Submitted:
03 October 2024
Posted:
04 October 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. Genomic DNA Extraction, PCR Amplifications and Sequencing
2.3. Sequencing Raw Data Processing and Data Analysis
2.4. Identification of Discriminatory Bacterial Fingerprints According to Host Species
3. Results
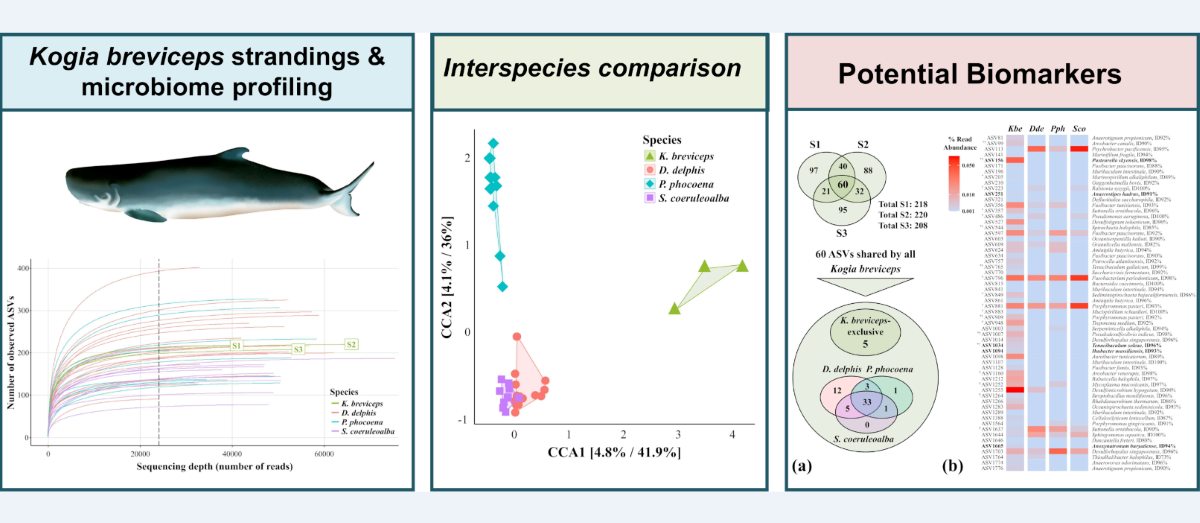
3.1. Structure of the Oral Community of Three Kogia Breviceps Specimens
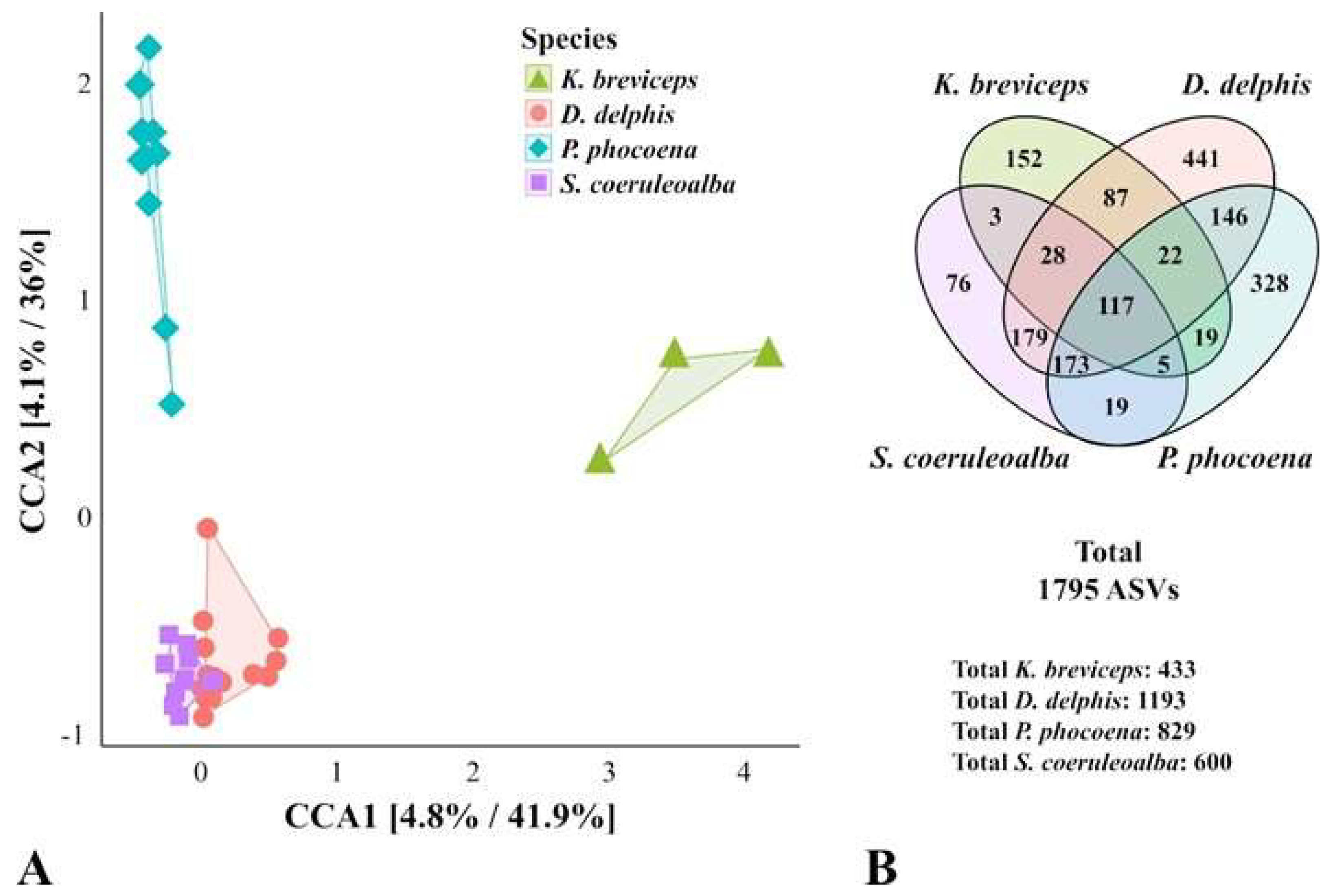
3.2. The Variable Composition of the Oral Microbiota Allows Discrimination of the Analyzed Cetacean Species
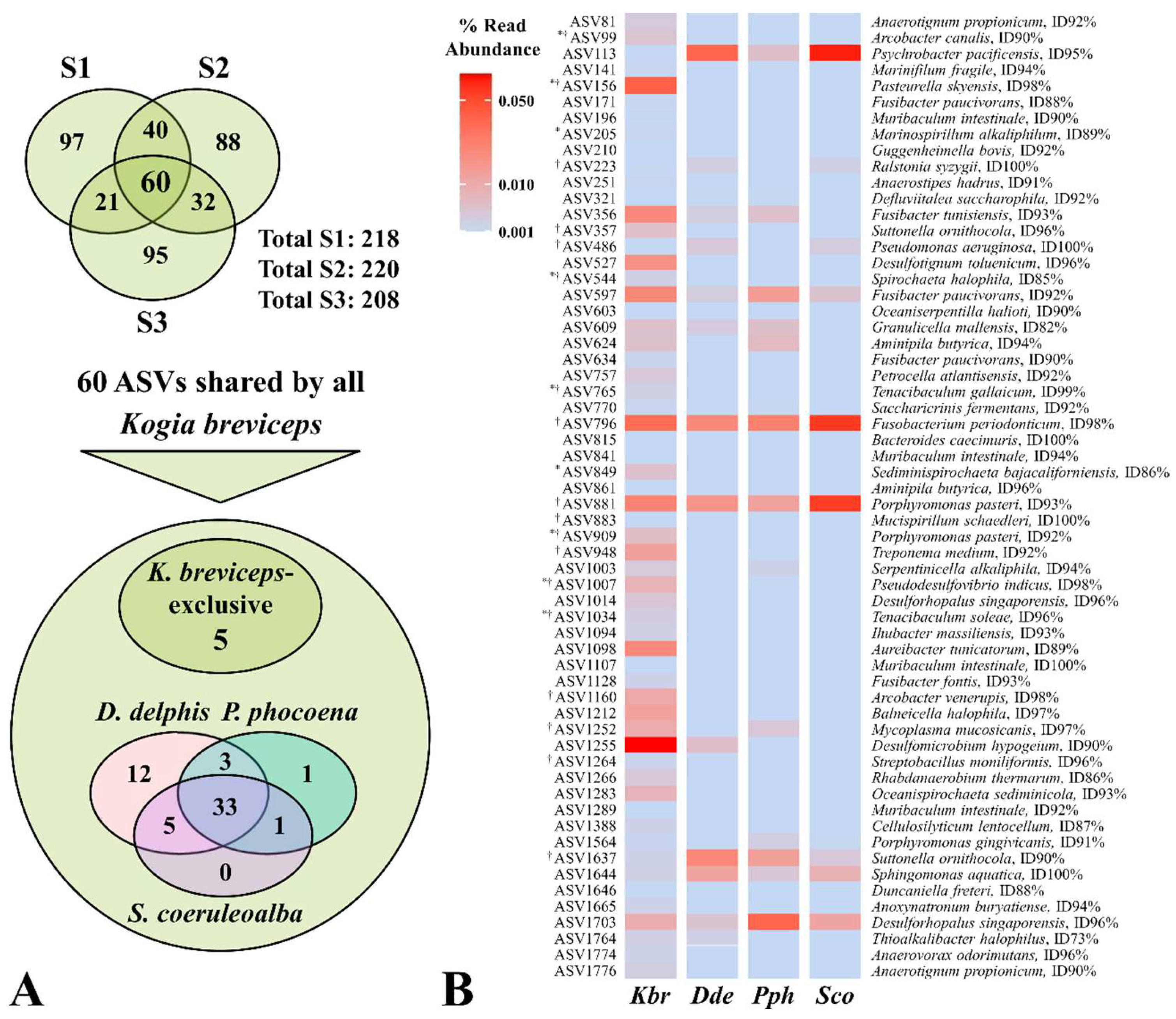
3.3. Potential Microbial Signatures within the Oral Cavity of Kogia breviceps
3.4. Hints of Disease-Association from Potential Pathogenic Features
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fleming, L.E.; Broad, K.; Clement, A.; Dewailly, E.; Elmir, S.; Knap, A.; Pomponi, S.A.; Smith, S.; Solo Gabriele, H.; Walsh, P. Oceans and Human Health: Emerging Public Health Risks in the Marine Environment. Mar Pollut Bull 2006, 53, 545–560. [CrossRef]
- Halpern, B.S.; Walbridge, S.; Selkoe, K.A.; Kappel, C. V.; Micheli, F.; D’Agrosa, C.; Bruno, J.F.; Casey, K.S.; Ebert, C.; Fox, H.E.; et al. A Global Map of Human Impact on Marine Ecosystems. Science (1979) 2008, 319, 948–952. [CrossRef]
- Bossart, G.D. Marine Mammals as Sentinel Species for Oceans and Human Health. Vet Pathol 2011, 48, 676–690. [CrossRef]
- Ferreira, M.; Monteiro, S.S.; Torres, J.; Oliveira, I.; Sequeira, M.; López, A.; Vingada, J.; Eira, C. Biological Variables and Health Status Affecting Inorganic Element Concentrations in Harbour Porpoises (Phocoena Phocoena) from Portugal (Western Iberian Peninsula). Environmental Pollution 2016, 210, 293–302. [CrossRef]
- Monteiro, S.S.; Torres, J.; Ferreira, M.; Marçalo, A.; Nicolau, L.; Vingada, J. V.; Eira, C. Ecological Variables Influencing Trace Element Concentrations in Bottlenose Dolphins (Tursiops Truncatus, Montagu 1821) Stranded in Continental Portugal. Science of The Total Environment 2016, 544, 837–844. [CrossRef]
- Vingada, J.; Eira, C. Conservation of Cetaceans and Seabirds in Continental Portugal. The LIFE+ MarPro Project. Final Project Report NAT/PT/00038; 2018;
- Bento, M.C.; Canha, R.; Eira, C.; Vingada, J.; Nicolau, L.; Ferreira, M.; Domingo, M.; Tavares, L.; Duarte, A. Herpesvirus Infection in Marine Mammals: A Retrospective Molecular Survey of Stranded Cetaceans in the Portuguese Coastline. Infection, Genetics and Evolution 2019, 67, 222–233. [CrossRef]
- Bento, M.C.R. de M.; Eira, C.I.C.S.; Vingada, J.V.; Marçalo, A.L.; Ferreira, M.C.T.; Fernandez, A.L.; Tavares, L.M.M.; Duarte, A.I.S.P. New Insight into Dolphin Morbillivirus Phylogeny and Epidemiology in the Northeast Atlantic: Opportunistic Study in Cetaceans Stranded along the Portuguese and Galician Coasts. BMC Vet Res 2016, 12, 1–12. [CrossRef]
- Soares-Castro, P.; Araújo-Rodrigues, H.; Godoy-Vitorino, F.; Ferreira, M.; Covelo, P.; López, A.; Vingada, J.; Eira, C.; Santos, P.M. Microbiota Fingerprints within the Oral Cavity of Cetaceans as Indicators for Population Biomonitoring. Scientific Reports 2019 9:1 2019, 9, 1–15. [CrossRef]
- Godoy-Vitorino, F.; Rodriguez-Hilario, A.; Alves, A.L.; Gonçalves, F.; Cabrera-Colon, B.; Mesquita, C.S.; Soares-Castro, P.; Ferreira, M.; Marçalo, A.; Vingada, J.; et al. The Microbiome of a Striped Dolphin (Stenella Coeruleoalba) Stranded in Portugal. Res Microbiol 2017, 168, 85–93. [CrossRef]
- Chivers, S.J.; Leduc, R.G.; Robertson, K.M.; Barros, N.B.; Dizon, A.E. Genetic Variation of Kogia Spp. with Preliminary Evidence for Two Species of Kogia Sima. Mar Mamm Sci 2005, 21, 619–634. [CrossRef]
- Hodge, L.E.W.; Baumann-Pickering, S.; Hildebrand, J.A.; Bell, J.T.; Cummings, E.W.; Foley, H.J.; McAlarney, R.J.; McLellan, W.A.; Pabst, D.A.; Swaim, Z.T.; et al. Heard but Not Seen: Occurrence of Kogia Spp. along the Western North Atlantic Shelf Break. Mar Mamm Sci 2018, 34, 1141–1153. [CrossRef]
- Moura, J.F.; Acevedo-Trejos, E.; Tavares, D.C.; Meirelles, A.C.O.; Silva, C.P.N.; Oliveira, L.R.; Santos, R.A.; Wickert, J.C.; Machado, R.; Siciliano, S.; et al. Stranding Events of Kogia Whales along the Brazilian Coast. PLoS One 2016, 11, e0146108. [CrossRef]
- Ferreira, M.; Eira, C.; López, A.; Sequeira, M. Kogia Breviceps Cachalote-Pigmeu. Livro Vermelho Dos Mamíferos de Portugal Continental. In; Mathias, M.D.L., Fonseca, C., Rodrigues, L., Grilo, C., Lopes-Fernandes, M., M. Palmeirim, J., Santos-Reis, M., Alves, P.C., Cabral, J.A., Ferreira, M., Mira, A., Eira, C., Negrões, N., Paupério, J., Pita, R., Rainho, A., Rosalino, L.M., Tapisso, J.T., Vingada, J., Eds.; FCiências.ID / ICNF, 2023; pp. 1–371 ISBN 978-989-53724-1-6.
- Sehnal, L.; Brammer-Robbins, E.; Wormington, A.M.; Blaha, L.; Bisesi, J.; Larkin, I.; Martyniuk, C.J.; Simonin, M.; Adamovsky, O. Microbiome Composition and Function in Aquatic Vertebrates: Small Organisms Making Big Impacts on Aquatic Animal Health. Front Microbiol 2021, 12, 567408. [CrossRef]
- Waltzek, T.B.; Cortés-Hinojosa, G.; Wellehan, J.F.X.; Gray, G.C. Marine Mammal Zoonoses: A Review of Disease Manifestations. Zoonoses Public Health 2012, 59, 521–535. [CrossRef]
- Li, C.; Tan, X.; Bai, J.; Xu, Q.; Liu, S.; Guo, W.; Yu, C.; Fan, G.; Lu, Y.; Zhang, H.; et al. A Survey of the Sperm Whale (Physeter Catodon) Commensal Microbiome. PeerJ 2019, 2019, e7257. [CrossRef]
- Marón, C.F.; Kohl, K.D.; Chirife, A.; Di Martino, M.; Fons, M.P.; Navarro, M.A.; Beingesser, J.; McAloose, D.; Uzal, F.A.; Dearing, M.D.; et al. Symbiotic Microbes and Potential Pathogens in the Intestine of Dead Southern Right Whale (Eubalaena Australis) Calves. Anaerobe 2019, 57, 107–114. [CrossRef]
- Terracciano, G.; Fichi, G.; Comentale, A.; Ricci, E.; Mancusi, C.; Perrucci, S. Dolphins Stranded along the Tuscan Coastline (Central Italy) of the “Pelagos Sanctuary”: A Parasitological Investigation. Pathogens 2020, Vol. 9, Page 612 2020, 9, 612. [CrossRef]
- Marangi, M.; Airoldi, S.; Beneduce, L.; Zaccone, C. Wild Whale Faecal Samples as a Proxy of Anthropogenic Impact. Scientific Reports 2021 11:1 2021, 11, 1–11. [CrossRef]
- Erwin, P.M.; Rhodes, R.G.; Kiser, K.B.; Keenan-Bateman, T.F.; McLellan, W.A.; Pabst, D.A. High Diversity and Unique Composition of Gut Microbiomes in Pygmy (Kogia breviceps) and Dwarf (K. sima) Sperm Whales. Scientific Reports 2017 7:1 2017, 7, 1–11. [CrossRef]
- Denison, E.R.; Rhodes, R.G.; McLellan, W.A.; Pabst, D.A.; Erwin, P.M. Host Phylogeny and Life History Stage Shape the Gut Microbiome in Dwarf (Kogia Sima) and Pygmy (Kogia Breviceps) Sperm Whales. Scientific Reports 2020 10:1 2020, 10, 1–13. [CrossRef]
- Kuiken, T.; García-Hartmann, M. Dissection Techniques and Tissue Sampling. Proceedings of the Proceedings of the first ECS workshop on Cetacean pathology, 1991.
- Pugliares, K.R.; Bogomolni, A.L.; Touhey, K.M.; Herzig, S.M.; Harry, C.T.; Moore, M.J. Marine Mammal Necropsy : An Introductory Guide for Stranding Responders and Field Biologists. 2007. [CrossRef]
- Joshi, N.; Fass, J. Sickle: A Windowed Adaptive Trimming for Fastq Files Using Quality Available online: https://github.com/najoshi/sickle (accessed on 13 May 2024).
- Nikolenko, S.I.; Korobeynikov, A.I.; Alekseyev, M.A. BayesHammer: Bayesian Clustering for Error Correction in Single-Cell Sequencing. BMC Genomics 2013, 14, 1–11. [CrossRef]
- Rognes, T.; Flouri, T.; Nichols, B.; Quince, C.; Mahé, F. VSEARCH: A Versatile Open Source Tool for Metagenomics. PeerJ 2016, 2016, e2584. [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, Interactive, Scalable and Extensible Microbiome Data Science Using QIIME 2. Nature Biotechnology 2019 37:8 2019, 37, 852–857. [CrossRef]
- Amir, A.; McDonald, D.; Navas-Molina, J.A.; Kopylova, E.; Morton, J.T.; Xu, Z.Z.; Kightley, E.P.; Thompson, L.R.; Hyde, E.R.; Gonzalez, A.; et al. Deblur Rapidly Resolves Single-Nucleotide Community Sequence Patterns. mSystems 2017, 2. [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA Ribosomal RNA Gene Database Project: Improved Data Processing and Web-Based Tools. Nucleic Acids Res 2013, 41, D590–D596. [CrossRef]
- Robeson, M.S.; O’Rourke, D.R.; Kaehler, B.D.; Ziemski, M.; Dillon, M.R.; Foster, J.T.; Bokulich, N.A. RESCRIPt: Reproducible Sequence Taxonomy Reference Database Management. PLoS Comput Biol 2021, 17, e1009581. [CrossRef]
- Chao, A.; Jost, L. Coverage-Based Rarefaction and Extrapolation: Standardizing Samples by Completeness Rather than Size. Ecology 2012, 93, 2533–2547. [CrossRef]
- .
- Maechler, M.; Rousseeuw, P.; Struyf, A.; Hubert, M.; Hornik, K. Cluster: Cluster Analysis Basics and Extensions. 2023.
- Andersen, K.S.; Kirkegaard, R.H.; Karst, S.M.; Albertsen, M. Ampvis2: An R Package to Analyse and Visualise 16S RRNA Amplicon Data. bioRxiv 2018, 299537. [CrossRef]
- Dixon, P. VEGAN, a Package of R Functions for Community Ecology. Journal of Vegetation Science 2003, 14, 927–930. [CrossRef]
- Dhariwal, A.; Chong, J.; Habib, S.; King, I.L.; Agellon, L.B.; Xia, J. MicrobiomeAnalyst: A Web-Based Tool for Comprehensive Statistical, Visual and Meta-Analysis of Microbiome Data. Nucleic Acids Res 2017, 45, W180–W188. [CrossRef]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic Biomarker Discovery and Explanation. Genome Biol 2011, 12, 1–18. [CrossRef]
- Amos, B.; Aurrecoechea, C.; Barba, M.; Barreto, A.; Basenko, E.Y.; Bażant, W.; Belnap, R.; Blevins, A.S.; Böhme, U.; Brestelli, J.; et al. VEuPathDB: The Eukaryotic Pathogen, Vector and Host Bioinformatics Resource Center. Nucleic Acids Res 2022, 50, D898–D911. [CrossRef]
- Venn-Watson, S.; Smith, C.R.; Jensen, E.D. Primary Bacterial Pathogens in Bottlenose Dolphins Tursiops Truncatus: Needles in Haystacks of Commensal and Environmental Microbes. Dis Aquat Organ 2008, 79, 87–93. [CrossRef]
- Martins, P.; Cleary, D.F.R.; Pires, A.C.C.; Rodrigues, A.M.; Quintino, V.; Calado, R.; Gomes, N.C.M. Molecular Analysis of Bacterial Communities and Detection of Potential Pathogens in a Recirculating Aquaculture System for Scophthalmus Maximus and Solea Senegalensis. PLoS One 2013, 8, e80847. [CrossRef]
- Apprill, A.; Miller, C.A.; Moore, M.J.; Durban, J.W.; Fearnbach, H.; Barrett-Lennard, L.G. Extensive Core Microbiome in Drone-Captured Whale Blow Supports a Framework for Health Monitoring. mSystems 2017, 2. [CrossRef]
- Raverty, S.A.; Rhodes, L.D.; Zabek, E.; Eshghi, A.; Cameron, C.E.; Hanson, M.B.; Schroeder, J.P. Respiratory Microbiome of Endangered Southern Resident Killer Whales and Microbiota of Surrounding Sea Surface Microlayer in the Eastern North Pacific. Scientific Reports 2017 7:1 2017, 7, 1–12. [CrossRef]
- Jurelevicius, D.; Cotta, S.R.; Montezzi, L.F.; Dias, A.C.F.; Mason, O.U.; Picão, R.C.; Jansson, J.K.; Seldin, L. Enrichment of Potential Pathogens in Marine Microbiomes with Different Degrees of Anthropogenic Activity. Environmental Pollution 2021, 268, 115757. [CrossRef]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and Applications. BMC Bioinformatics 2009, 10, 1–9. [CrossRef]
- Frantzis, A.; Herzing, D.L. Mixed-Species Associations of Striped Dolphins (Stenella Coeruleoalba), Short-Beaked Common Dolphins (Delphinus Delphis), and Risso’s Dolphins (Grampus Griseus) in the Gulf of Corinth (Greece, Mediterranean Sea). Aquat Mamm 2002, 28, 188–197.
- Quérouil, S.; Silva, M.A.; Cascão, I.; Magalhães, S.; Seabra, M.I.; Machete, M.A.; Santos, R.S. Why Do Dolphins Form Mixed-Species Associations in the Azores? Ethology 2008, 114, 1183–1194. [CrossRef]
- McGowen, M.R.; Spaulding, M.; Gatesy, J. Divergence Date Estimation and a Comprehensive Molecular Tree of Extant Cetaceans. Mol Phylogenet Evol 2009, 53, 891–906. [CrossRef]
- Apprill, A.; Robbins, J.; Eren, A.M.; Pack, A.A.; Reveillaud, J.; Mattila, D.; Moore, M.; Niemeyer, M.; Moore, K.M.T.; Mincer, T.J. Humpback Whale Populations Share a Core Skin Bacterial Community: Towards a Health Index for Marine Mammals? PLoS One 2014, 9, e90785. [CrossRef]
- Chiarello, M.; Villéger, S.; Bouvier, C.; Auguet, J.C.; Bouvier, T. Captive Bottlenose Dolphins and Killer Whales Harbor a Species-Specific Skin Microbiota That Varies among Individuals. Scientific Reports 2017 7:1 2017, 7, 1–12. [CrossRef]
- Robles-Malagamba, M.J.; Walsh, M.T.; Ahasan, M.S.; Thompson, P.; Wells, R.S.; Jobin, C.; Fodor, A.A.; Winglee, K.; Waltzek, T.B. Characterization of the Bacterial Microbiome among Free-Ranging Bottlenose Dolphins (Tursiops Truncatus). Heliyon 2020, 6, e03944. [CrossRef]
- Toro-Valdivieso, C.; Toro, F.; Stubbs, S.; Castro-Nallar, E.; Blacklaws, B. Patterns of the Fecal Microbiota in the Juan Fernández Fur Seal (Arctocephalus Philippii). Microbiologyopen 2021, 10, e1215. [CrossRef]
- Glaeser, S.P.; Silva, L.M.R.; Prieto, R.; Silva, M.A.; Franco, A.; Kämpfer, P.; Hermosilla, C.; Taubert, A.; Eisenberg, T. A Preliminary Comparison on Faecal Microbiomes of Free-Ranging Large Baleen (Balaenoptera Musculus, B. Physalus, B. Borealis) and Toothed (Physeter Macrocephalus) Whales. Microb Ecol 2022, 83, 18–33. [CrossRef]
- Webb, K.; Zain, N.M.M.; Stewart, I.; Fogarty, A.; Nash, E.F.; Whitehouse, J.L.; Smyth, A.R.; Lilley, A.K.; Knox, A.; Williams, P.; et al. Porphyromonas Pasteri and Prevotella Nanceiensis in the Sputum Microbiota Are Associated with Increased Decline in Lung Function in Individuals with Cystic Fibrosis. J Med Microbiol 2022, 71, 001481. [CrossRef]
- Forster, S.C.; Clare, S.; Beresford-Jones, B.S.; Harcourt, K.; Notley, G.; Stares, M.D.; Kumar, N.; Soderholm, A.T.; Adoum, A.; Wong, H.; et al. Identification of Gut Microbial Species Linked with Disease Variability in a Widely Used Mouse Model of Colitis. Nature Microbiology 2022 7:4 2022, 7, 590–599. [CrossRef]
- López, J.R.; Piñeiro-Vidal, M.; García-Lamas, N.; de la Herran, R.; Navas, J.I.; Hachero-Cruzado, I.; Santos, Y. First Isolation of Tenacibaculum Soleae from Diseased Cultured Wedge Sole, Dicologoglossa Cuneata (Moreau), and Brill, Scophthalmus Rhombus (L.). J Fish Dis 2010, 33, 273–278. [CrossRef]
- Sabdono, A.; Radjasa, O.K. Anti-Bacterial Property of a Coral-Associated Bacterium Bacillus Sp. against Coral Pathogenic Bbd (Black Band Disease). J. Coastal Dev 2006, 9, 175–182.
- Gu, H.J.; Sun, Q.L.; Luo, J.C.; Zhang, J.; Sun, L. A First Study of the Virulence Potential of a Bacillus Subtilis Isolate From Deep-Sea Hydrothermal Vent. Front Cell Infect Microbiol 2019, 9. [CrossRef]
- Birkbeck, T.H.; Laidler, L.A.; Grant, A.N.; Cox, D.I. Pasteurella Skyensis Sp. Nov., Isolated from Atlantic Salmon (Salmo Salar L.). Int J Syst Evol Microbiol 2002, 52, 699–704. [CrossRef]
- Allen-Vercoe, E.; Daigneault, M.; White, A.; Panaccione, R.; Duncan, S.H.; Flint, H.J.; O’Neal, L.; Lawson, P.A. Anaerostipes Hadrus Comb. Nov., a Dominant Species within the Human Colonic Microbiota; Reclassification of Eubacterium Hadrum Moore et al. 1976. Anaerobe 2012, 18, 523–529. [CrossRef]
- Ndongo, S.; Lagier, J.C.; Fournier, P.E.; Raoult, D.; Khelaifia, S. “Ihubacter Massiliensis”: A New Bacterium Isolated from the Human Gut. New Microbes New Infect 2016, 13, 104–105. [CrossRef]
- Collado, L.; Inza, I.; Guarro, J.; Figueras, M.J. Presence of Arcobacter Spp. in Environmental Waters Correlates with High Levels of Fecal Pollution. Environ Microbiol 2008, 10, 1635–1640. [CrossRef]
- Pérez-Cataluña, A.; Salas-Massó, N.; Figueras, M.J. Arcobacter Canalis Sp. Nov., Isolated from a Water Canal Contaminated with Urban Sewage. Int J Syst Evol Microbiol 2018, 68, 1258–1264. [CrossRef]
- Lee, C.; Agidi, S.; Marion, J.W.; Lee, J. Arcobacter in Lake Erie Beach Waters: An Emerging Gastrointestinal Pathogen Linked with Human-Associated Fecal Contamination. Appl Environ Microbiol 2012, 78, 5511–5519. [CrossRef]
- Stoddard, R.A.; Atwill, E.R.; Gulland, F.M.D.; Miller, M.A.; Dabritz, H.A.; Paradies, D.M.; Worcester, K.R.; Jang, S.; Lawrence, J.; Byrne, B.A.; et al. Risk Factors for Infection with Pathogenic and Antimicrobial-Resistant Fecal Bacteria in Northern Elephant Seals in California. Public Health Rep 2008, 123, 360–370. [CrossRef]
- Fera, M.T.; Maugeri, T.L.; Gugliandolo, C.; Beninati, C.; Giannone, M.; La Camera, E.; Carbone, M. Detection of Arcobacter Spp. in the Coastal Environment of the Mediterranean Sea. Appl Environ Microbiol 2004, 70, 1271–1276. [CrossRef]
- Goldman, C.G.; Matteo, M.J.; Loureiro, J.D.; Almuzara, M.; Barberis, C.; Vay, C.; Catalano, M.; Heredia, S.R.; Mantero, P.; Boccio, J.R.; et al. Novel Gastric Helicobacters and Oral Campylobacters Are Present in Captive and Wild Cetaceans. Vet Microbiol 2011, 152, 138–145. [CrossRef]
- Almuzara, M.N.; Cittadini, R.; Ocampo, C.V.; Bakai, R.; Traglia, G.; Ramirez, M.S.; Del Castillo, M.; Vay, C.A. Intra-Abdominal Infections Due to Comamonas Kerstersii. J Clin Microbiol 2013, 51, 1998–2000. [CrossRef]
- Peng, W.; Zhao, X.; Li, X. Helicobacter Bilis Contributes to the Occurrence of Inflammatory Bowel Disease by Inducing Host Immune Disorders. Biomed Res Int 2022, 2022, 1837850. [CrossRef]
- Simpson, V.R.; Davison, N.J.; Dagleish, M.P. Causes of Mortality and Lesions Observed Post Mortem in European Moles (Talpa Europaea) in Cornwall, South-West England: Disease in Wildlife or Exotic Species. J Comp Pathol 2019, 167, 18–25. [CrossRef]
- Sarzosa, M.S.; Duignan, P.; Derango, E.J.; Field, C.; Ríos, C.; Sanchez, S.; Espinoza, E.; Loyola, A.; Rueda, D.; Páez-Rosas, D. Occurrence of Mycoplasmas in Galapagos Sea Lions (Zalophus Wollebaeki) and Their Association with Other Respiratory Pathogens. J Wildl Dis 2021, 57, 623–627. [CrossRef]
- Herp, S.; Durai Raj, A.C.; Salvado Silva, M.; Woelfel, S.; Stecher, B. The Human Symbiont Mucispirillum Schaedleri: Causality in Health and Disease. Med Microbiol Immunol 2021, 210, 173–179. [CrossRef]
- Venn-Watson, S.; Daniels, R.; Smith, C. Thirty Year Retrospective Evaluation of Pneumonia in a Bottlenose Dolphin Tursiops Truncatus Population. Dis Aquat Organ 2012, 99, 237–242. [CrossRef]
- Fernández-Álvarez, C.; Santos, Y. Identification and Typing of Fish Pathogenic Species of the Genus Tenacibaculum. Appl Microbiol Biotechnol 2018, 102, 9973–9989. [CrossRef]
- Nowlan, J.P.; Lumsden, J.S.; Russell, S. Advancements in Characterizing Tenacibaculum Infections in Canada. Pathogens 2020, Vol. 9, Page 1029 2020, 9, 1029. [CrossRef]
- Burioli, E.A.V.; Varello, K.; Trancart, S.; Bozzetta, E.; Gorla, A.; Prearo, M.; Houssin, M. First Description of a Mortality Event in Adult Pacific Oysters in Italy Associated with Infection by a Tenacibaculum Soleae Strain. J Fish Dis 2018, 41, 215–221. [CrossRef]
- Rungrassamee, W.; Klanchui, A.; Maibunkaew, S.; Chaiyapechara, S.; Jiravanichpaisal, P.; Karoonuthaisiri, N. Characterization of Intestinal Bacteria in Wild and Domesticated Adult Black Tiger Shrimp (Penaeus Monodon). PLoS One 2014, 9, e91853. [CrossRef]
- Nelson, T.M.; Rogers, T.L.; Brown, M. V. The Gut Bacterial Community of Mammals from Marine and Terrestrial Habitats. PLoS One 2013, 8. [CrossRef]
- Archie, E.A.; Tung, J. Social Behavior and the Microbiome. Curr Opin Behav Sci 2015, 6, 28–34. [CrossRef]
- Delport, T.C.; Power, M.L.; Harcourt, R.G.; Webster, K.N.; Tetu, S.G. Colony Location and Captivity Influence the Gut Microbial Community Composition of the Australian Sea Lion (Neophoca Cinerea). Appl Environ Microbiol 2016, 82, 3440–3449. [CrossRef]
| Samples | S1 | S2 | S3 | |
|---|---|---|---|---|
| Metadata | Gender | Male | Male | Male |
| Sexual maturity | Mature | Mature | Immature | |
| Location | Praia de Mira | Praia do Navio | Praia da Rainha | |
| Geographic region | Western Atlantic Iberian coast | Western Atlantic Iberian coast | Western Atlantic Iberian coast | |
| Cause of death | Disease | Disease | Disease | |
| Sequencing metrics | No. of raw reads | 94.650 | 135.918 | 142.087 |
| No. of filtered reads | 67.509 | 117.826 | 98.055 | |
| No. of classified sequences | 39.523 | 64.461 | 52.972 | |
| Total no. of ASVs | 218 | 220 | 208 | |
| No. of unique ASVs | 38 | 24 | 62 | |
| No. of phyla | 12 | 10 | 13 | |
| No. of classes | 21 | 19 | 23 | |
| No. of orders | 34 | 29 | 35 | |
| No. of families | 57 | 55 | 56 | |
| No. of genera | 67 | 51 | 55 |
| ASV | Presence in other Odontoceti species | Best BLASTn hit | % Identity |
||
|---|---|---|---|---|---|
| Dde | Pph | Sco | |||
| ASVs contributing solely to K. breviceps cluster | |||||
| ASV32 | - | - | - | Balneicella halophila | 98 |
| ASV77 | - | - | - | Thioalkalibacter halophilus | 74 |
| ASV156 *† | - | - | - | Pasteurella skyensis | 98 |
| ASV170 | - | - | - | Balneicella halophila | 96 |
| ASV189 | - | - | - | Marinospirillum alkaliphilum | 88 |
| ASV251 | - | - | - | Anaerostipes hadrus | 91 |
| ASV383 † | - | - | - | Actinobacillus delphinicola | 98 |
| ASV519 *† | - | - | - | Arcobacter marinus | 92 |
| ASV545 † | - | - | - | Suttonella indologenes | 91 |
| ASV570 | - | - | - | Defluviitalea saccharophila | 93 |
| ASV658 | - | - | - | Altericista lacusladogae | 76 |
| ASV725 *† | - | - | - | Pasteurella skyensis | 98 |
| ASV753 † | - | - | - | Arcobacter aquimarinus | 89 |
| ASV777 † | - | - | - | Streptobacillus moniliformis | 96 |
| ASV817 | - | - | - | Crocinitomix algicola | 88 |
| ASV888 * | - | - | - | Moraxella lincolnii | 96 |
| ASV915 | - | - | - | Anaerostipes hadrus | 87 |
| ASV1034 *† | - | - | - | Tenacibaculum soleae | 96 |
| ASV1036 | - | - | - | Anoxynatronum buryatiense | 94 |
| ASV1084 | - | - | - | Fructobacillus pseudoficulneus | 75 |
| ASV1094 | - | - | - | Ihubacter massiliensis | 93 |
| ASV1104 *† | - | - | - | Pasteurella skyensis | 99 |
| ASV1148 | - | - | - | Labilibacter aurantiacus | 91 |
| ASV1155 *† | - | - | - | Pasteurella skyensis | 99 |
| ASV1497 | - | - | - | Deinococcus petrolearius | 74 |
| ASV1603 | - | - | - | Lutibacter oceani | 97 |
| ASV1647 | - | - | - | Nodosilinea alaskaensis | 78 |
| ASV1665 | - | - | - | Anoxynatronum buryatiense | 94 |
| ASVs mainly contributing to K. breviceps cluster (not exclusively) | |||||
| ASV81 | + | - | + | Anaerotignum propionicum | 92 |
| ASV99 *† | + | - | + | Arcobacter canalis | 90 |
| ASV111 † | + | - | - | Sneathia sanguinegens | 98 |
| ASV141 | + | - | - | Marinifilum fragile | 94 |
| ASV148 * | - | - | + | Oceanispirochaeta sediminicola | 92 |
| ASV171 | + | - | - | Fusibacter paucivorans | 88 |
| ASV205 * | + | - | - | Marinospirillum alkaliphilum | 89 |
| ASV218 | + | - | + | Ruminiclostridium cellobioparum | 86 |
| ASV301 | + | - | - | Spongiimonas flava | 86 |
| ASV356 | + | + | + | Fusibacter tunisiensis | 93 |
| ASV357 † | + | - | - | Suttonella ornithocola | 96 |
| ASV402 | + | - | - | Faecalicatena contorta | 94 |
| ASV426 | + | - | - | Salinivirga cyanobacteriivorans | 92 |
| ASV458 † | + | - | - | Sneathia sanguinegens | 95 |
| ASV492 | + | - | - | Polymorphobacter multimanifer | 82 |
| ASV527 | + | - | + | Desulfotignum toluenicum | 96 |
| ASV544 *† | + | - | - | Spirochaeta halophila | 85 |
| ASV603 | + | - | - | Oceaniserpentilla haliotis | 90 |
| ASV635 *† | + | - | - | Helicobacter bilis | 90 |
| ASV676 | + | - | - | Acholeplasma modicum | 90 |
| ASV754 | + | + | - | Fusibacter paucivorans | 91 |
| ASV757 | + | - | - | Petrocella atlantisensis | 92 |
| ASV765 *† | - | + | - | Tenacibaculum gallaicum | 99 |
| ASV778 | + | - | - | Hydrogenophaga soli | 98 |
| ASV839 | + | - | - | Methylophaga thalassica | 92 |
| ASV849 * | + | + | + | Sediminispirochaeta bajacaliforniensis | 86 |
| ASV861 | + | + | + | Aminipila butyrica | 96 |
| ASV885 | + | - | + | Treponema zuelzerae | 90 |
| ASV898 | - | + | - | Ignavibacterium album | 88 |
| ASV909 *† | + | + | + | Porphyromonas pasteri | 92 |
| ASV948 † | + | + | + | Treponema medium | 92 |
| ASV1007 *† | + | + | + | Pseudodesulfovibrio indicus | 98 |
| ASV1098 | + | - | - | Aureibacter tunicatorum | 89 |
| ASV1121 * | + | - | - | Oceanispirochaeta sediminicola | 94 |
| ASV1128 | + | - | - | Fusibacter fontis | 93 |
| ASV1160 † | + | + | + | Arcobacter venerupis | 98 |
| ASV1252 † | + | + | - | Mycoplasma mucosicanis | 97 |
| ASV1255 | + | + | + | Desulfomicrobium hypogeium | 90 |
| ASV1264 † | + | - | - | Streptobacillus moniliformis | 96 |
| ASV1266 | + | + | - | Rhabdanaerobium thermarum | 86 |
| ASV1283 | + | - | + | Oceanispirochaeta sediminicola | 93 |
| ASV1298 † | + | - | + | Comamonas kerstersii | 99 |
| ASV1342 | + | - | - | Ruminiclostridium cellobioparum | 90 |
| ASV1388 | + | - | + | Cellulosilyticum lentocellum | 87 |
| ASV1404 † | + | - | - | Fusobacterium russii | 98 |
| ASV1410 † | + | - | - | Mycoplasma opalescens | 98 |
| ASV1486 | + | - | - | Labilibacter sediminis | 88 |
| ASV1519 | + | - | - | Paenalcaligenes hermetiae | 97 |
| ASV1535 | - | + | - | Edaphobacter acidisoli | 75 |
| ASV1546 | + | - | - | Defluviitalea raffinosedens | 92 |
| ASV1624 *† | + | + | - | Pasteurella skyensis | 96 |
| ASV1641 | + | - | + | Acholeplasma morum | 88 |
| ASV1698 | + | - | - | Balneicella halophila | 99 |
| ASV1701 | + | - | - | Lachnotalea glycerini | 90 |
| ASV1774 | + | - | - | Anaerovorax odorimutans | 96 |
| ASV1776 | + | - | - | Anaerotignum propionicum | 90 |
| ASV1777 | + | - | - | Crocinitomix algicola | 88 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





