1. Introduction
The eukaryotic protein 14-3-3 assumes several multifaceted cellular tasks, manifesting its characteristic moonlighting behavior. Within fungal kingdom, the functional repertoire of 14-3-3 encompasses various pivotal roles; these include involvement in several processes as follow: virulence modulation (Rodríguez-Romero et al. 2019; Marcos et al. 2016), adhesion mechanisms (Assato et al. 2015; Marcos et al. 2016), orchestration of growth dynamics (Sun et al. 2018), regulation of cell signaling pathways and cycles (Lottersberger et al. 2006), modulation of carbon metabolism (Y. Wang et al. 2004), facilitation of filamentation and ascospore formation (Herod et al. 2022), DNA duplication (Kumar 2018), cellular survival mechanisms under stressful conditions (Sun et al. 2018), cell secretion (Gelperin et al. 1995), coordination of cell wall composition and chitin synthesis (Lottersberger et al. 2006), aggresomal assembly (Xu et al. 2013), regulation of the spindle assembly checkpoint during cell division (Caydasi et al. 2014), modulation of retrograde signaling pathways (Trendeleva and Zvyagilskaya 2018), and participation in the intricate regulation of apoptosis (Tohru Ichimura et al. 2004).
The 14-3-3 protein exhibits diverse isoform variability across various fungi genera and species and can be expressed through various gene translations. Notably, this protein can range from a singular isoform in fungi like Paracoccidioides brasiliensis (Assato et al. 2015) and Candida albicans (Cognetti, Davis, and Sturtevant 2002; Palmer et al. 2004), seven isoforms in Homo sapiens (T Ichimura et al. 1988), to a more extensive repertoire, as observed in the plant species Arabidopsis thaliana, which boasts a total of 13 distinct isoforms (Wilson, Swatek, and Thelen 2016), highlighting the considerable diversity and complexity present within the 14-3-3 protein family across different organisms.
The Onygenales order within the Ajellomycetaceae family encompasses several pathogenic dimorphic fungi such as Histoplasma, Paracoccidioides, Emergomyces, Emmonsiellopsis, Emmonsia, and Blastomyces, all of which share similar characteristics concerning host-pathogen interactions and possess a distinctive ability to undergo morphological shifts in response to environmental cues during their transition from the environment to the host system (Gauthier 2015). This classification underscores commonalities among these fungi in terms of their behavior and interactions with host organisms (Muñoz et al. 2018), in which 14-3-3 protein plays a pivotal role.
Recently, it has become evident that climate change due to human activity is creating conditions conducive to the emergence of new human fungal pathogens. These changes increase virulence, fungal dispersal, vectors, geographic range, and host susceptibility (Nnadi and Carter 2021). These scenarios, coupled with the growing resistance of fungi to available antifungal drugs (Fisher et al. 2022), underscore the urgent need for new specific targets.
In this context, phylogenetic analysis is a crucial bioinformatic tool enabling a comprehensive view of organisms' evolutionary trajectories over time in silico. This analytical approach unveils genetic relationships and similarities among different species, shedding light on their evolutionary history and providing valuable insights into their genetic makeup (Burr 2010). It is possible to evaluate similarities and differences among species that can be applied as diagnostic markers, sequences used in broad-spectrum fungal identification analysis, and the development of new broad-spectrum drugs and vaccines (Simonson et al. 2021; Van Burik et al. 1998; Taborda and Camargo 1994).
Thus, in this study we aimed to compare 14-3-3 orthologs within the genome of the Ajellomycetaceae family and Homo sapiens’ isoforms seeking to yield novel insights that could be show in the protein functions of a new shared and specific targeting for broad-spectrum fungal sequences, drug-specific targets, and vaccine development to infections caused by dimorphic fungi.
2. Results
We conducted a comparative analysis of 21 species, examining a total of 193,630 genes. Our investigation revealed the presence of 181,719 orthogroup genes, constituting 93.8% of the total, distributed across 13,770 orthogroups. Additionally, we identified 11,911 unassigned genes, comprising 6.2% of the dataset. Within this dataset, 369 orthogroups were specific to particular species, encompassing 1,554 species-specific genes. The average size of orthogroups was 13.2, with a median size of 15.0. Furthermore, our analysis identified 3,839 orthogroups shared among all species, and 2,383 single-copy orthogroups.
In the genomes of the
Ajellomycetaceae family, we identified two copies of the 14-3-3 gene family, except for
Histoplasma ohiensis G217B, which possesses only one gene copy, and
Emergomyces africanus CBS 136260, which harbors a third gene copy (
Table 1). The human genome exhibits 17 copies of the gene family (
Table 2).
The first 14-3-3 gene copy within the
Ajellomycetaceae family exhibited a closer phylogenetic relationship to the human 14-3-3 protein ε (
Figure 1) (Ortholog 9,
Table 2). Upon alignment of 14-3-3 orthologs, the sequence displayed a conserved peptide binding site from the human 14-3-3 protein ε across all fungal proteins. However, polymorphic sites were observed in the dimer interface of the second gene copy (
Figure 2).
Regarding phylogenetic analysis, the first 14-3-3 gene copy (
Figure 1, blue) formed a clade comprising orthologs from the genera
Histoplasma,
Blastomyces,
Emergomyces, and
Emmonsia, with the
Paracoccidioides genus as the external group. Conversely, the second gene copy (
Figure 1, red) formed a clade with the
Paracoccidioides genus and other genera, while the
Histoplasma genus served as the external group. Furthermore, the second copy of the 14-3-3 gene found in the
Ajellomycetaceae family also demonstrated a common ancestor with
Homo sapiens 14-3-3 ε isoform.
When the resulting 14-3-3 alignment was visualized in Geneious Prime V 2023.0 (
Figure 2), conserved and distinct amino acids were identified in the
Ajellomycetaceae fungi and
Homo sapiens sequences. Additionally, the fungal copies were distinct from each other and differed when compared to the
Homo sapiens copy. Nevertheless, an extensive conserved sequence was also visible among all sequences. The mean length of the 14-3-3 protein was 268 amino acids, with 29.9% (104) identical sites among all species analyzed and a pairwise identity of 71.9%. Furthermore, amino acid groups comprised 18.2% acidic, 13.4% basic, 31.6% charged, 25.5% polar uncharged, 43.7% hydrophobic, 24.4% GC-rich, and 21.5% AT-rich amino acids. Finally, the frequency of Serine (Ser) was 7.7% (1,178 frequency), and Threonine (Thr) was 4.7% (712 frequency). The mean molecular weight was 29.936 kDa, the mean isoelectric point was 4.47, and the mean absorbance of 280 nm of 1mg/mL was 0.93 AU.
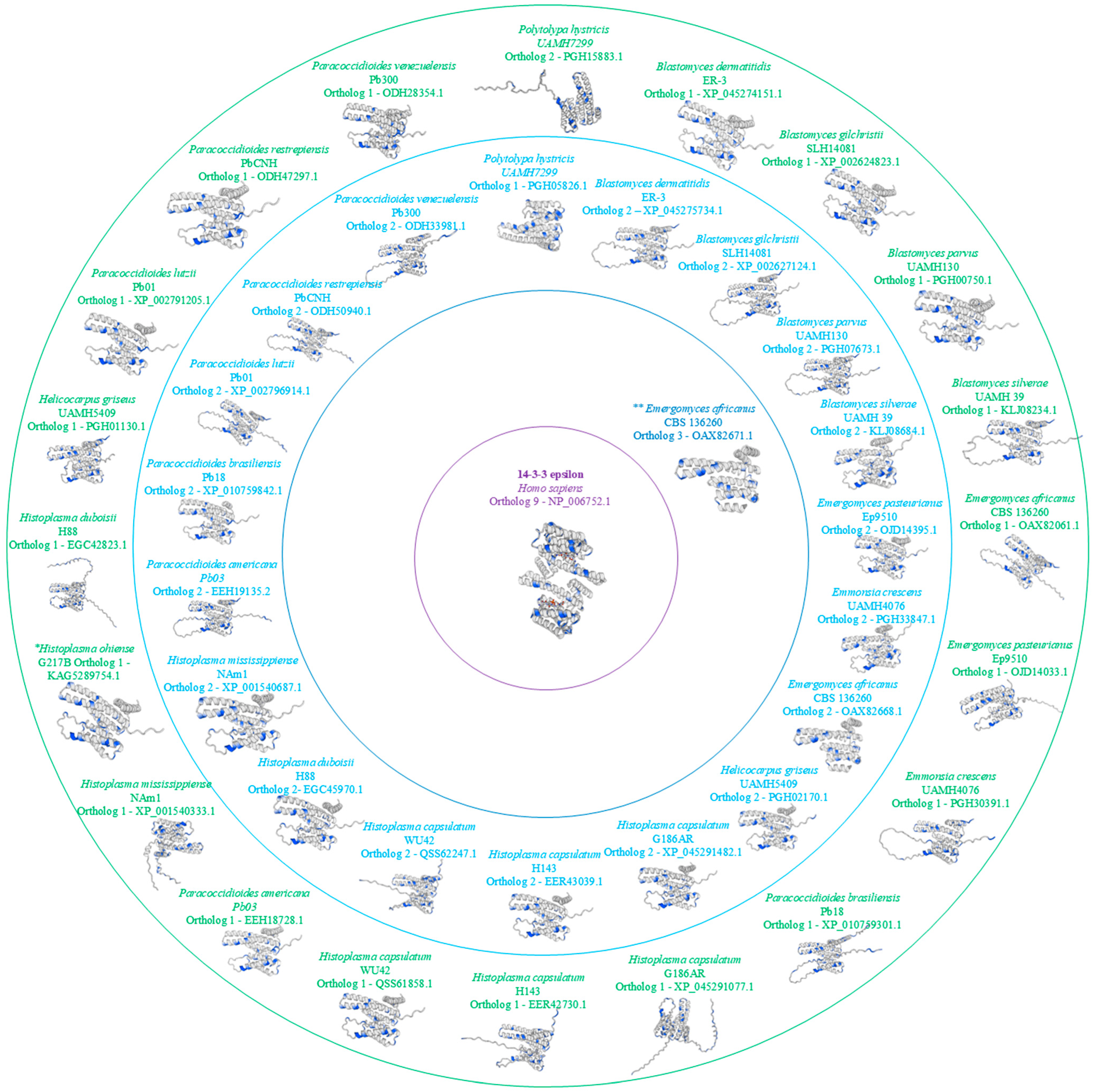
Protein modeling was created with Swiss-model considering oligo state as a monomer, GMQE (Global Model Quality Estimate) ≥ 0.80, 0.85 < coverage < 1.0, QMEAN Z-score < 1, and 86.72.0 < sequence identity < 100.
Figure 3 represents protein 3D models for each
Ajellomycetaceae family fungus.
Figure 2.
(continuation). Sequence alignment of human 14-3-3 proteins withAjellomycetaceaefamily. Sequence alignments were performed using the Clustal Omega program and visualized in Geneious. The dimer interface sites [polypeptide binding] from human 14-3-3ɛ protein are highlighted.
Figure 2.
(continuation). Sequence alignment of human 14-3-3 proteins withAjellomycetaceaefamily. Sequence alignments were performed using the Clustal Omega program and visualized in Geneious. The dimer interface sites [polypeptide binding] from human 14-3-3ɛ protein are highlighted.
Figure 2.
(continuation). Sequence alignment of human 14-3-3 proteins withAjellomycetaceaefamily. Sequence alignments were performed using the Clustal Omega program and visualized in Geneious. The dimer interface sites [polypeptide binding] from human 14-3-3ɛ protein are highlighted..
Figure 2.
(continuation). Sequence alignment of human 14-3-3 proteins withAjellomycetaceaefamily. Sequence alignments were performed using the Clustal Omega program and visualized in Geneious. The dimer interface sites [polypeptide binding] from human 14-3-3ɛ protein are highlighted..
Figure 3.
3D Model ofHomo sapiens14-3-3ɛ Protein and Orthologs from theAjellomycetaceaeFamily. Serine/threonine (Ser/Thr) residues are highlighted in blue in the 3D model, with ortholog 1 shown in green, ortholog 2 in light blue, ortholog 3 in dark blue, and the Homo sapiens 14-3-3ɛ protein depicted in purple. * Only one ortholog. ** Three orthologs.
Figure 3.
3D Model ofHomo sapiens14-3-3ɛ Protein and Orthologs from theAjellomycetaceaeFamily. Serine/threonine (Ser/Thr) residues are highlighted in blue in the 3D model, with ortholog 1 shown in green, ortholog 2 in light blue, ortholog 3 in dark blue, and the Homo sapiens 14-3-3ɛ protein depicted in purple. * Only one ortholog. ** Three orthologs.
Through the 3D analysis of the 14-3-3 protein in fungi from the Ajellomycetaceae family, a high degree of similarity with the human 14-3-3ɛ protein is observed, with similarity exceeding 70%, depending on the species analyzed. However, one noticeable difference is the more significant number of amino acids at the C- and N-terminal ends of the fungal 14-3-3 protein. This difference has not been demonstrated to have any relation to the pathogenicity of these fungi.
3. Discussion
The 14-3-3 protein is categorized as a prominent class of molecular chaperones, interacting with over 200 protein targets and engaging a diverse array of binding partners involved in cellular signaling pathways (Hondermarck 2010), exhibiting closer evolutionary proximity among human protein to yeasts and plants than to other animals, suggesting a shared ancestral protein function (Wang and Shakes 1996). The multiple binding partners of 14-3-3 are due to the phosphorylation majority, therefore the interaction with phosphatases and kinases modulates its phosphorylation, which has been related to various cell pathways of differentiation, growth, migration, and survival (Hondermarck 2010).
All isoforms of 14-3-3 proteins contain conserved serine/threonine (Ser/Thr) sequence motifs, with phosphorylation necessary to activate and bind f the protein C-termini locus. While certain assumptions can be inferred from the presence of 14-3-3 motifs, the sequences of binding proteins alone cannot serve as a sole predictive factor (Liu et al. 2021; Fu, Subramanian, and Masters 2000).
In human cells, among 14-3-3 isoforms, the 14-3-3ɛ, known as tyrosine 3/ tryptophan 5-monooxygenase activation protein epsilon due to its activity, is mainly expressed in lymphoblasts, brain, heart, testes, and adipocyte cells, been also related to a negative androgen regulator mostly found in mitochondria during steroidogenesis, although this isoform can be found in almost all tissues (Muslin et al. 1996; Aitken et al. 2002; Qu et al. 2022; Zhang et al. 2024).
The specificity of 14-3-3ε is intricately linked to various physiological mechanisms, including cellular proliferation, signal transduction, cell cycle regulation, apoptosis, autophagy, intracellular electrolyte balance, cardiac morphogenesis and repolarization, neurodevelopment, and innate immune responses. As such, 14-3-3ε assumes significance in the progression of cardiovascular diseases, neurodegenerative disorders, cancer, and inflammatory conditions (Zhang et al. 2024). Although the 14-3-3ɛ is a cytoplasm protein, it can also be present in the nucleus, Golgi apparatus, mitochondria, and plasm membrane (Celis et al. 1990; Guo et al. 2014).
Noteworthy, the human 14-3-3ε regulates cell innate immunity through T cells activation. The SH2 domain-containing leukocyte protein of 76 kD (SLP-76) during viral infections regulates T-cell signaling. The binding of phosphorylated- SLP-76 to 14-3-3ε results in a negative signal to regulate T cells activation (Bartolo et al. 2007; Iyer et al. 2022), leading to a dumped immune response.
Similarly, this protein has a moonlighting characteristic in fungi cells due to its wide range of cell signaling pathways. For fungi, the 14-3-3 closed to human 14-3-3ɛ protein (Fig. 1) exhibits an important role during host-pathogen interaction, acting as a critical virulence factor. Studies conducted by silencing 55% of 14-3-3 protein (NCBI XP_010759842.1) in Paracoccidioides (Pb14-3-3 aRNA) have demonstrated significant enhancement of survival of pneumocytes, implying that the improved survival is a result of reduced interaction between the fungi and the host plasma membrane, increasing Galleria mellonella larval survival in 35% when infected with silenced strain (Marcos et al. 2016; 2019). Furthermore, recent studies highlighted its importance during biofilm formation in P. brasiliensis (Sardi et al. 2024). According to this study, wild P. brasiliensis (Pb18) increases its enolase, 14-3-3, and GAPDH gene expression, while the Pb14-3-3 aRNA strain shows a decrease in the expression of these genes, demonstrating that 14-3-3 could influence the expression of other biofilm-related genes (Sardi et al. 2024).
The reduction in its interaction with host cells appears to be mediated by the modulation of the 14-3-3 protein in the TLR signaling pathway (Bonfim, Mamoni, and Lima Blotta 2009). Specifically, it binds to TLR2, TLR3, TLR4, TLR7/8, and TLR9, iproducing pro-inflammatory cytokines such as IL-6, TNFα, and IFN-β. This binding dampens the pro-inflammatory response, inhibits nitric oxide (NO) production, and suppresses the activation of IFNγ/CD8/T cells and IL-17/CD8/T cells, along with cytotoxic functions, including the downregulation of granzyme B and perforin. Notably, the binding of 14-3-3 to TLR3 has recently been proposed as an evasion mechanism (Jannuzzi et al. 2019; Bonfim, Mamoni, and Lima Blotta 2009).
It is crucial to emphasize that, in the case of other members within the Ajellomycetaceae family, existing literature lacks data showcasing the binding of the 14-3-3 protein to TLR receptors. Despite observed differences in peptide sequences (Fig. 2), notable similarity persists among the studied 14-3-3ɛ-like protein of dimorphic fungi, indicating maybe a shared target, indicated by conservative Ser/Thr residues among these fungi family (Fig. 3).
Consequently, we hypothesize that moonlight proteins, such as enolase, Hsp60 (de Matos Silva et al. 2024), and 14-3-3, are virulence factors in dimorphic fungi related to evasion mechanisms and biofilm formation (Marcos et al. 2016; Sardi et al. 2015; Silva et al. 2013). The phylogenetic analysis of the second copy of the 14-3-3 protein within the Ajellomycetaceae family revealed a shared ancestor with human isoforms (Fig. 1, red), and sequence alignment further highlighted significant distinctions among the sequences identified in the Ajellomycetaceae family.
Despite Paracoccidioides and Histoplasma belonging to different clades, this divergence may lead to variations in the evolution of these microorganisms, influencing pathogenicity, host-pathogen interaction specificity, and genetic diversity within the 14-3-3 protein family. It is known that population and comparative studies among the Ajellomycetaceae family have demonstrated gene expansions and contractions resulted in enhanced or decreased virulence (Muñoz et al. 2016; 2015; Desjardins et al. 2011), creating the hypothesis that these events could be related to evolutionary mechanisms of adaptation in dimorphic fungi (Muñoz et al. 2018).
In our analysis, we utilized both pathogenic and rarely pathogenic dimorphic fungi from the Ajellomycetaceae family, along with non-dimorphic species such as P. hysterics and H. griseus were also present. The results demonstrated the presence of two copies in most analyzed species, suggesting that the 14-3-3 protein is essential not only for host-pathogen interactions and as an evasion mechanism, as hypothesized, but also might function as a non-exclusive pathogenic factor during infection.
Similar observations were made in studies comparing pathogenic and non-pathogenic fungi within the Ajellomycetaceae family, indicating the conservation of many switch-related proteins in non-pathogenic fungi, suggesting that these proteins are not solely required for survival in mammalian hosts, thereby not specifying pathogenicity, but also renders them necessary for environmental survival during the mycelial phase (Muñoz et al. 2018). A parallel scenario is observed in Cryptococcus neoformans, where the alpha mating factor contributes to mouse virulence and amoebae survival (Nielsen et al. 2005).
The divergence in both gene and protein expression of the 14-3-3 second copy may suggest an evolutionary mechanism of differentiation among species within the Ajellomycetaceae family. Various mechanisms contribute to genetic differentiation in fungi, including recombination events, gene gain and loss through duplication or excision, gene family expansion and contraction, genome rearrangements, and horizontal gene transfer. Notably, gene duplications tend to occur more frequently in proteins responding to stress rather than those related to metabolism, leading to differential expression rather than the emergence of new protein functions. Consequently, duplication events in proteins associated with multiple pathways ensure comprehensive coverage (Taylor et al. 2017) , conferring survival advantages to fungi possessing multiple gene copies of a moonlighting protein compared to those without one.
Furthermore, the duplication combined of a contracted third copy of the gene observed in Emergomyces africanus might result from environmental pressures favoring the survival of this soil fungus to migrate to mammal hosts (Muñoz et al. 2018).
The 3D similarities and conserved Ser/Thr phosphorylation sites demonstrated among Ajellomycetaceae 14-3-3 proteins could be employed as an immunogenic protein for the developing new specific drug targets and vaccines. Further studies are necessary to validate our hypothesis and explore these conserved regions’ potential in therapeutic applications. Investigating the functional roles of these phosphorylation sites and their interactions with other cellular proteins will be crucial in understanding their contribution to fungal pathogenicity and their viability as targets for antifungal strategies.
4. Methods
To perform the orthologs analysis, we download all predicted proteins of the
Ajellomycetaceae family (
Table 1) and Human (GRCh38.p14) (
Table 2) from the representative genomes NCBI database. All the data used here have been previously published, except the
P. restrepiensis 60855. We analyzed homology using OrthoFinder version 2.5.4 by default settings (Emms and Kelly 2015). In the ortho-group output, we found the 14-3-3 orthogroup searching the human protein 14-3-3. Then, the resolved gene tree for the protein orthogroup was visualized using Treegrap2. The protein sequences reported in the 14-3-3 orthogroup were aligned using Clustal Omega v1.2.2. Finally, the resultant alignment was visualized in Geneious Prime V 2023.0 (Biomatters, NZ) and 3D protein modeling was obtained with SWISS-MODEL (Waterhouse et al. 2018)
Non-dimorphic species of Ajellomycetaceae, such as Polytolypa hysterics and Helicocarpus griseus, and dimorphic pathogenic fungi including Blastomyces spp., Histoplasma spp., Paracoccidioides spp., Emergomyces spp., and Emmonsia spp. were used in the analysis. The NCBI protein accession number is referenced in Tables 1 (Ajellomycetaceae family) and 2 (Homo sapiens).
Author Contributions
Samanta de Matos Silva, Oscar Mauricio Guzman, Marco Antonio Rodrigues, Henrique Gomes Martins, and Angel Gonzalez conceived the idea and wrote the first draft of the manuscript; Maria José Soares Mendes-Giannini, Orville Hernandez Ruiz, and Ana Marisa Fusco-Almeida provided critical revision of the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP) [grant 2022/15826-0 REGULAR, 2022/15233-0 (MAR), 2024/03014-7 (HGM)]; and Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES) [finance code 001, 88887.600612/2021-00 (SMS), 88887.839588/2023-00 (SMS), 88887.949391/2024-00 (HGM)].
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Aitken, A., H. Baxter, T. Dubois, S. Clokie, S. Mackie, K. Mitchell, A. Peden, and E. Zemlickova. 2002. “Specificity of 14-3-3 Isoform Dimer Interactions and Phosphorylation.” Biochemical Society Transactions 30 (4): 351–60. [CrossRef]
- Assato, Patricia Akemi, Julhiany de Fátima da Silva, Haroldo Cesar de Oliveira, Caroline Maria Marcos, Danuza Rossi, Sandro Roberto Valentini, Maria José Soares Mendes-Giannini, Cleslei Fernando Zanelli, and Ana Marisa Fusco-Almeida. 2015. “Functional Analysis of Paracoccidioides Brasiliensis 14-3-3 Adhesin Expressed in Saccharomyces Cerevisiae.” BMC Microbiology 15 (1): 256. [CrossRef]
- Bartolo, Vincenzo Di, Benjamin Montagne, Mogjiborahman Salek, Britta Jungwirth, Florent Carrette, Julien Fourtane, Nathalie Sol-Foulon, et al. 2007. “A Novel Pathway Down-Modulating T Cell Activation Involves HPK-1–Dependent Recruitment of 14-3-3 Proteins on SLP-76.” The Journal of Experimental Medicine 204 (3): 681–91. [CrossRef]
- Bonfim, Camila Vicente, Ronei Luciano Mamoni, and Maria Heloisa Souza Lima Blotta. 2009. “TLR-2, TLR-4 and Dectin-1 Expression in Human Monocytes and Neutrophils Stimulated by Paracoccidioides Brasiliensis.” Medical Mycology 47 (7): 722–33. [CrossRef]
- Burik, Jo-Anne Van, David Myerson, Randall W. Schreckhise, and Raleigh A. Bowden. 1998. “Panfungal PCR Assay for Detection of Fungal Infection in Human Blood Specimens.” Journal of Clinical Microbiology 36 (5): 1169–75. [CrossRef]
- Burr, Tom. 2010. “Phylogenetic Trees in Bioinformatics.” Current Bioinformatics 5 (1): 40–52. [CrossRef]
- Caydasi, Ayse Koca, Yagmur Micoogullari, Bahtiyar Kurtulmus, Saravanan Palani, and Gislene Pereira. 2014. “The 14-3-3 Protein Bmh1 Functions in the Spindle Position Checkpoint by Breaking Bfa1 Asymmetry at Yeast Centrosomes.” Molecular Biology of the Cell 25 (14): 2143–51. [CrossRef]
- Celis, Julio E., Borbala Gesser, Hanne Holm Rasmussen, Peder Madsen, Henrik Leffers, Kurt Dejgaard, Bent Honore, et al. 1990. “Comprehensive Two-dimensional Gel Protein Databases Offer a Global Approach to the Analysis of Human Cells: The Transformed Amnion Cells (AMA) Master Database and Its Link to Genome DNA Sequence Data.” ELECTROPHORESIS 11 (12): 989–1071. [CrossRef]
- Cognetti, David, Dana Davis, and Joy Sturtevant. 2002. “The Candida Albicans 14-3-3 Gene, BMH1, Is Essential for Growth.” Yeast 19 (1): 55–67. [CrossRef]
- Desjardins, Christopher A., Mia D. Champion, Jason W. Holder, Anna Muszewska, Jonathan Goldberg, Alexandre M. Bailão, Marcelo Macedo Brigido, et al. 2011. “Comparative Genomic Analysis of Human Fungal Pathogens Causing Paracoccidioidomycosis.” PLoS Genetics 7 (10): e1002345. [CrossRef]
- Emms, David M., and Steven Kelly. 2015. “OrthoFinder: Solving Fundamental Biases in Whole Genome Comparisons Dramatically Improves Orthogroup Inference Accuracy.” Genome Biology 16 (1): 157. [CrossRef]
- Fisher, Matthew C., Ana Alastruey-Izquierdo, Judith Berman, Tihana Bicanic, Elaine M. Bignell, Paul Bowyer, Michael Bromley, et al. 2022. “Tackling the Emerging Threat of Antifungal Resistance to Human Health.” Nature Reviews Microbiology 20 (9): 557–71. [CrossRef]
- Fu, Haian, Romesh R. Subramanian, and Shane C. Masters. 2000. “14-3-3 Proteins: Structure, Function, and Regulation.” Annual Review of Pharmacology and Toxicology 40 (1): 617–47. [CrossRef]
- Gauthier, Gregory M. 2015. “Dimorphism in Fungal Pathogens of Mammals, Plants, and Insects.” PLOS Pathogens 11 (2): e1004608. [CrossRef]
- Gelperin, D, J Weigle, K Nelson, P Roseboom, K Irie, K Matsumoto, and S Lemmon. 1995. “14-3-3 Proteins: Potential Roles in Vesicular Transport and Ras Signaling in Saccharomyces cerevisiae.” Proceedings of the National Academy of Sciences 92 (25): 11539–43. [CrossRef]
- Guo, Zhendong, Chao Han, Jiajun Du, Siyan Zhao, Yingying Fu, Guanyu Zheng, Yucheng Sun, et al. 2014. “Proteomic Study of Differential Protein Expression in Mouse Lung Tissues after Aerosolized Ricin Poisoning.” International Journal of Molecular Sciences 15 (5): 7281–92. [CrossRef]
- Herod, S. Grace, Annie Dyatel, Stefanie Hodapp, Marko Jovanovic, and Luke E. Berchowitz. 2022. “Clearance of an Amyloid-like Translational Repressor Is Governed by 14-3-3 Proteins.” Cell Reports 39 (5): 110753. [CrossRef]
- Hondermarck, Hubert. 2010. “14-3-3 Proteins.” In Handbook of Cell Signaling, 1367–74. Elsevier. [CrossRef]
- Ichimura, T, T Isobe, T Okuyama, N Takahashi, K Araki, R Kuwano, and Y Takahashi. 1988. “Molecular Cloning of CDNA Coding for Brain-Specific 14-3-3 Protein, a Protein Kinase-Dependent Activator of Tyrosine and Tryptophan Hydroxylases.” Proceedings of the National Academy of Sciences 85 (19): 7084–88. [CrossRef]
- Ichimura, Tohru, Hiroyuki Kubota, Takeshi Goma, Noboru Mizushima, Yoshinori Ohsumi, Maki Iwago, Kazue Kakiuchi, et al. 2004. “Transcriptomic and Proteomic Analysis of a 14-3-3 Gene-Deficient Yeast.” Biochemistry 43 (20): 6149–58. [CrossRef]
- Iyer, Vaishnavi Srinivasan, Sanjaykumar V. Boddul, Anna-Karin Johnsson, Bruno Raposo, Ravi K. Sharma, Yunbing Shen, Zsolt Kasza, et al. 2022. “Modulating T-Cell Activation with Antisense Oligonucleotides Targeting Lymphocyte Cytosolic Protein 2.” Journal of Autoimmunity 131 (July):102857. [CrossRef]
- Jannuzzi, Grasielle Pereira, José Roberto Fogaça de Almeida, Gustavo P. Amarante-Mendes, Lavínia Maria Dal’Mas Romera, Gilberto Hideo Kaihami, José Ronnie Vasconcelos, Camila Pontes Ferreira, Sandro Rogério de Almeida, and Karen Spadari Ferreira. 2019. “TLR3 Is a Negative Regulator of Immune Responses Against Paracoccidioides brasiliensis.” Frontiers in Cellular and Infection Microbiology 8 (January). [CrossRef]
- Kumar, Ravinder. 2018. “Differential Abundance and Transcription of 14-3-3 Proteins during Vegetative Growth and Sexual Reproduction in Budding Yeast.” Scientific Reports 8 (1): 2145. [CrossRef]
- Liu, Jiaqi, Shengliang Cao, Guofei Ding, Bin Wang, Yingchao Li, Yuzhong Zhao, Qingyuan Shao, et al. 2021. “The Role of 14-3-3 Proteins in Cell Signalling Pathways and Virus Infection.” Journal of Cellular and Molecular Medicine 25 (9): 4173–82. [CrossRef]
- Lottersberger, Francisca, Andrea Panza, Giovanna Lucchini, Simonetta Piatti, and Maria Pia Longhese. 2006. “The Saccharomyces Cerevisiae 14-3-3 Proteins Are Required for the G1/S Transition, Actin Cytoskeleton Organization and Cell Wall Integrity.” Genetics 173 (2): 661–75. [CrossRef]
- Marcos, Caroline Maria, Haroldo Cesar de Oliveira, Patricia Akemi Assato, Cleverton Roberto de Andrade, Ana Marisa Fusco-Almeida, and Maria José Soares Mendes-Giannini. 2019. “Paracoccidioides brasiliensis 14-3-3 Protein Is Important for Virulence in a Murine Model.” Medical Mycology 57 (7): 900–904. [CrossRef]
- Marcos, Caroline Maria, Julhiany de Fátima da Silva, Haroldo Cesar de Oliveira, Patrícia Akemi Assato, Junya de Lacorte Singulani, Angela Maria Lopez, Diana Patricia Tamayo, et al. 2016. “Decreased Expression of 14-3-3 in Paracoccidioides Brasiliensis Confirms Its Involvement in Fungal Pathogenesis.” Virulence 7 (2): 72–84. [CrossRef]
- Matos Silva, Samanta de, Carolina Rodriguez Echeverri, Maria José Soares Mendes-Giannini, Ana Marisa Fusco-Almeida, and Angel Gonzalez. 2024. “Common Virulence Factors between Histoplasma and Paracoccidioides: Recognition of Hsp60 and Enolase by CR3 and Plasmin Receptors in Host Cells.” Current Research in Microbial Sciences 7:100246. [CrossRef]
- Muñoz, José F., Rhys A. Farrer, Christopher A. Desjardins, Juan E. Gallo, Sean Sykes, Sharadha Sakthikumar, Elizabeth Misas, et al. 2016. “Genome Diversity, Recombination, and Virulence across the Major Lineages of Paracoccidioides.” MSphere 1 (5). [CrossRef]
- Muñoz, José F., Gregory M. Gauthier, Christopher A. Desjardins, Juan E. Gallo, Jason Holder, Thomas D. Sullivan, Amber J. Marty, et al. 2015. “The Dynamic Genome and Transcriptome of the Human Fungal Pathogen Blastomyces and Close Relative Emmonsia.” PLOS Genetics 11 (10): e1005493. [CrossRef]
- Muñoz, José F., Juan G. McEwen, Oliver K. Clay, and Christina A. Cuomo. 2018. “Genome Analysis Reveals Evolutionary Mechanisms of Adaptation in Systemic Dimorphic Fungi.” Scientific Reports 8 (1): 4473. [CrossRef]
- Muslin, Anthony J, J.William Tanner, Paul M Allen, and Andrey S Shaw. 1996. “Interaction of 14-3-3 with Signaling Proteins Is Mediated by the Recognition of Phosphoserine.” Cell 84 (6): 889–97. [CrossRef]
- Nielsen, Kirsten, Gary M. Cox, Anastasia P. Litvintseva, Eleftherios Mylonakis, Stephanie D. Malliaris, Daniel K. Benjamin, Steven S. Giles, et al. 2005. “Cryptococcus Neoformans α Strains Preferentially Disseminate to the Central Nervous System during Coinfection.” Infection and Immunity 73 (8): 4922–33. [CrossRef]
- Nnadi, Nnaemeka Emmanuel, and Dee A. Carter. 2021. “Climate Change and the Emergence of Fungal Pathogens.” PLOS Pathogens 17 (4): e1009503. [CrossRef]
- Palmer, Glen E., Kevin J. Johnson, Sumana Ghosh, and Joy Sturtevant. 2004. “Mutant Alleles of the Essential 14-3-3 Gene in Candida albicans Distinguish between Growth and Filamentation.” Microbiology 150 (6): 1911–24. [CrossRef]
- Qu, Jia-Hua, Kirill V. Tarasov, Khalid Chakir, Yelena S. Tarasova, Daniel R. Riordon, and Edward G. Lakatta. 2022. “Proteomic Landscape and Deduced Functions of the Cardiac 14-3-3 Protein Interactome.” Cells 11 (21): 3496. [CrossRef]
- Rodríguez-Romero, Julio, Marco Marconi, Víctor Ortega-Campayo, Marie Demuez, Mark D. Wilkinson, and Ane Sesma. 2019. “Virulence- and Signaling-associated Genes Display a Preference for Long 3′ UTRs during Rice Infection and Metabolic Stress in the Rice Blast Fungus.” New Phytologist 221 (1): 399–414. [CrossRef]
- Sardi, Janaina de Cássia Orlandi, Jaqueline Derissi Braz Carlton, Caroline Maria Marcos, Ana Marisa Fusco Almeida, and Maria José Soares Mendes Giannini. 2024. “Unveiling the Functional Significance of the 14.3.3 Protein: A Key Player in Paracoccidioides brasiliensis Biofilm Formation.” Microbial Pathogenesis 188 (March):106537. [CrossRef]
- Sardi, Janaina de Cássia Orlandi, Nayla de Souza Pitangui, Aline Raquel Voltan, Jaqueline Derissi Braz, Marcelo Pelajo Machado, Ana Marisa Fusco Almeida, and Maria Jose Soares Mendes Giannini. 2015. “In Vitro Paracoccidioides Brasiliensis Biofilm and Gene Expression of Adhesins and Hydrolytic Enzymes.” Virulence 6 (6): 642–51. [CrossRef]
- Silva, Julhiany de Fatima da, Haroldo César de Oliveira, Caroline Maria Marcos, Rosângela Aparecida Moraes da Silva, Tania Alves da Costa, Vera Lucia García Calich, Ana Marisa Fusco Almeida, and Maria José Soares Mendes-Giannini. 2013. “Paracoccidoides brasiliensis 30 KDa Adhesin: Identification as a 14-3-3 Protein, Cloning and Subcellular Localization in Infection Models.” PLoS ONE 8 (4): e62533. [CrossRef]
- Simonson, Andrew W., Agustey S. Mongia, Matthew R. Aronson, John N. Alumasa, Dennis C. Chan, Atip Lawanprasert, Michael D. Howe, et al. 2021. “Pathogen-Specific Antimicrobials Engineered de Novo through Membrane-Protein Biomimicry.” Nature Biomedical Engineering 5 (5): 467–80. [CrossRef]
- Sun, Zhongfeng, Jiabin Song, Xi’an Xin, Xianan Xie, and Bin Zhao. 2018. “Arbuscular Mycorrhizal Fungal 14-3-3 Proteins Are Involved in Arbuscule Formation and Responses to Abiotic Stresses During AM Symbiosis.” Frontiers in Microbiology 9 (March). [CrossRef]
- Taborda, C P, and Z P Camargo. 1994. “Diagnosis of Paracoccidioidomycosis by Dot Immunobinding Assay for Antibody Detection Using the Purified and Specific Antigen Gp43.” Journal of Clinical Microbiology 32 (2): 554–56. [CrossRef]
- Taylor, John W., Sara Branco, Cheng Gao, Chris Hann-Soden, Liliam Montoya, Iman Sylvain, and Pierre Gladieux. 2017. “Sources of Fungal Genetic Variation and Associating It with Phenotypic Diversity.” Microbiology Spectrum 5 (5). [CrossRef]
- Trendeleva, T. A. , and R. A. Zvyagilskaya. 2018. “Retrograde Signaling as a Mechanism of Yeast Adaptation to Unfavorable Factors.” Biochemistry (Moscow) 83 (2): 98–106. [CrossRef]
- Wang, Wenfu, and Diane C. Shakes. 1996. “Molecular Evolution of the 14-3-3 Protein Family.” Journal of Molecular Evolution 43 (4): 384–98. [CrossRef]
- Wang, Ying-Kai, Biswadip Das, David H. Huber, Melanie Wellington, M. Anaul Kabir, Fred Sherman, and Elena Rustchenko. 2004. “Role of the 14–3–3 Protein in Carbon Metabolism of the Pathogenic Yeast Candida Albicans.” Yeast 21 (8): 685–702. [CrossRef]
- Waterhouse, Andrew, Martino Bertoni, Stefan Bienert, Gabriel Studer, Gerardo Tauriello, Rafal Gumienny, Florian T Heer, et al. 2018. “SWISS-MODEL: Homology Modelling of Protein Structures and Complexes.” Nucleic Acids Research 46 (W1): W296–303. [CrossRef]
- Wilson, Rashaun S., Kirby N. Swatek, and Jay J. Thelen. 2016. “Regulation of the Regulators: Post-Translational Modifications, Subcellular, and Spatiotemporal Distribution of Plant 14-3-3 Proteins.” Frontiers in Plant Science 7 (May). [CrossRef]
- Xu, Zhe, Kourtney Graham, Molly Foote, Fengshan Liang, Raed Rizkallah, Myra Hurt, Yanchang Wang, Yuying Wu, and Yi Zhou. 2013. “14-3-3 Targets Chaperone-Associated Misfolded Proteins to Aggresomes.” Journal of Cell Science, January. [CrossRef]
- Zhang, Yue, Man Yan, Yongjun Yu, Jiangping Wang, Yuqi Jiao, Minying Zheng, and Shiwu Zhang. 2024. “14–3-3ε: A Protein with Complex Physiology Function but Promising Therapeutic Potential in Cancer.” Cell Communication and Signaling 22 (1): 72. [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).