLOS ONE 2024, 19(2), e0298928. [CrossRef]
- Sipaviciute, A.; Sileika, E.; Burneckis, A.; Dulskas, A. Late gastrointestinal toxicity after radiotherapy for rectal cancer: a systematic review. International Journal of Colorectal Disease 2020, 35(6), 977-983. [CrossRef]
- Baltussen, J.C.; de Glas, N.A.; van Holstein, Y.; van der Elst, M.; Trompet, S.; Uit den Boogaard, A.; van der Plas-Krijgsman, W.; Labots, G.; Holterhues, C.; van der Bol, J.M.; et al. Chemotherapy-Related Toxic Effects and Quality of Life and Physical Functioning in Older Patients. JAMA Network Open 2023, 6(10), e2339116-e2339116. [CrossRef]
- Podolski, A.J.; Gucalp, R. GI Toxicities from Cancer Therapy. In Geriatric Gastroenterology; Springer: 2021; pp. 341-379. [CrossRef]
- Dumic, I.; Nordin, T.; Jecmenica, M.; Stojkovic Lalosevic, M.; Milosavljevic, T.; Milovanovic, T. Gastrointestinal tract disorders in older age. Can J Gastroentero Hepato. 2019, 2019, 6757524. [CrossRef]
- Busby-Whitehead, J.; Whitehead, W.E.; Sperber, A.D.; Palsson, O.S.; Simrén, M. The aging gut: Symptoms compatible with disorders of gut-brain interaction (DGBI) in older adults in the general population. J Am Geriatr Soc. 2024, 72(2), 479-489. [CrossRef]
- Rentscher, K.E.; Bethea, T.N.; Zhai, W.; Small, B.J.; Zhou, X.; Ahles, T.A.; Ahn, J.; Breen, E.C.; Cohen, H.J.; Extermann, M. Epigenetic aging in older breast cancer survivors and noncancer controls: preliminary findings from the Thinking and Living with Cancer Study. Cancer 2023, 129(17), 2741-2753. [CrossRef]
- Gallicchio, L.; Guida, J.L.; Green, P.A. Introduction to the special section on cancer survivors and treatment-related accelerated aging. J Cancer Surviv. 2024, 1-4. [CrossRef]
- Wang, H.; Chen, X.; Wang, S.; Zhang, H. Exploration of the causal effects of leukocyte telomere length and four gastrointestinal diseases: a two-sample bidirectional Mendelian randomization study. BMC Gastroenterol. 2023, 23(1), 446. [CrossRef]
- Wang, S.; El Jurdi, N.; Thyagarajan, B.; Prizment, A.; Blaes, A.H. Accelerated Aging in Cancer Survivors: Cellular Senescence, Frailty, and Possible Opportunities for Interventions. Int J Mol Sci. 2024, 25(6), 3319. [CrossRef]
- Brown, J.C.; Sturgeon, K.; Sarwer, D.B.; Troxel, A.B.; DeMichele, A.M.; Denlinger, C.S.; Schmitz, K.H. The effects of exercise and diet on oxidative stress and telomere length in breast cancer survivors. Breast Cancer Res Treat. 2023;199(1):109-117. [CrossRef]
- Xu, X.; Qu, K.; Pang, Q.; Wang, Z.; Zhou, Y.; Liu, C. Association between telomere length and survival in cancer patients: a meta-analysis and review of literature. Front Med 2016, 10(2), 191-203. [CrossRef]
- Vince, R.A.; Jiang, R.; Bank, M.; Quarles, J.; Patel, M.; Sun, Y.; Hartman, H.; Zaorsky, N.G.; Jia, A.; Shoag, J. Evaluation of social determinants of health and prostate cancer outcomes among black and white patients: a systematic review and meta-analysis. JAMA Netw Open. 2023, 6(1), e2250416-e2250416. [CrossRef]
- Han, C.J.; Tounkara, F.; Kalady, M.; Noonan, A.M.; Burse, N.R.; Paskett, E.D.; Von Ah, D. Risk factors of health-related quality of life among gastrointestinal cancer survivors in the US: with a focus on social and behavioral determinants of health (SBDH). Int J Environ Res Public Health. 2023;20(17):6676. [CrossRef]
- Yao, J.; Chen, X.; Meng, F.; Cao, H.; Shu, X. Combined influence of nutritional and inflammatory status and depressive symptoms on mortality among US cancer survivors: Findings from the NHANES. Brain Behav Immun. 2024;115:109-117. [CrossRef]
- Antoni, M.H.; Moreno, P.I.; Penedo, F.J. Stress management interventions to facilitate psychological and physiological adaptation and optimal health outcomes in cancer patients and survivors. Annu Rev Psychol. 2023;74:423-455. [CrossRef]
- Needham, B.L.; Adler, N.; Gregorich, S.; Rehkopf, D.; Lin, J.; Blackburn, E.H.; Epel, E.S. Socioeconomic status, health behavior, and leukocyte telomere length in the National Health and Nutrition Examination Survey, 1999-2002. Soc Sci Med 2013, 85, 1-8. [CrossRef]
- Kumar, K.; Kumar, P.; Deb, D.; Unguresan, M.-L.; Muresan, V. Artificial intelligence and machine learning based intervention in medical infrastructure: a review and future trends. In Proceedings of the Healthcare, 2023;11(2):207. Published 2023 Jan 10. [CrossRef]
- Oyebode, O.; Fowles, J.; Steeves, D.; Orji, R. Machine learning techniques in adaptive and personalized systems for health and wellness. Int J Human-Comp Int. 2022;39(9):1938-1962. [CrossRef]
- Black, J.E.; Kueper, J.K.; Williamson, T.S. An introduction to machine learning for classification and prediction. Fam Pract. 2023;40(1):200-204. [CrossRef]
- Shu, X.; Ye, Y. Knowledge Discovery: Methods from data mining and machine learning. Soc Sci Res. 2023;110:102817. [CrossRef]
- Zhang, B.; Shi, H.; Wang, H. Machine learning and AI in cancer prognosis, prediction, and treatment selection: a critical approach. J Multidiscip Healthc. 2023;16:1779-1791. Published 2023 Jun 26. [CrossRef]
- Dye, B.; Barker, L.; Selwitz, R.; Lewis, B.; Wu, T.; Fryar, C.; Ostchega, Y.; Beltran, E.; Ley, E. Overview and quality assurance for the National Health and Nutrition Examination Survey (NHANES) oral health component, 1999–2002. Community Dent Oral Epidemiol. 2007;35(2):140-151. [CrossRef]
- Johnson, C.L.; Paulose-Ram, R.; Ogden, C.L.; Carroll, M.D.; Kruszon-Moran, D.; Dohrmann, S.M.; Curtin, L.R. National health and nutrition examination survey: analytic guidelines, 1999-2010. Vital Health Stat 2. 2013;(161):1-24.
- Wirth, R.; Hipp, J. CRISP-DM: Towards a standard process model for data mining. In Proceedings of the Proceedings of the 4th international conference on the practical applications of knowledge discovery and data mining, 2000; pp. 29-39. Manchester.
- Liu, B.; Taioli, E. Seasonal variations of complete blood count and inflammatory biomarkers in the US population-analysis of NHANES data. PloS one 2015, 10, e0142382. 2015;10(11):e0142382. [CrossRef]
- Mazidi, M.; Kengne, A.P.; Sahebkar, A.; Banach, M. Telomere Length Is Associated With Cardiometabolic Factors in US Adults. Angiology 2018, 69, 164-169. [CrossRef]
- Mazidi, M.; Kengne, A.P.; Vatanparast, H. Food Security and Leukocyte Telomere Length in Adult Americans. Oxid Med Cell Longev 2017, 2017, 5427657. [CrossRef]
- Mazidi, M.; Penson, P.; Banach, M. Association between telomere length and complete blood count in US adults. Arch Med Sci 2017, 13(3), 601-605. [CrossRef]
- Shen, G.; Huang, J.Y.; Huang, Y.Q.; Feng, Y.Q. The Relationship between Telomere Length and Cancer Mortality: Data from the 1999-2002 National Healthy and Nutrition Examination Survey (NHANES). J Nutr Health Aging 2020, 24(1), 9-15. [CrossRef]
- NHANES - National Health and Nutrition Examination Survey Homepage. www.cdc.gov. Published June 24, 2024. Accessed July 16, 2024. http://cdc.gov/nchs/nhanes.
- Gómez, C.A.; Kleinman, D.V.; Pronk, N.; Gordon, G.L.W.; Ochiai, E.; Blakey, C.; Johnson, A.; Brewer, K.H. Addressing health equity and social determinants of health through healthy people 2030 J Public Health Manag Pract. 2021;27(Suppl 6):S249-S257. [CrossRef]
- Committee on the Recommended Social and Behavioral Domains and Measures for Electronic Health Records; Board on Population Health and Public Health Practice; Institute of Medicine. Capturing Social and Behavioral Domains and Measures in Electronic Health Records: Phase 2. Washington (DC): National Academies Press (US); January 8, 2015.
- Giuse, N.B.; Koonce, T.Y.; Kusnoor, S.V.; Prather, A.A.; Gottlieb, L.M.; Huang, L.C.; Phillips, S.E.; Shyr, Y.; Adler, N.E.; Stead, W.W. Institute of Medicine Measures of Social and Behavioral Determinants of Health: A Feasibility Study. Am J Prev Med 2017, 52, 199-206. [CrossRef]
- Byhoff, E.; Tripodis, Y.; Freund, K.M.; Garg, A. Gender Differences in Social and Behavioral Determinants of Health in Aging Adults. J Gen Intern Med 2019, 34(11), 2310-2312. [CrossRef]
- Kendall, A.; Olson, C.M.; Frongillo, E.A., Jr. Validation of the Radimer/Cornell measures of hunger and food insecurity. J Nutr 1995, 125(11), 2793-2801. [CrossRef]
- Bush, K.; Kivlahan, D.R.; McDonell, M.B.; Fihn, S.D.; Bradley, K.A. The AUDIT alcohol consumption questions (AUDIT-C): an effective brief screening test for problem drinking. Ambulatory Care Quality Improvement Project (ACQUIP). Alcohol Use Disorders Identification Test. Arch Intern Med 1998, 158(16), 1789-1795. [CrossRef]
- Zhang, L.; Ferguson, T.F.; Simonsen, N.; Chen, L.; Tseng, T.S. Racial/Ethnic Disparities in Health-Related Quality of Life among Participants with Self-Reported Diabetes from NHANES 2001-2010. Diabetes Educ 2014, 40(4), 496-506. [CrossRef]
- Cabello-Solorzano K, De Araujo IO, Peña M, Correia L, Tallón-Ballesteros AJ. The impact of data normalization on the accuracy of machine learning algorithms: a comparative analysis. In: Lecture Notes in Networks and Systems. ; 2023:344-353. [CrossRef]
- Gobbens, R.J.; Luijkx, K.G.; Wijnen-Sponselee, M.T.; Schols, J.M. Towards an integral conceptual model of frailty. J Nutr Health Aging 2010, 14(3), 175-181. [CrossRef]
- Swanson, K.; Wu, E.; Zhang, A.; Alizadeh, A.A.; Zou, J. From patterns to patients: Advances in clinical machine learning for cancer diagnosis, prognosis, and treatment. Cell 2023, 186(8), 1772-1791. [CrossRef]
- Cruz, J.A.; Wishart, D.S. Applications of machine learning in cancer prediction and prognosis. Cancer informatics 2006, 2, 59-77, 117693510600200030.
- Rafique, R.; Islam, S.R.; Kazi, J.U. Machine learning in the prediction of cancer therapy. Comput Struct Biotechnol J. 2021, 19, 4003-4017. [CrossRef]
- Haun, M.W.; Simon, L.; Sklenarova, H.; Zimmermann-Schlegel, V.; Friederich, H.C.; Hartmann, M. Predicting anxiety in cancer survivors presenting to primary care–A machine learning approach accounting for physical comorbidity. Cancer Medicine 2021, 10(14), 5001-5016. [CrossRef]
- Badger, T.A.; Segrin, C.; Crane, T.E.; Chalasani, P.; Arslan, W.; Hadeed, M.; Sikorskii, A. Social determinants of health and symptom burden during cancer treatment. Nursing research 2023, 72(2), 103-113. [CrossRef]
- McCall, M.K.; Connolly, M.; Nugent, B.; Conley, Y.P.; Bender, C.M.; Rosenzweig, M.Q. Symptom experience, management, and outcomes according to race and social determinants including genomics, epigenomics, and metabolomics (SEMOARS+ GEM): an explanatory model for breast cancer treatment disparity. J Cancer Educ. 2020, 35(3), 428-440. [CrossRef]
- Westrick, A.C.; Langa, K.M.; Eastman, M.; Ospina-Romero, M.; Mullins, M.A.; Kobayashi, L.C. Functional aging trajectories of older cancer survivors: a latent growth analysis of the US Health and Retirement Study. J Cancer Surviv. 2023, 17(5), 1499-1509. [CrossRef]
- Chi, Z.; Bai, X.; Zhang, Z. Risk relationship between leukocyte telomere length and constipation: a Mendelian randomization study. Frontiers in Medicine 2023, 10, 1177785. [CrossRef]
- Singh, S.; Giron, L.B.; Shaikh, M.W.; Shankaran, S.; Engen, P.A.; Bogin, Z.R.; Bambi, S.A.; Goldman, A.R.; Azevedo, J.L.; Orgaz, L. Distinct intestinal microbial signatures linked to accelerated systemic and intestinal biological aging. Microbiome 2024, 12(1), 31. [CrossRef]
- Hooten, N.N.; Pacheco, N.L.; Smith, J.T.; Evans, M.K. The accelerated aging phenotype: the role of race and social determinants of health on aging. Ageing Res Rev. 2022, 73, 101536. [CrossRef]
- Androulakis, I.P. From cells to society: untangling the web of stress, inflammation, and social determinants of health. 2024, 2, 1358784. [CrossRef]
- Tarver, W.L.; Justice, Z.; Jonnalagadda, P.; Rahurkar, S.; Obeng-Gyasi, S.; Krok-Schoen, J.L.; Petrecca, A.; Paskett, E.D. A scoping review of the evidence on survivorship care plans among minority, rural, and low-income populations. J Cancer Surviv. 2024, 1-39. [CrossRef]
- Vardy, J.; Liew, A.; Turner, J.; Kerin-Ayres, K.; Butler, S.; Deguchi, C.; Khatri, S.; Wildbore, C.; Mo, C.; Hamayun, M. What happens to cancer survivors attending a structured cancer survivorship clinic? Symptoms, quality of life and lifestyle changes over the first year at the Sydney Cancer Survivorship Centre clinic. Support Care Cancer. 2021, 29(3), 1337-1345. [CrossRef]
- Addison, S.; Shirima, D.; Aboagye-Mensah, E.B.; Dunovan, S.G.; Pascal, E.Y.; Lustberg, M.B.; Arthur, E.K.; Nolan, T.S. Effects of tandem cognitive behavioral therapy and healthy lifestyle interventions on health-related outcomes in cancer survivors: a systematic review. J Cancer Surviv. 2022, 16(5), 1023-1046. [CrossRef]
- Nouve, Y.; Zhao, S.; Zheng, Y. Decoding the misperception: Exploring measurement error in self-rated assessments of diet quality. Food Quality and Preference 2024, 105234. [CrossRef]
- Tucker, A.C.; Bresnahan, C.; John, S.; Johnson, J.; Leung, C.W.; Mui, Y.; Hager, E.R.; Wolfson, J.A. Food (in) security in relation to nutrition (in) security in a national cross-sectional sample of Supplemental Nutrition Assistance Program participants: considerations of an emerging construct. Am J Clin Nutr. 2024, 119(6), 1475-1484. [CrossRef]
- Robien, K.; Clausen, M.; Sullo, E.; Ford, Y.R.; Griffith, K.A.; Le, D.; Wickersham, K.E.; Wallington, S.F. Prevalence of food insecurity among cancer survivors in the United States: a scoping review. J Acad Nutr Diet. 2023, 123(2), 330-346. [CrossRef]
- Christian, V.J.; Miller, K.R.; Martindale, R.G. Food insecurity, malnutrition, and the microbiome. Curr Nutr Rep. 2020, 9(4), 356-360. [CrossRef]
- Hussaini, S.Q.; Chen, K.Y.; Blackford, A.L.; Chino, F.; Gupta, A. Food insecurity and gastrointestinal (GI) cancer mortality in the United States, 2015 to 2019. J Clin Oncol. 2023, 41(4), 788. [CrossRef]
| Total cancer survivors (N = 645) |
Training seta (n = 75% of total sample, n = 484 |
Test setb (n = 25% of total sample, n = 161) |
p | GI health (n, %) | ||
|---|---|---|---|---|---|---|
| Better | Worse | p | ||||
| Age (years) mean ± SE, range |
66.3 ± 14.7 (21-85) | 65.5 ± 16.2 (22-85) | .102 | 63.3 (10.9) | 66.4 (11.2) | 47.4, .031 |
| Female (n,%) | 235 (49.5) | 84 (50.7) | .311 | 153 (47) | 103(65) | 6.1, .013 |
| Modified Comorbidities(>2)(n,%) | 168 (42.3) | 66 (43.2) | .122 | 133(41) | 71 (45) | 5.4, .043 |
| Types of Cancers (n,%) |
Skin: 152 (21.2) GU: 102 (21.0) Breast: 75 (15.6) Ovary-Uterine: 45 (9.3) Head & Neck: 42 (8.6) GI: 41 (8.4) Lung: 15 (3.1) Hematological: 12(2.5) |
Skin: 44 (27.3) Breast: 35 (21.7) GU: 30 (18.6) Head & Neck: 21(13.0) GI: 15 (9.3) Ovary-Uterine: 8 (5.0) Lung: 5 (3.1) Hematological: 3 (1.9) |
.143 | Skin: 65 (20.1) GU: 62 (19) Breast: 53 (16.3) Ovary-Uterine: 37 (11.3) Head & Neck: 31 (9.5) GI: 27 (8.5) Lung:13 (4.1) Hematological: 36 (11.2) |
Skin: 31(19.8) GU: 26 (16.2) Breast:27(17.3) Ovary-Uterine: 18(11.5) Head & Neck: 17 (10.9) GI: 15 (9.3) Lung: 8 (5.2) Hematological 15 (9.8) |
12.1, .100 |
| WBC (k/ul), normal (4 -11k/ul), mean ± SE |
7.0 (2.1) | 7.04 (2.0) | .192 | 5.4 (1.1) | 8.5 (1.5) | 146.3, .046 |
| CRP (mg/dl), normal (<0.3mg/dl), mean ± SE | 0.5 (0.9) | 0.6 (1.4) | .124 | 0.4 (0.8) | 1.0 (1.1) | 238.4, .001 |
| Telomere Lengths (kb) mean ± SE |
0.93 (0.2) | 0.93 (0.2) | .823 | 0.97 (0.2) | 0.64 (0.3) | 85.1, .013 |
| Gastrointestinal Health (n, %) |
Worse: 158 (32.5) Better: 324 (66.7) |
Worse: 59 (36.6) Better: 101 (62.7) |
.412 | Not applicable | ||
| Total cancer survivors (N = 645). n (%) otherwise specified |
Training seta (n = 75% of total sample, n = 484 |
Test setb (n = 25% of total sample, n = 161) |
p | GI Health (n, %) | ||||
|---|---|---|---|---|---|---|---|---|
| Better | Worse | p | ||||||
| Race/Ethnicity | .413 | 24.2, .039 | ||||||
| Non-Hispanic White | 356 (73.3) | 121(75.0) | 260 (80.3) | 122 (77.3) | ||||
| Non-Hispanic Black | 53 (10.9) | 18 (11.0) | 35 (10.7) | 17 (10.5) | ||||
| Non-Hispanic Other | 6 (1.2) | 2 (1.2) | 5 (1.5) | 2 (1.5) | ||||
| Hispanic | 69(14.2) | 20 (12.8) | 24 (7.5) | 17 (10.7) | ||||
| Marital status | .541 | 3.6, .730 | ||||||
| Married/Partnered | 329 (68.1) | 110 (68.3) | 220(67.9) | 104 (65.6) | ||||
| Divorced/Widowed/Single | 155 (31.9) | 51 (31.7) | 104(32.1) | 54 (34.4) | ||||
| Education | .112 | 16.6, .502 | ||||||
| High school or less | 247 (51.0) | 80 (49.7) | 158(48.8) | 81 (51.1) | ||||
| College of technical school | 130 (26.9) | 44 (27.3) | 88 (27.1) | 41 (25.8) | ||||
| Graduate school | 107 (22.1) | 37 (23.0) | 78 (24.1) | 36 (23.1) | ||||
| Household Income (yr.) | .353 | 8.43, .038 | ||||||
| Less than $25,000 | 169 (34.9) | 57 (35.4) | 114(35.3) | 58 (36.8) | ||||
| $25,000 to <$55,000 | 150 (31.0) | 51 (31.7) | 100(31.0) | 45 (28.3) | ||||
| $55,000 to <$75,000 | 45 (9.2) | 17 (10.6) | 50 (15.4) | 26 (16.4) | ||||
| $75,000 and over | 107 (22.1) | 33 (20.5) | 59 (18.3) | 29 (18.5) | ||||
| Poverty-income ratio (PIR) <1 indicating a high poverty level (Yes): Annual household income below the poverty level. | 193 (39.7) | 60 (37.3) | .423 | 113(34.9) | 59 (37.6) | 18.01,<.001 | ||
| Food Insecurity (Yes) | 42 (8.6) | 13 (8.1) | .879 | 18 (5.6) | 13 (8.0) | 17.01, .021 | ||
| Cancer Health Behaviors (Yes) | ||||||||
| Current Smoking Status | 86 (17.7) | 31 (19.3) | .114 | 53 (16.3) | 31 (19.5) | 13.1, .080 | ||
| Current Heavy Alcohol Use | 86 (17.7) | 21 (13.0) | .198 | 49 (15.2) | 34 (21.3) | 37.01,<.001 | ||
| Regular physical activity | 286 (58.8) | 76 (47.2) | .108 | 189(58.3) | 61 (38.5) | .52.4, .035 | ||
| Diet quality(HEI-2015 Score, 0-100, mean ± SE) |
48.8 (12.3) | 48.9 (8.3) | .103 | 52.5 (5.6) | 47.3 (7.5) | 56.1, .038 | ||
| Model | AUC | Accuracy | Precision | Sensitivity (Recall) |
Specificity | F1 Score |
| Training Dataset | ||||||
| LR | 0.7918 | 0.7192 | 0.7214 | 0.8978 | 0.4197 | 0.8111 |
| SVM | 0.7994 | 0.7112 | 0.7753 | 0.7585 | 0.6321 | 0.7668 |
| Decision Tree | 0.9758 | 0.9089 | 0.9340 | 0.9195 | 0.8912 | 0.9267 |
| RF | 0.9842 | 0.9341 | 0.9213 | 0.9783 | 0.8601 | 0.9489 |
| GBM | 0.8952 | 0.7907 | 0.7867 | 0.9133 | 0.5855 | 0.8453 |
| XGBoost | 0.7829 | 0.7755 | 0.9195 | 0.5544 | 0.5544 | 0.8414 |
| Test Dataset | ||||||
| LR | 0.7904 | 0.7287 | 0.7447 | 0.8642 | 0.5312 | 0.8312 |
| SVM | 0.7774 | 0.7054 | 0.7792 | 0.7407 | 0.6458 | 0.7595 |
| Decision Tree | 0.6480 | 0.6512 | 0.7093 | 0.7531 | 0.4792 | 0.7305 |
| RF | 0.7760 | 0.7364 | 0.7640 | 0.8395 | 0.5625 | 0.8000 |
| GBM | 0.8035 | 0.7442 | 0.7609 | 0.8642 | 0.5417 | 0.8092 |
| XGBoost | 0.7834 | 0.7287 | 0.7500 | 0.8519 | 0.5208 | 0.7977 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





