1. Introduction
Throughout their lifecycle, plants are subjected to various forms of abiotic and biological stresses. These stresses promptly induce the synthesis of reactive oxygen species (ROS) within the plants, subsequently leading to an accumulation of aldehydes [
1]. Aldehydes, which are metabolic byproducts of carbohydrates, lipids, and amino acids, can, in appropriate quantities, positively influence the physiological metabolism of plants. However, excessive concentrations of aldehydes can cause cellular damage, disrupt metabolic processes, and lead to the formation of toxic substances detrimental to plant health, compromising their normal growth [
2]. Hence, the selective removal and regulation of aldehyde concentrations within plants hold significant implications for their growth and development, mandating strict regulation [
3].
Aldehyde dehydrogenase (ALDH), often referred to as the "aldehyde scavenger," is effective in eliminating active aldehyde molecules in plants [
4]. ALDH is a category of enzymes that encode NAD(P)+-dependent reactions, showcasing a diversity of amino acid sequences and incorporating different motifs such as the cysteine active site (PS00070), glutamic acid active site (PS00687), and the Rossmann fold [
5]. To mitigate the damage inflicted by excessive aldehyde accumulation during plant growth, the ALDH gene is induced to express differently under stress, subsequently leading to a substantial increase in aldehyde dehydrogenase proteins [
6]. Consequently, the toxic aldehydes generated due to stress are irreversibly oxidized into their non-toxic carboxylic acid counterparts, thus enhancing the plant's stress tolerance and reducing stress-induced damage. This indicates a critical detoxification mechanism in plants [
7]. Furthermore, ALDH also plays a pivotal role in plant metabolism, such as glycolysis, amino acid metabolism, and carnitine biosynthesis [
8]. Under osmotic stress, ALDH helps maintain cellular osmotic equilibrium in plants through the synthesis of osmoregulators [
9].
Aldehyde dehydrogenase (ALDH) is predominantly found in both eukaryotes and prokaryotes. As the genome sequencing information for an ever-increasing number of species becomes available, the presence of ALDH genes in more species is being discovered. The ALDH Gene Naming Committee (ANGC) defines a classification system based on protein sequence similarity between a new gene and an identified gene. If this similarity is less than 40%, the gene is considered as part of a new family; if the similarity ranges between 40% and 60%, it is classified within a family; if the similarity exceeds 60%, it is allocated to a subfamily [
10]. Currently, the identified ALDH genes are categorized into 24 families, namely ALDH1-ALDH24, with 14 families found in plants. These 14 include ALDH2, ALDH3, ALDH5, ALDH6, ALDH7, ALDH10, ALDH11, ALDH12, ALDH18, ALDH19, ALDH21, ALDH22, ALDH23, and ALDH24, with the last seven exclusive to plants [
11,
12]. Over recent years, we have seen an increasing number of ALDH genes being identified in various plants, each with varying quantities, such as 14, 20, 28, 29, 53, 19, 27, 27, 23, 22, and 39 in
Arabidopsis [
5], rice [
13], maize [
14], tomato [
15], soybean [
16], sorghum [
17], Chinese cabbage [
18], grape [
19], potato [
20], and apple [
21] respectively. These genes play a significant role in the regulation of stress response, growth, and development in plants, hence critical for normal plant growth [
22,
23]. For instance,
AtALDH3I1 and
AtALDH7B4 in
Arabidopsis are seen to reduce MDA (malondialdehyde) and lipid peroxidation when their expression is up-regulated, improving the plant's tolerance to drought and salt stress [
24]. Overexpression of
BrALDH7B2 in tobacco can enhance photosynthesis, reduce ROS production, thereby improving salt tolerance [
25]. When wheat is subjected to drought and salt stress,
TraeALDH7B1-5A is highly expressed in various parts of the plant, and overexpression of this gene in transgenic
Arabidopsis significantly improves plant tolerance to drought stress [
26]. Similarly, rice exhibited elevated expression levels of
OsALDH2-4,
OsALDH3-4,
OsALDH7,
OsALDH18-2, and
OsALDH12 when subjected to drought stress, surpassing control levels by twofold [
13]. Virus-Induced Gene Silencing (VIGS) experiment performed on the cotton genes
Gohir.A11G040800 and
Gohir.
D06G046200 demonstrated increased sensitivity of the silenced plants to salt stress as compared to the control, indicating the potential involvement of these genes in cotton's salt stress response [
27]. In soybeans, acetaldehyde dehydrogenase can reduce the content of toxic malondialdehyde (MDA), and overexpression of
ALDH3H1 increases soybean resistance to salt stress [
28].
ZmALDH9,
ZmALDH13, and
ZmALDH17 have been reported to participate in the maize response to drought stress and are involved in plant acid tolerance and pathogen infection response [
14].
Cucumis melo L., a member of the genus Cucumis in the Cucurbitaceae family, is an annual horticultural crop of high economic worth, primarily due to its flavorful fruits and substantial nutritional value [29-31]. In 2012, the genome of Muskmelon DHL92 was sequenced by the Spanish Agricultural Genomics Research Center, paving the way for bioinformatics research related to muskmelon [
32]. As bioinformatics technology continued to advance, Ruggieri carried out an upgrade of the DHL92 genome to version V3.6.1 in 2018, resulting in an enhanced sequence and annotation information [
33]. Later in 2019, Castanera further improved the V3.6.1 version of DHL92 to version V4.0 [
34]. Leveraging the available melon genomic information, researchers worldwide have conducted numerous gene family studies, focusing on gene families such as SUAR[
35], ARF[
36], LBD[
37], SUN[
38], and JMJ-C[
39]. However, no research has yet been reported on the ALDH gene family in melon. In this study, we identified 17 ALDH genes in the melon genome using the available genomic data and conducted a thorough biogenic analysis, examining aspects such as chromosome localization, phylogeny, gene structure, and collinearity. To explore the expression patterns of the ALDH gene in different melon tissues under various stresses, we utilized published transcriptome data to analyze ALDH gene expression levels. The findings of this study establish a basis for future research on the ALDH gene family in melon and provide potential target candidate genes for resistance breeding in melon.
2. Results
2.1. Identification and Physicochemical Characteristics of the ALDH Family Member in Melon
This research identified 17 ALDH genes in the melon genome (DHL92V4) through the use of the hidden Markov model PF00171, and discovered conserved ALDH domains in Pfam, SMART, and NCBI conserved domain databases. An analysis of the physical and chemical properties of the ALDH family members in melon revealed that the smallest CDS gene was MELO3C004383 (1149 bp), while the largest was MELO3C007705 (3132 bp). The encoded amino acid length ranged from 382aa (MELO3C004383) to 1043aa (MELO3C007705); molecular weights fell between 41812.14 kD (MELO3C004383) and 114376.9 kD (MELO3C007705); and theoretical isoelectric point values varied from 5.32 (MELO3C017125) to 8.89 (MELO3C010493). Our instability coefficient analysis indicated that only one ALDH gene in melon, MELO3C007705, yielded a stable protein (instability coefficient >40), while the rest were classified as unstable proteins (instability coefficient <40). The fat coefficients for the 17 ALDH genes spanned a range from 77.52 (MELO3C007705) to 105.54 (MELO3C008245). An evaluation of their average hydrophilic values revealed that 12 ALDH genes had discernible hydrophilic values (mean hydrophilic value <0), with the remaining 5 classified as hydrophobic proteins. The mitochondria housed the most ALDH genes, with a total of six. This was followed by the chloroplast with five genes, the cytoplasm with four, and the nuclear and plasma membrane regions each containing one.
Table 1.
Information and physicochemical properties of 17 CmALDH members.
Table 1.
Information and physicochemical properties of 17 CmALDH members.
| Gene ID |
CDS size (bp) |
Number of
Amino
Acid(aa) |
Molecular
Weight
(kDa) |
Theoretical
pI |
Instability Index |
Aliphatic Index |
Grand Average of Hydropathicity |
Prediction of Subcellular Location |
| MELO3C018583 |
1614 |
537 |
57554.02 |
6.47 |
34.29 |
90.63 |
0.041 |
Mitochondrial |
| MELO3C024345 |
1365 |
454 |
48695.21 |
6.19 |
37.34 |
96.17 |
0.085 |
Chloroplast |
| MELO3C025328 |
1701 |
566 |
61733.01 |
8.81 |
31.75 |
87.05 |
-0.069 |
Mitochondrial |
| MELO3C017125 |
1512 |
503 |
54555.85 |
5.32 |
25.17 |
92.94 |
-0.001 |
Cytoplasmic |
| MELO3C017100 |
1665 |
554 |
60067.44 |
7.23 |
33.22 |
89.10 |
-0.084 |
Chloroplast |
| MELO3C008245 |
2154 |
717 |
77669.64 |
6.20 |
32.92 |
105.54 |
-0.096 |
Mitochondrial |
| MELO3C009229 |
1941 |
646 |
69861.68 |
5.45 |
34.89 |
104.78 |
-0.032 |
Cytoplasmic |
| MELO3C014601 |
1446 |
481 |
54034.52 |
8.55 |
39.02 |
95.90 |
-0.043 |
Plasma Membrane |
| MELO3C004383 |
1149 |
382 |
41812.14 |
8.30 |
26.07 |
93.19 |
-0.022 |
Chloroplast |
| MELO3C004430 |
1620 |
539 |
58481.63 |
7.60 |
37.92 |
91.37 |
-0.002 |
Chloroplast |
| MELO3C010494 |
1296 |
431 |
47243.77 |
8.11 |
34.49 |
104.29 |
0.046 |
Cytoplasmic |
| MELO3C010493 |
1560 |
519 |
57189.43 |
8.89 |
35.71 |
99.79 |
0.02 |
Mitochondrial |
| MELO3C017542 |
1395 |
464 |
51874.30 |
8.76 |
37.35 |
97.93 |
-0.06 |
Cytoplasmic |
| MELO3C007705 |
3132 |
1043 |
114376.90 |
8.10 |
48.04 |
77.52 |
-0.38 |
Nuclear |
| MELO3C019622 |
1626 |
541 |
57939.60 |
6.98 |
38.09 |
95.21 |
0.039 |
Chloroplast |
| MELO3C021272 |
1815 |
604 |
66876.10 |
7.20 |
36.64 |
92.33 |
-0.032 |
Mitochondrial |
| MELO3C002203 |
1620 |
539 |
60367.76 |
6.33 |
39.65 |
95.45 |
-0.09 |
Mitochondrial |
2.2. Chromosome Distribution of Melon ALDH Genes
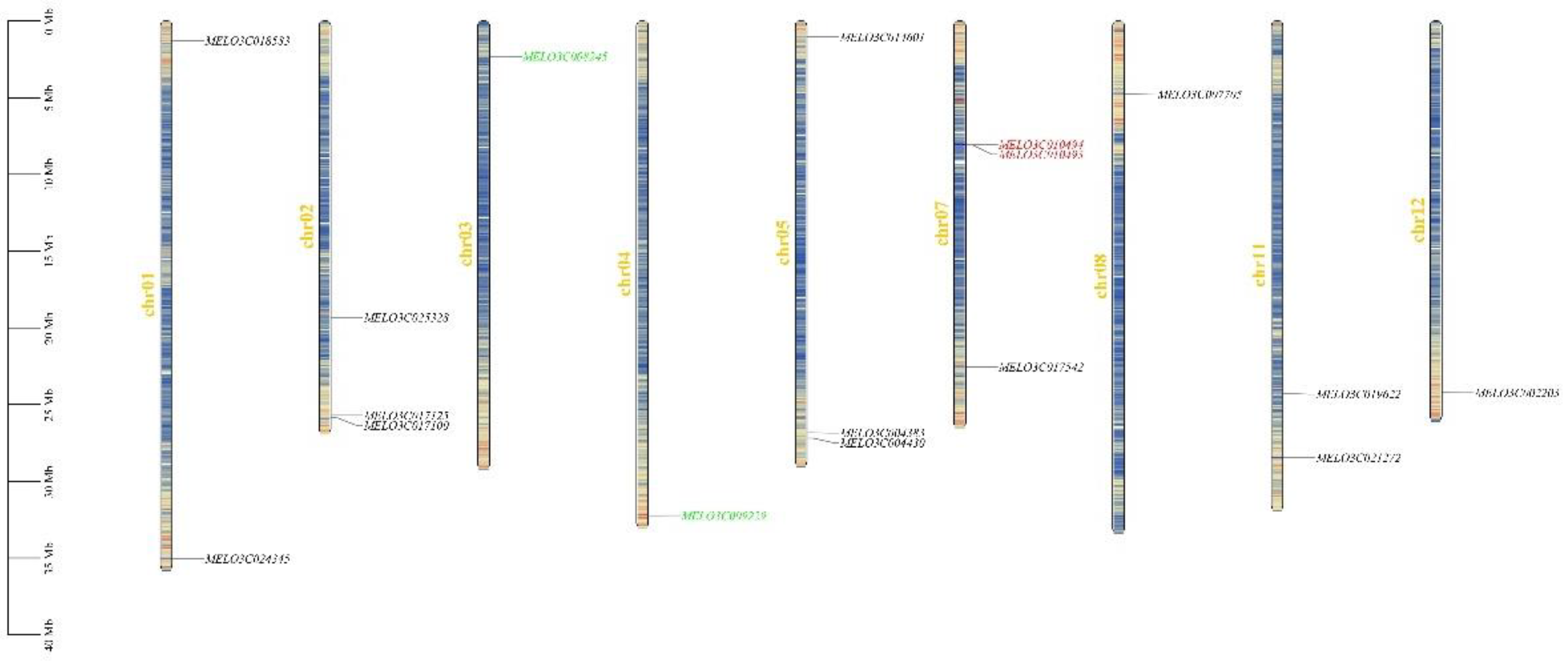
Utilizing the location information of ALDH gene family members on melon chromosomes, a distribution map of the ALDH gene in melon was constructed using TBtools software (
Figure 1). The findings revealed that ALDH genes were present on all chromosomes except for 6, 9, and 10, which lacked these genes, and a certain degree of variation in distribution quantity. For instance, the highest number of distributed genes were found on chromosomes 2 and 7, with three on each. Conversely, chromosomes 3, 4, 8, and 12 harbored the fewest genes, each with only one. This suggests that chromosomes 2 and 7 may play a significant role in the evolution of ALDH in melon. By observing gene positioning on the chromosomes, it was observed that ALDH genes in melon predominantly resided on the termini of chromosomes, with some located centrally. Furthermore, one pair of tandem repeat genes and fragment repeat genes, namely
MELO3C010494/
MELO3C010493 and
MELO3C008245/
MELO3C009229 respectively, were identified within the 17 ALDH genes of melon.
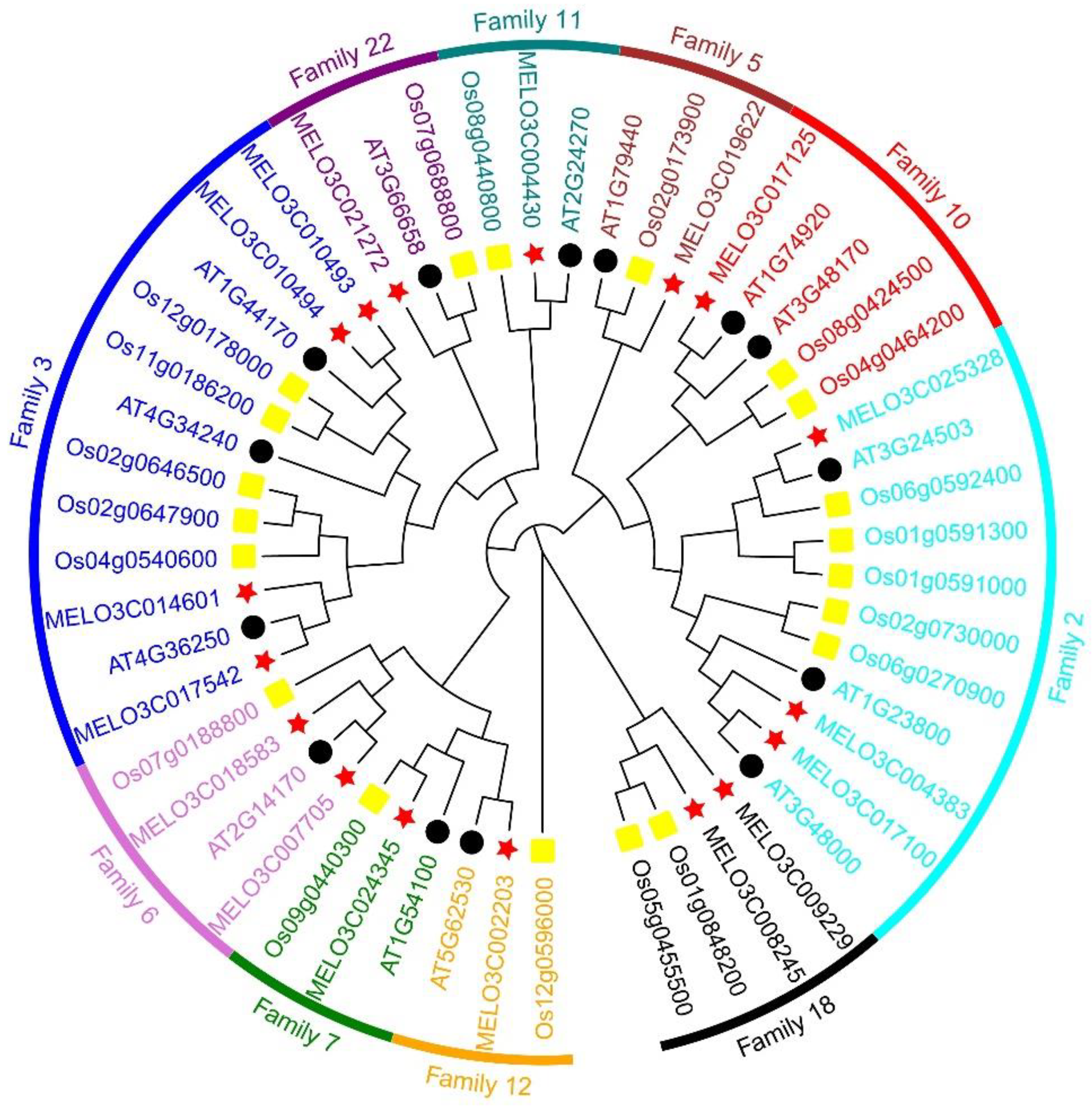
2.3. Phylogenetic Tree Analysis of ALDH Family Genes
To elucidate the phylogenetic relationship of ALDH genes in melon, we procured the protein sequences of acknowledged ALDH genes from
Arabidopsis and rice genome database. These sequences were aligned with the 17 ALDH protein sequences in melon to facilitate phylogenetic analysis and construct a phylogenetic tree (
Figure 2). Subsequently, melon's ALDH genes were partitioned into 10 subfamilies based on
Arabidopsis and rice groupings. Family 3 was identified as the most abundant, containing four melon ALDH genes, followed by Family 2 with three. Aside from Family 6 and 18, which each comprised of two melon ALDH genes, the remaining six subfamilies each held a single ALDH gene.
We discovered seven pairs of direct homologous genes between the ALDH family of muskmelon and Arabidopsis, namely MELO3C004430/AT2G24270, MELO3C017125/AT1G74920, MELO3C025328/AT3G24503, MELO3C017100/AT3G48000, MELO3C002203/AT5G62530, MELO3C007705/AT2G14170, and MELO3C017542/AT4G36250. Additionally, we found one family of orthologous genes between melon and rice ALDH, specifically MELO3C024345/Os09g0440300. In the melon ALDH gene family, only one pair of paralogs genes, MELO3C010493/MELO3C010494, was identified. Evolutionarily related ALDH genes, which exhibit similar biological functions and gene structures, were noted. Future studies can predict the biological function of the melon ALDH gene based on previously reported ALDH genes in Arabidopsis and rice.
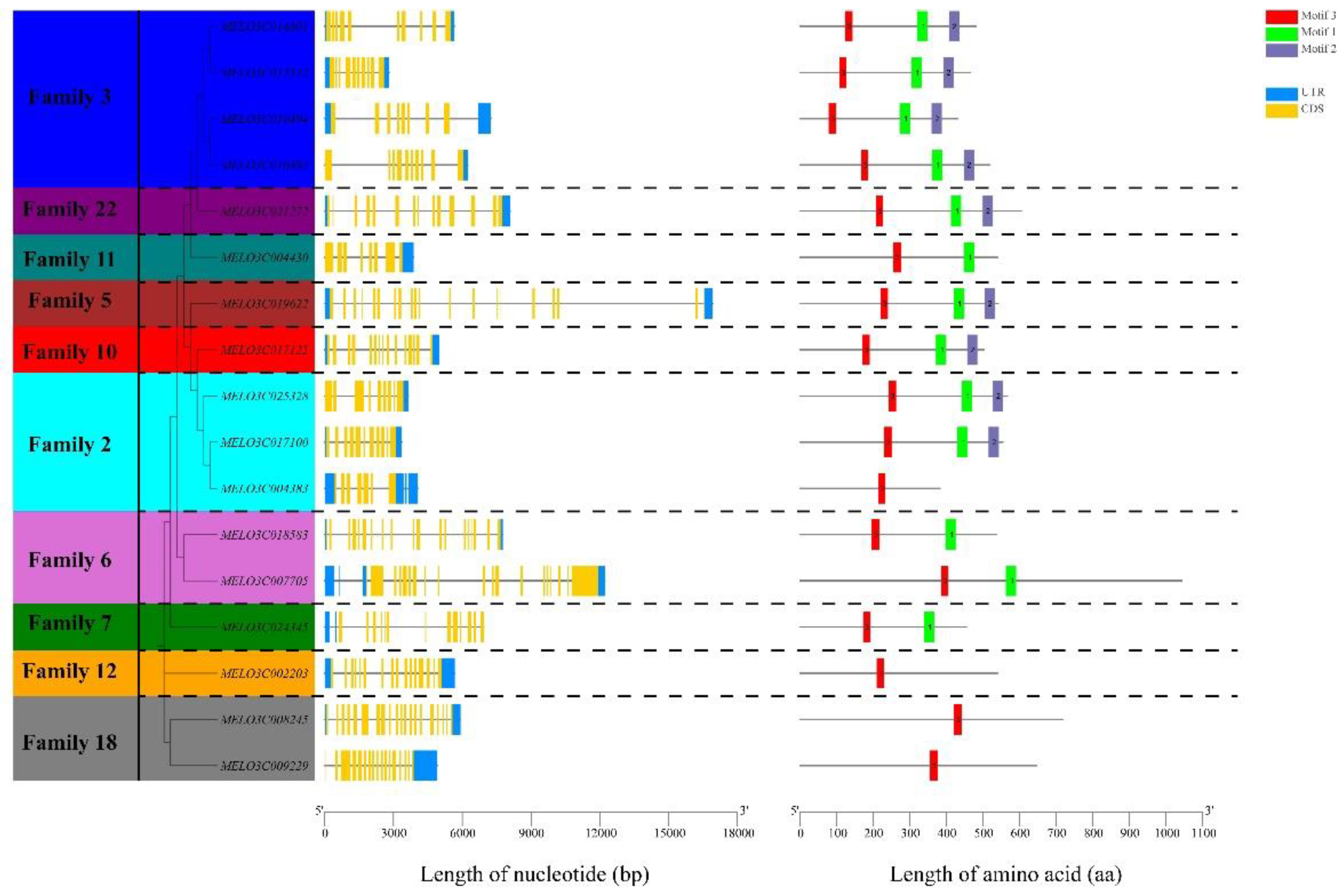
2.4. Gene Structure and Conserved Motif Analysis of Melon ALDH Genes
We conducted a phylogenetic tree construction using the protein sequences from 17 melon ALDH genes, underpinned by gene structure and conserved motif analysis in correlation with melon genome files (
Figure 3). The findings demonstrated that melon's ALDH gene family could be segregated into 10 distinct subfamilies. ALDH genes originating from the same subfamily exhibited a tendency to cluster, which essentially aligns with the ALDH gene clustering outcomes from melon,
Arabidopsis, and rice (
Figure 2).
In the context of multi-gene families, the structural disparities in exon-introns within the same family serve as pivotal factors in the evolutionary process. As per the gene structure of ALDH genes in the melon family, the exon count fluctuates between 7 to 20. The genes MELO3C019622 and MELO3C008245 are characterized by the highest number of exons, tallying at 20, succeeded by MELO3C018583 and MELO3C007705 from the same subfamily, each having 19 exons. The lowest count is attributed to MELO3C004383, with a mere 7 exons. Genes within the same subfamily generally exhibit similarities in their exon-intron structure. The resemblance in these gene structures, coupled with high sequence homology, suggests that gene duplication events transpired in the evolution of the melon's ALDH gene family.
Conserved motif analysis revealed that the motif sequences of Family 3, 22, 11, 5, 10, and 2 were nearly identical, in the sequence of 3, 1, and 2. The motifs in Family 6 and Family 7 displayed the same sequence, 3, 1. Similarly, the motifs in Family 12 and Family 18 followed the same sequence, incorporating only motif 3. This suggests that ALDH genes within these similar subfamilies could potentially share similar biological functions.
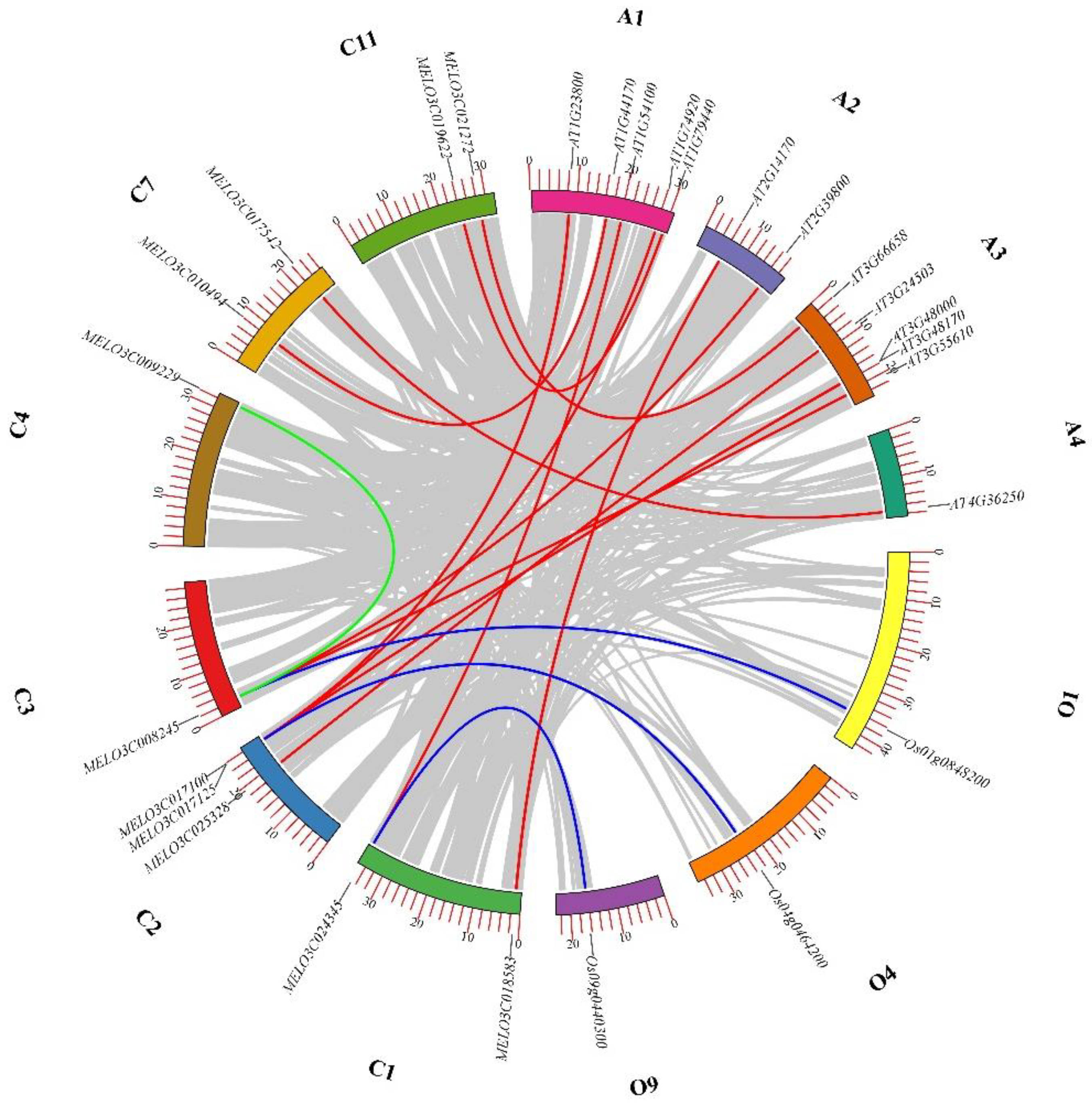
2.5. Synteny Analysis of ALDH Genes among Melon, Arabidopsis, and Rice
To deepen our understanding of the evolutionary details of ALDH genes in melon, we conducted a synteny analysis of these genes (
Figure 4). Our findings revealed that, out of the 17 ALDH genes in melon, there was only one pair of repeated gene fragments, specifically
MELO3C008245/
MELO3C009229. Furthermore, we analyzed ALDH orthologous genes between melon,
Arabidopsis, and rice.Among the melon's ALDH family, there were nine genes (
MELO3C018583,
MELO3C024345,
MELO3C025328,
MELO3C017100,
MELO3C008245,
MELO3C010494,
MELO3C0117542,
MELO3C019622,
MELO3C021272) and twelve in
Arabidopsis (
AT1G23800,
AT1G44170,
AT1G54100,
AT1G74920,
AT1G79440,
AT2G14170,
AT2G39800,
AT3G66658,
AT3G24503,
AT3G48170,
AT3G55610,
AT4G36250). We identified 12 collinearity relationships among these. We also discovered three collinearity relationships between three melon ALDH genes (
MELO3C024345,
MELO3C017125,
MELO3C008245) and three rice ALDH genes (
Os01g0848200,
Os04g0464200,
Os09g0440300). However, the remaining six ALDH genes in melon (
MELO3C014601,
MELO3C004383,
MELO3C004430,
MELO3C010493,
MELO3C007705,
MELO3C002203) exhibited no collinearity with those in
Arabidopsis and rice. Indicated that these genes demonstrate a certain degree of conservation within the ALDH family.
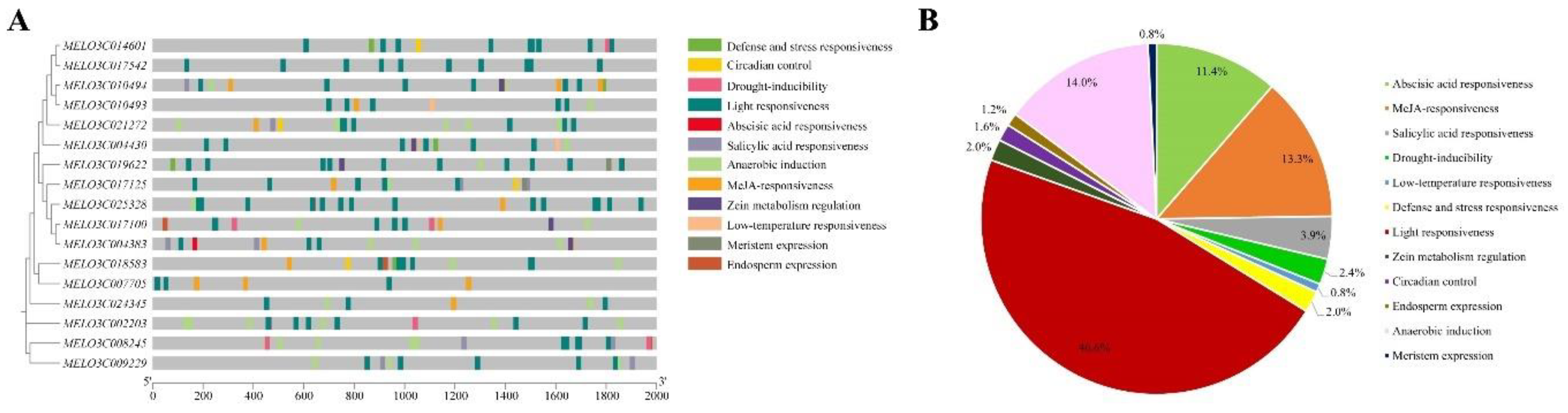
2.6. Analysis of the Cis-Acting Elements in Melon ALDH Genes
Based on the
cis-acting element analysis, we identified 12 distinct
cis-acting elements in the promoter sequences of the melon ALDH genes (
Figure 5A). Light responsiveness elements constituted the majority, accounting for 45.6% of the total and were uniformly distributed among all 17 ALDH genes in melon. The second most prevalent were the hormone-related
cis-acting elements (abscisic acid responsiveness, MeJA-responsiveness, and salicylic acid responsiveness), contributing to 28.6% of the total, following photoresponsive elements (
Figure 5B). Additionally, we examined elements related to stress response and endosperm expression. Our findings suggest that the ALDH gene could respond to a range of hormones and stress conditions. Thus, the identified response elements could significantly impact the stress response capability of the ALDH gene under stress conditions.
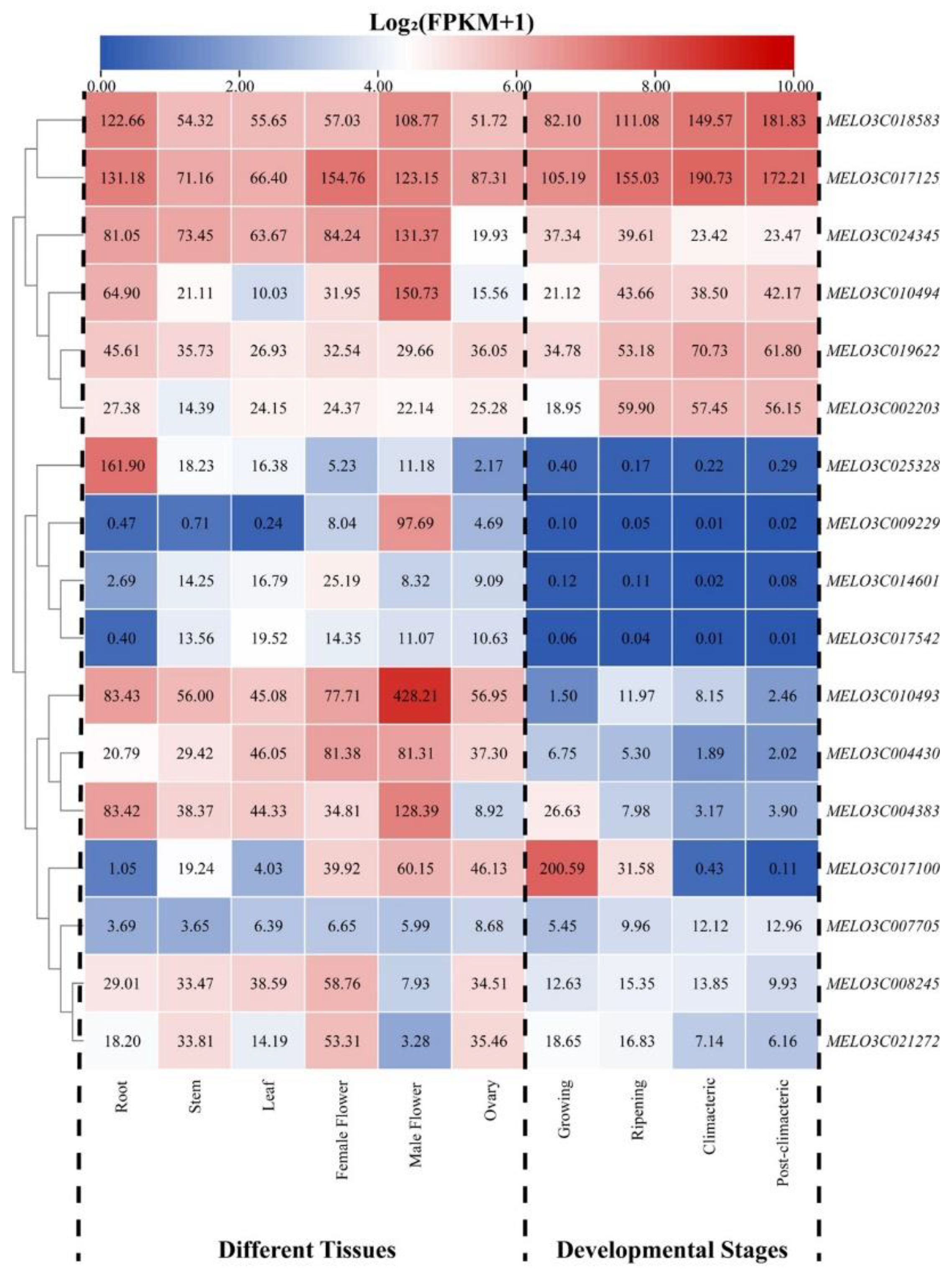
2.7. Tissue-Specific Expression Analysis of Melon ALDH Genes
Utilizing previously published transcriptome data from an array of melon tissues and fruit at different developmental stages, we were able to analyze the tissue-specific expression profiles of the melon's ALDH gene family members during various stages of development (
Figure 6). The histospecificity analysis of the ALDH gene family revealed high expression levels of
MELO3C018583 and
MELO3C017125 across all organs. Notably,
MELO3C010493 is expressed specifically in male flowers ang showed nearly an 8-fold greater expression level compared to other organs. Similar to
MELO3C025328 and
MELO3C009229, which presented specific expression only in the roots and male flowers. Conversely,
MELO3C008245 and
MELO3C021272 exhibited low expression in male flowers, but high expression in other organs.
MELO3C024345 and
MELO3C004383 were highly expressed in all organs except the ovary, where expression levels were significantly lower. The expression of
MELO3C019622 and
MELO3C002203 were more evenly distributed across all organs. Interestingly,
MELO3C004430 displayed significantly higher expression levels in both male and female flowers than other organs, which suggests it may play a role in the formation of melon flower organs and fruits. Among the 17 ALDH genes, only
MELO3C007705 showed no specific organ expression and maintained low expression levels, suggesting this gene is not involved in the growth and development of melon organs.
The expression profiles of ALDH family genes during different stages of fruit development showed that MELO3C018583 and MELO3C017125 were highly expressed at all four stages (growing, ripening, climacteric, and post-climacteric). This pattern, combined with their organ-specific expression, suggests these genes participate in melon organ development and have a role in fruit ripening and development. MELO3C024345, MELO3C010494, MELO3C019622, and MELO3C002203 also displayed high expression levels in all four stages, second only to MELO3C018583 and MELO3C017125. However, MELO3C0225328, MELO3C009229, MELO3C014601, and MELO3C017542 exhibited very low or no expression throughout the entire fruit development stage. MELO3C010493 and MELO3C004383 showed stage-specific expression during the ripening and growing stages, respectively, while MELO3C017100 was significantly more expressed during the growing stage than at other stages. No expression was observed during the climacteric and post-climacteric stages, suggesting this gene may be involved only in the growth and ripening of muskmelon fruits.
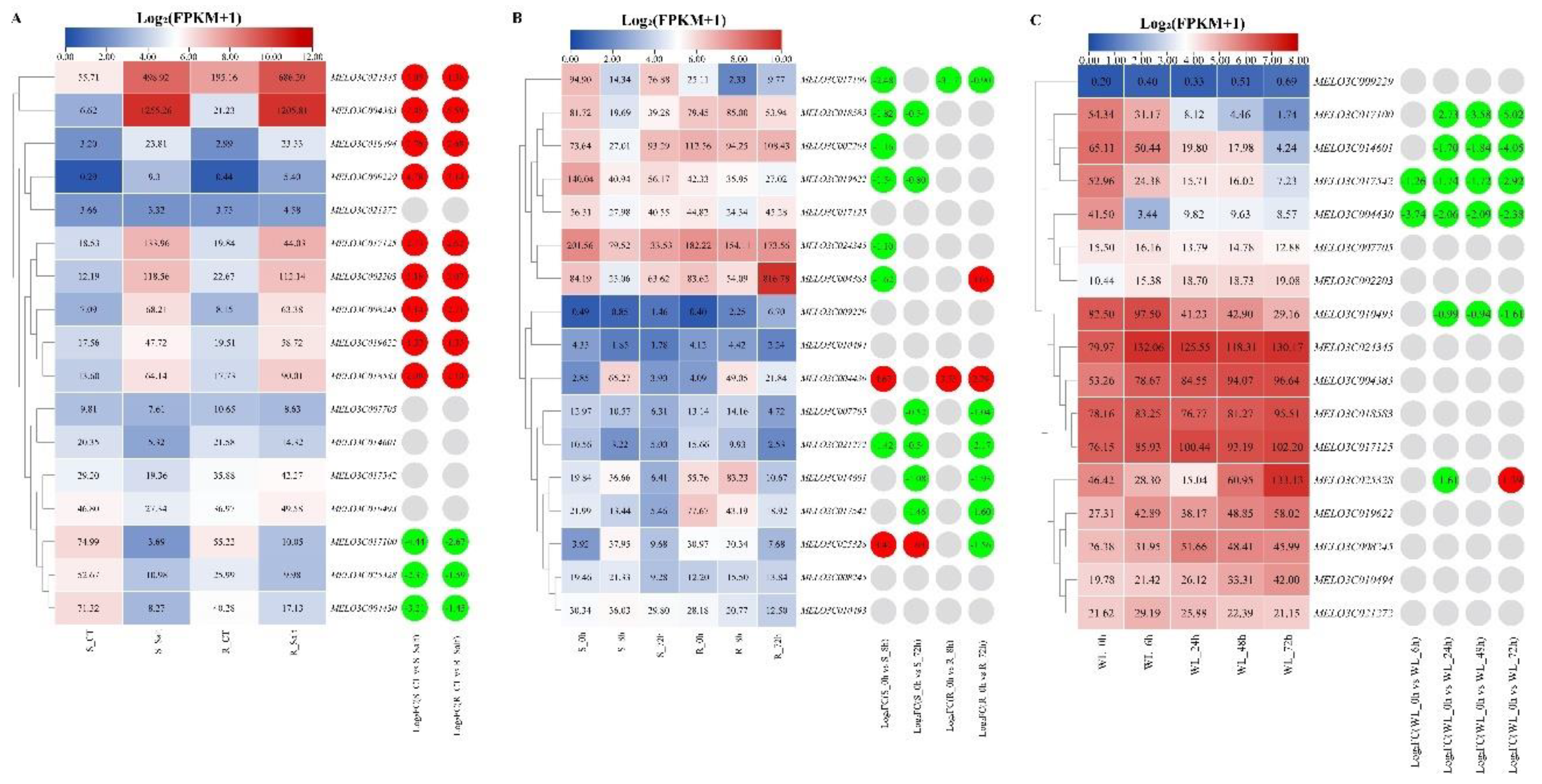
2.8. Expression Patterns Analysis of Melon ALDH Genes under Abiotic Stresses
To investigate the expression patterns of the ALDH gene in melon under varying abiotic stress conditions, we used published transcriptome sequencing data from melon studies conducted under salt, chilling, and waterlogged stress. This allowed us to analyze ALDH gene expression levels under these stresses (
Figure 7).
Under salt stress (
Figure 7A), only five ALDH genes –
MELO3C021272,
MELO3C007705,
MELO3C014601,
MELO3C017542, and
MELO3C010493 – were not differentially expressed compared to the control. Conversely,
MELO3C017100,
MELO3C025328, and
MELO3C004430 showed significant down-regulation in both salt-sensitive and salt-resistant materials. Other genes, particularly
MELO3C004383 and
MELO3C024345, were significantly up-regulated in these same materials.
In response to chillingstress (
Figure 7B), only
MELO3C004430 was significantly up-regulated in both chilling sensitive and resistant materials when compared with the control. MELO3C004383 showed a down-regulation in sensitive materials and an up-regulation in resistant materials, whereas
MELO3C025328 exhibited an opposed pattern – up-regulation in sensitive and down-regulation in resistant materials. Other genes –
MELO3C017100,
MELO3C007705,
MELO3C021272,
MELO3C014601, and
MELO3C017542 – were significantly down-regulated in both types of materials. However, down-regulated expressions of
MELO3C018583,
MELO3C002203,
MELO3C019622, and
MELO3C024345 were only observed in sensitive materials.
When under waterlogging stress (
Figure 7C),
MELO3C025328 was significantly down-regulated after 24 hours of treatment, but showed a significant up-regulation after 72 hours. Both
MELO3C017542 and
MELO3C004430 showed significant down-regulation in all four treatments.
MELO3C017100,
MELO3C014601, and
MELO3C010493 did not present differential expression at the 6-hour mark but demonstrated down-regulation at 24, 48, and 72 hours.
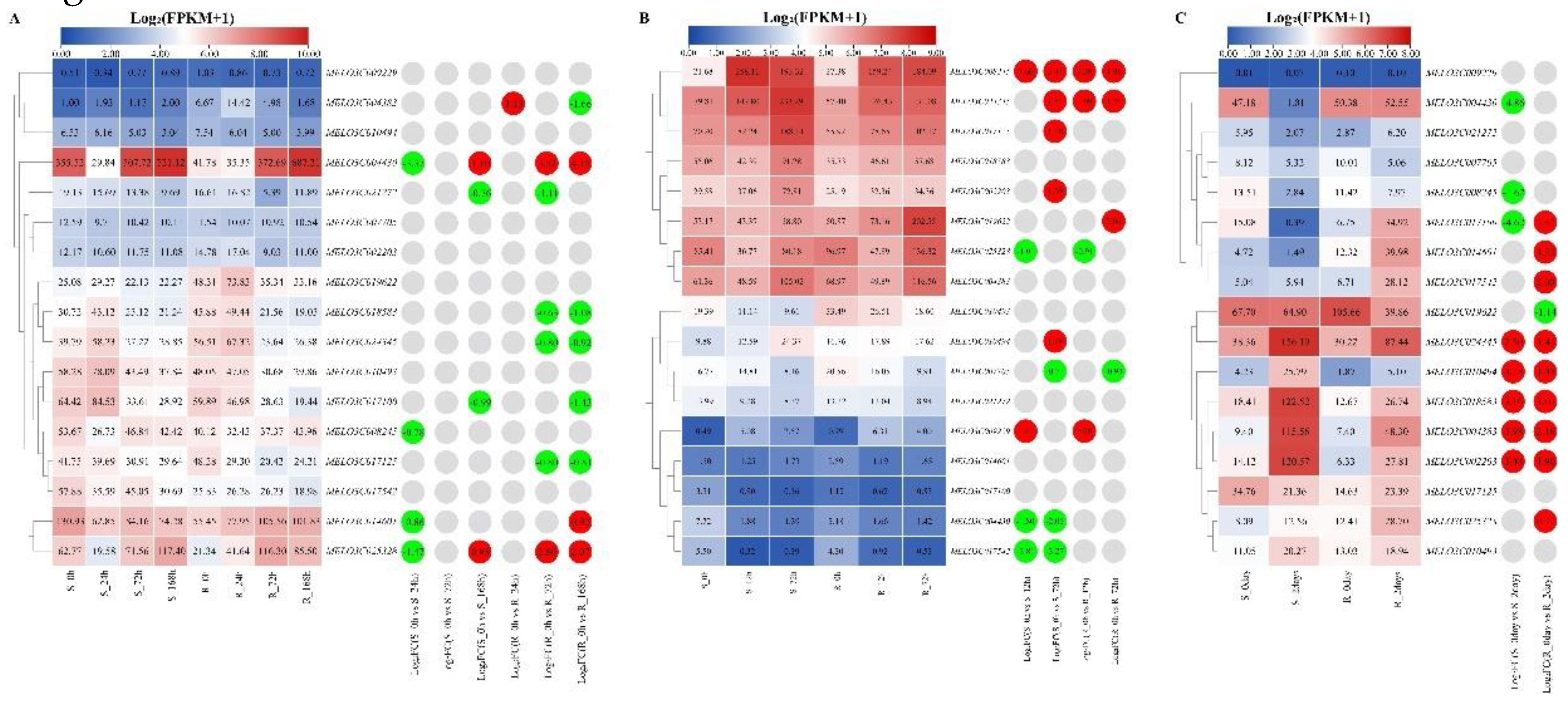
2.9. Expression Patterns Analysis of Melon ALDH Genes under Biotic Stresses
To comprehend the expression patterns of muskmelon under various biological stresses, we reanalyzed the published transcriptomic data of muskmelon under powdery mildew,
fusarium wilt, and gummy stem blight stress. This reanalysis was performed in conjunction with the melon genome data (DHL92 V4) using TBtools software to generate expression heat maps (
Figure 8).
During powdery mildew stress (
Figure 8A),
MELO3C004430 and
MELO3C025328 exhibited identical differential expression patterns. Their expression levels significantly decreased and were downregulated after 24 hours of treatment in the sensitive material, then increased significantly and were upregulated following 168 hours of treatment.
MELO3C014601 showed downregulation in sensitive materials but upregulation in resistant materials.
MELO3C004383 demonstrated a conflux of both upregulated and downregulated expressions in resistant materials. Both
MELO3C021272 and
MELO3C017100 were significantly downregulated in both sensitive and resistant materials, while
MELO3C008245 showed significant downregulation only in sensitive materials. Conversely,
MELO3C018583,
MELO3C024345, and
MELO3C017125 were significantly downregulated in resistant materials.
Under
fusarium wilt stress (
Figure 8B), compared with the control materials,
MELO3C008245,
MELO3C024345, and
MELO3C017125 showed the highest expression and significantly upregulated in both sensitive and resistant materials. Similarly,
MELO3C009229 was significantly upregulated in both sensitive and resistant materials. In contrast,
MELO3C025328 and
MELO3C007705 demonstrated significant downregulation in both types of materials.
MELO3C017125,
MELO3C002203, and
MELO3C010494 were significantly upregulated in sensitive materials, while
MELO3C004430 and
MELO3C017542 were significantly downregulated. Notably,
MELO3C019622 was only significantly upregulated in resistant materials.
Under gummy stem blight stress (
Figure 8C), five ALDH genes –
MELO3C024345,
MELO3C010494,
MELO3C018583,
MELO3C004383, and
MELO3C002203 – were significantly upregulated in both sensitive and resistant materials compared with control materials. Excluding
MELO3C010494, the expression levels of the other four ALDH genes showed significant changes in sensitive materials.
MELO3C017100 was downregulated in sensitive materials yet significantly upregulated in resistant ones. Conversely,
MELO3C004430 and
MELO3C008245 were significantly downregulated in sensitive materials.
MELO3C014601,
MELO3C017542, and
MELO3C025328 were significantly upregulated in resistant materials. Lastly,
MELO3C019622 demonstrated a significant downregulation in resistant materials.
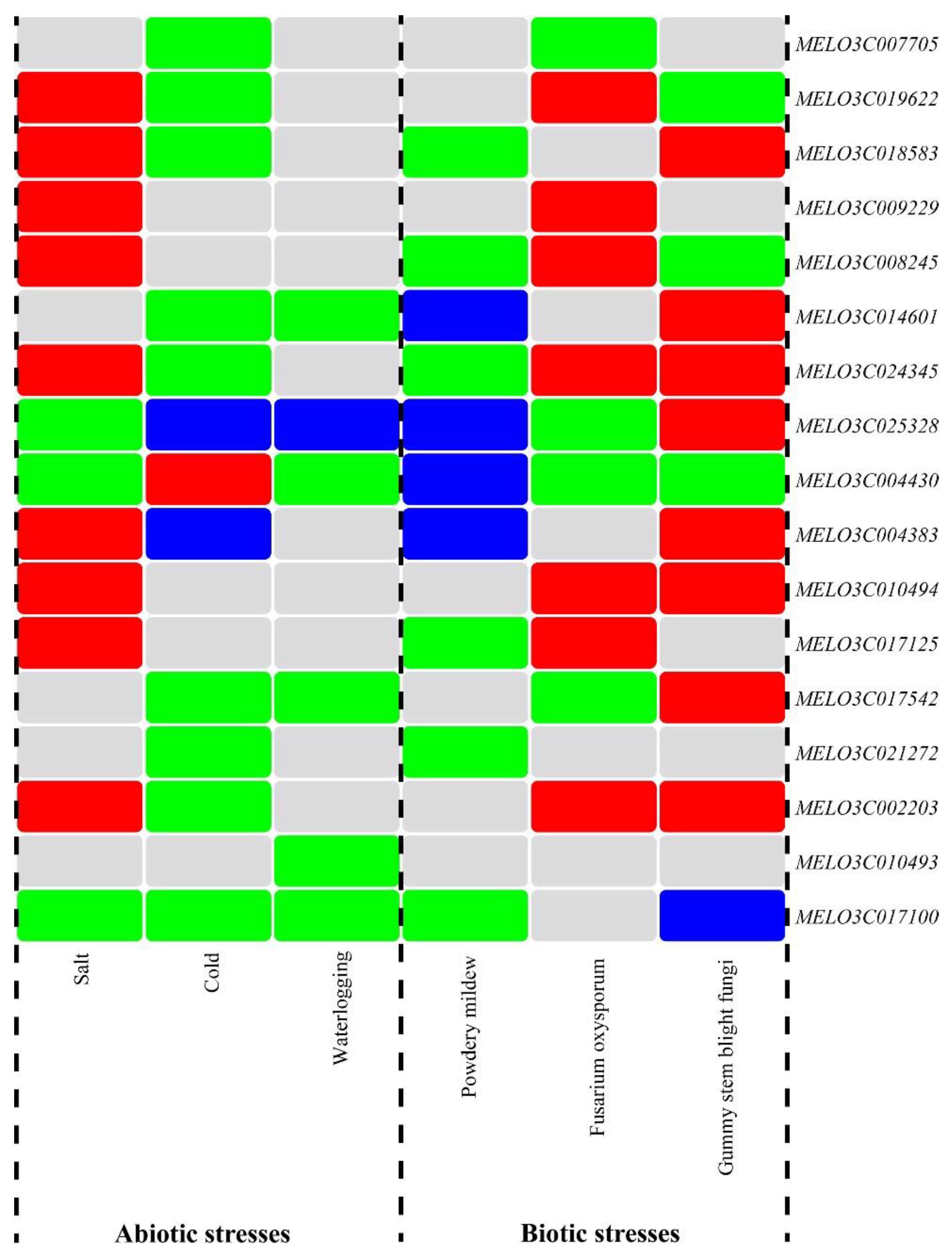
2.10. Regulation Patterns of Melon ALDH Genes under Stresses
The expression profiles of the melon ALDH gene family under various stresses were utilized to summarize and label the stress response of the ALDH gene family, and a heat map of ALDH genes under differing stresses was created using TBtools software (
Figure 9). As indicated in the figure, 17 ALDH genes in melon participate in the plant's stress response to varying extents. Notably, the ALDH genes
MELO3C025328 and
MELO3C004430 exhibited significant differential expression in both abiotic and biological stress scenarios. This indicates that these two genes are capable of initiating a positive response under stress conditions. Hence, they may serve as key candidate genes for future studies examining melon's stress responses. Interestingly, while
MELO3C017100 did not exhibit different expression under
fusarium wilt stress, it was significantly downregulated under all other five forms of stress. Some ALDH genes in melon, such as
MELO3C010493, only showed downregulation under waterlogging stress conditions. In the context of abiotic stress, most of the 17 ALDH genes were upregulated under salt stress, but downregulated under chilling and waterlogging stresses. Under biological stress, the majority of these ALDH genes were downregulated in response to powdery mildew stress but upregulated in response to
fusarium wilt and gummy stem blight stress.These findings highlight the differential expression of melon's ALDH gene family under abiotic and biological stresses, providing a favorable starting point for further research into the molecular biological functions of the ALDH gene family in melons.
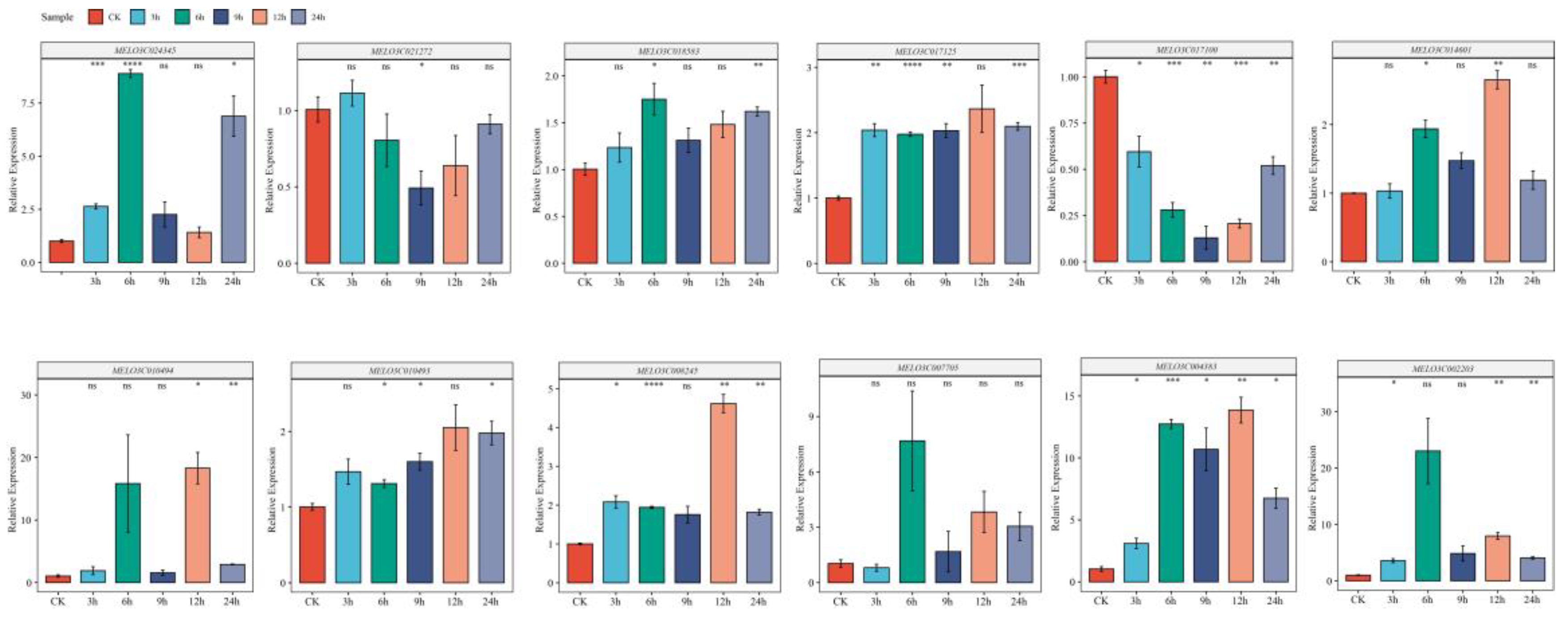
2.11. RT-qPCR Analysis of the Melon ALDH Gene Family
To evaluate the precision of our transcriptomic data analysis, we selected 10 genes from the CmALDH gene family. We assessed the impact of salt stress treatment (300mM) at varying time intervals (0h, 3h, 6h, 12h, and 24h) on their expression levels using the RT-qPCR method (
Figure 10). In this investigation, the transcriptomic data was salt stress treatment after 24 hours. According to our RT-qPCR test results, the expression patterns were generally consistent for the two sets of data we focused on. Such as
MELO3C024345,
MELO3C018583,
MELO3C007705, and
MELO3C002203, peaked in expression after 6 hours of NaCl treatment (with CK serving as the control), showing significant differences in comparison to the control. Conversely, other genes, including
MELO3C017125,
MELO3C014601,
MELO3C010494,
MELO3C010493,
MELO3C008245, and
MELO3C004383, exhibited their highest expression levels and significant differences at the 12-hour mark. Of these, the gene
MELO3C017100 displayed a substantial decrease in expression following stress treatment, reaching its lowest point after 9 hours, before demonstrating an increasing trend. The expression level of
MELO3C004383 showed a rising pattern post-stress, exhibiting significant differences compared to the control. This increase was particularly notable at the 12-hour point, after which it started to decrease.
3. Discussion
Research into plant responses to both abiotic and biological stresses has consistently attracted the attention of scientists. Stress has the potential to not only diminish the yield and quality of food crops [
59] but also, in severe instances, result in plant death, thereby incurring substantial losses in agricultural production [
60]. The ALDH enzyme, aside from its protective and detoxifying influence, also plays a crucial role in plant stress response [
61]. The most prominently studied gene within the ALDH gene family is the betaine aldehyde dehydrogenase gene. This gene, belonging to the ALDH10 family, is a well-established osmoregulator and has demonstrated an effective response to adversity. It becomes upregulated in response to both drought and salt stress, enhancing plant resilience to these stresses [
62]. As scientific and technological progress has allowed for the publication of an increasing amount of genome and transcriptome sequencing information, it's become particularly valuable to utilize this data to identify stress-related genes. This approach not only increases the usefulness of the existing data but also holds significant implications for related research [
63]. Melon, the second cucurbit crop to have its whole genome sequenced (as early as 2012) [
32], has yet to have any studies published on its ALDH gene family. This lack of information significantly hinders research into melon's biological functions linked to ALDH genes. Therefore, this study aims to fill this gap by conducting a whole genome identification and expression pattern analysis of the ALDH gene family in melons. The results will provide a theoretical framework for future ALDH gene family studies in melons and contribute to the identification of advantageous genes for resistance breeding in melons.
In this study, we identified 17 ALDH genes in the melon genome using the latest genomic information and advanced bioinformatics analysis methods. These findings differ from the number of ALDH genes identified in other species, which may be due to variations in the number of ALDH gene family members across different species and distinct stages of evolution [
64]. It could also be linked to the specific characteristics of the melon's chromosomes and genome [
65]. In our phylogenetic analysis, these 17 melon ALDH genes were classified into 10 subfamilies, each with a largely consistent gene structure and motif arrangement. Comparative analysis with the model plants
Arabidopsis and rice revealed that there are 7 pairs of directly homologous genes with
Arabidopsis, and only one pair with rice's ALDH gene family. This suggests potential similarities in biological functions, allowing for future melon studies to refer to the homologous genes in
Arabidopsis and rice. Our gene duplication analysis within the melon's ALDH gene family revealed a pair of tandem and fragment repeats, suggesting that the amplification of the melon's ALDH gene was solely due to these repeats, a finding that aligns with reports on gene families in other plants [
66].
Transcriptomics forms a critical component of genomics research. Bolstered by the ongoing advancements in high-throughput sequencing technology, transcriptome sequencing is becoming increasingly prevalent in plant-related studies. This has led to deepened insights into gene expression regulatory mechanisms and cellular functional diversity in plants [
67,
68]. Numerous transcriptome sequencing projects have used melons as their subject, with the resulting data undergoing validation via qRT-PCR and rigorous peer review before publication [
69,
70]. This process has led to an extensive database of melon transcriptomes. This study leverages this wealth of published transcriptome sequencing data to analyze the expression patterns of 17 ALDH genes in melons across different tissues, developmental stages, and stress conditions.
ALDH's role in plant organ development has been widely reported. For instance, the
ALDH2B2 (
Rf2a) gene is linked to cytoplasmic male sterility in maize and plays a key role in anther development [
71]. ALDH is also highly expressed in rice anthers during early meiosis to microspore [
72]. The
ALDH3F1 mutant triggers early flowering in plants, while its overexpression causes delayed flowering [
73]. In grapes, the expression levels of three genes (
ALDH2B8,
ALDH3H5, and
ALDH18B1) noticeably increase during fruit development and maturation, while the levels of
ALDH2B4 and
ALDH5F1 significantly decrease [
11]. In this study, we found that all 17 ALDH genes in melons were expressed to varying degrees in different melon tissues, with some genes exhibiting tissue-specific expression. This specific expression pattern across various melon tissues contributes to the synergistic regulation of melon growth and development. Only four ALDH genes were not expressed at different developmental stages of melon fruit. Notably, the
MELO3C017100 gene exhibited the highest expression level during the fruit growth stage and in male and female flowers and the ovary. However, its expression level dropped dramatically as the fruit matured, indicating that this gene primarily contributes to melon fruit growth.
The ALDH gene family's response to stress in plants has become a key focus of recent studies, suggesting that this gene family is crucial for plant stress resistance. In European sweet cherry, fluorescence quantitative experiments confirmed that
PaALDH17's expression under salt stress was significantly higher than that of other genes.
PaALDH17 was subsequently cloned and transferred to
Arabidopsis, where transgenic
PaALDH17 plants exhibited stronger stress resistance and fewer changes in various physiological indices under salt stress, compared to wild-type plants [
74]. In grapes,
VvALDH2B4 and
VvALDH2B8 expression levels significantly increased under drought and salt stress [
11]. Moreover, overexpressing
VvALDH2B4 in wild Chinese grapes led to a decrease in MDA levels, significantly improving resistance to pathogenic bacteria [
75]. In bamboo, ALDH gene expression significantly increased under drought stress compared to the control, with
PeALDH2B2 responding to drought stress by interacting with
PeGPB1[
76]. Wang et al. [
16] discovered that the wheat ALDH gene
TraeALDH7B1-
5A exhibited significant differences under both drought and salt stress. Its soybean homologous gene,
GmALDH7B1, was significantly upregulated in both roots and leaves under 20% PEG treatment. To understand the melon ALDH gene family's role in abiotic stress, this study analyzed the ALDH family genes' expression patterns under salt, chilling, and waterlogging stress using published melon transcriptome sequencing data. It was revealed that over half of the melon ALDH genes displayed significantly upregulated expression under salt stress, in line with previous reports on plant ALDH genes' upregulated expression under salt and drought stress [
20]. In contrast, most melon ALDH genes' expression was significantly downregulated under chilling and waterlogging stress. This suggests that these ALDH genes are negatively regulated when transcribed into mRNA under stress, leading to reduced expression [
77]. Interestingly, some melon ALDH genes were differentially expressed under all three abiotic stresses, while others were only differentially expressed under one stress. For instance,
MELO3C017125,
MELO3C009229,
MELO3C010494, and
MELO3C008245 were differentially expressed solely under salt stress, while their expression levels remained unchanged under chilling and waterlogged stress.
MELO3C021272 and
MELO3C007705 were differentially expressed only under chilling stress, and
MELO3C010493 was differentially expressed only under waterlogging stress. These results suggest that while some ALDH genes can be induced under various abiotic stresses, others are only induced by a specific abiotic stress. In addition to abiotic stress analysis, this study also examined the ALDH gene family's expression patterns in melons under biological stress. It was observed that the 17 melon ALDH genes were primarily downregulated under powdery mildew stress and primarily upregulated under
fusarium wilt stress, with a higher number of differentially expressed genes than under powdery mildew stress. These ALDH genes were positively responsive to these stresses and played a role in plant disease resistance [
78]. Among these,
MELO3C021272 was significantly downregulated under powdery mildew stress but not expressed under
fusarium wilt stress. Similarly,
MELO3C007705 and
MELO3C009229 were differentially expressed under
fusarium wilt stress. However,
MELO3C010493 was not differentially expressed under any of these three biological stresses, suggesting that this gene did not contribute to melon's response to these biological stresses. The functional characteristics of melon ALDH genes showed that 17 ALDH genes were differentially expressed under both abiotic and biological stresses, with
MELO3C024328 and
MELO3C004430 being differentially expressed under all stresses (salt, chilling, waterlogging, powdery mildew,
fusarium wilt, and gummy stem blight). Interestingly,
MELO3C024328 was upregulated and downregulated in chilling, waterlogged, and powdery mildew stress, suggesting that these two melon ALDH genes play a role in plants facing these stresses.
Figure 1.
Distribution of the melon ALDH family genes on the chromosomes. The genes marked in red color were tandem duplication gene pairs, and the genes marked in green color were segmental duplication gene.
Figure 1.
Distribution of the melon ALDH family genes on the chromosomes. The genes marked in red color were tandem duplication gene pairs, and the genes marked in green color were segmental duplication gene.
Figure 2.
Phylogenetic relationships of melon, rice, and Arabidopsis ALDH proteins.
Figure 2.
Phylogenetic relationships of melon, rice, and Arabidopsis ALDH proteins.
Figure 3.
Gene structures and protein motifs of the melon ALDH gene family.
Figure 3.
Gene structures and protein motifs of the melon ALDH gene family.
Figure 4.
Assessment of the ALDH gene duplication between melon, Arabidopsis, and rice. The green lines represented the segmentally duplicated ALDH genes in melon. The red lines represented the orthologous relationships of the ALDH genes between melon and Arabidopsis. The blue lines represent the orthologous relationships of the ALDH genes between melon and rice.
Figure 4.
Assessment of the ALDH gene duplication between melon, Arabidopsis, and rice. The green lines represented the segmentally duplicated ALDH genes in melon. The red lines represented the orthologous relationships of the ALDH genes between melon and Arabidopsis. The blue lines represent the orthologous relationships of the ALDH genes between melon and rice.
Figure 5.
Cis-regulatory elements in promoter region of melon ALDH genes (A) and their proportions (B).
Figure 5.
Cis-regulatory elements in promoter region of melon ALDH genes (A) and their proportions (B).
Figure 6.
Expression analysis of melon ALDH genes in different tissue and developmental stages.
Figure 6.
Expression analysis of melon ALDH genes in different tissue and developmental stages.
Figure 7.
The expression patterns of melon ALDH genes in response to abiotic stresses. (A)The expression heatmap of melon ALDH genes in response to salt stress. S: susceptible plant; R: resistant plant; CT: control treatment; Salt: salt treatment. (B)The expression heatmap of melon ALDH genes in response to chilling stress. S: susceptible plant; R: resistant plant; 0h, 8h and 72h were treatment for 0h, 8h and 72 hours. (C)The expression heatmap of melon ALDH genes in response to waterlogging stress. 0h, 6h, 24h, 48h and 72h were treatment for 0h, 6h, 24h, 48h and 72h hours. The data in the left expression heatmaps were the original FPKM values; the data in the right boxes were log2 (fold change) values highlighted by red (upregulation) and green (downregulation) colors.
Figure 7.
The expression patterns of melon ALDH genes in response to abiotic stresses. (A)The expression heatmap of melon ALDH genes in response to salt stress. S: susceptible plant; R: resistant plant; CT: control treatment; Salt: salt treatment. (B)The expression heatmap of melon ALDH genes in response to chilling stress. S: susceptible plant; R: resistant plant; 0h, 8h and 72h were treatment for 0h, 8h and 72 hours. (C)The expression heatmap of melon ALDH genes in response to waterlogging stress. 0h, 6h, 24h, 48h and 72h were treatment for 0h, 6h, 24h, 48h and 72h hours. The data in the left expression heatmaps were the original FPKM values; the data in the right boxes were log2 (fold change) values highlighted by red (upregulation) and green (downregulation) colors.
Figure 8.
The expression patterns of melon ALDH genes in response to biotic stresses. (A)The expression heatmap of melon ALDH genes in response to downy mildew stress. S: susceptible plant; R: resistant plant; 0h, 24h, 72h and 168h were 0, 24, 72, and 168 hours post-inoculation, respectively. (B)The expression heatmap of melon ALDH genes in response to Fusarium will stress. S: susceptible plant; R: resistant plant; 0h, 12h, and 72h were 0, 12, and 72 hours post-inoculation, respectively. (C)The expression heatmap of melon ALDH genes in response to gummy stem blight stress. S: susceptible plant; R: resistant plant; 0 day and 2days were 0, and 2 days post-inoculation, respectively. The data in the left expression heatmaps were the original FPKM values; the data in the right boxes were log2 (fold change) values highlighted by red (upregulation) and green (downregulation) colors.
Figure 8.
The expression patterns of melon ALDH genes in response to biotic stresses. (A)The expression heatmap of melon ALDH genes in response to downy mildew stress. S: susceptible plant; R: resistant plant; 0h, 24h, 72h and 168h were 0, 24, 72, and 168 hours post-inoculation, respectively. (B)The expression heatmap of melon ALDH genes in response to Fusarium will stress. S: susceptible plant; R: resistant plant; 0h, 12h, and 72h were 0, 12, and 72 hours post-inoculation, respectively. (C)The expression heatmap of melon ALDH genes in response to gummy stem blight stress. S: susceptible plant; R: resistant plant; 0 day and 2days were 0, and 2 days post-inoculation, respectively. The data in the left expression heatmaps were the original FPKM values; the data in the right boxes were log2 (fold change) values highlighted by red (upregulation) and green (downregulation) colors.
Figure 9.
An expression pattern heatmap of the melon ALDH genes under abiotic and biotic stresses. The gray color represented unchanged expression, red represented upregulated expression, green represented downregulated expression, and blue represented both upregulated and downregulated expression.
Figure 9.
An expression pattern heatmap of the melon ALDH genes under abiotic and biotic stresses. The gray color represented unchanged expression, red represented upregulated expression, green represented downregulated expression, and blue represented both upregulated and downregulated expression.
Figure 10.
Relative expression level of 10 CmALDH genes in response to salt treatments. Error bars are standard deviations of three biological replicates. Asterisks were used to indicate the significant degree of the expression level compared to the value of the control (*P < 0.05, **P < 0.01, ***P < 0.001).
Figure 10.
Relative expression level of 10 CmALDH genes in response to salt treatments. Error bars are standard deviations of three biological replicates. Asterisks were used to indicate the significant degree of the expression level compared to the value of the control (*P < 0.05, **P < 0.01, ***P < 0.001).