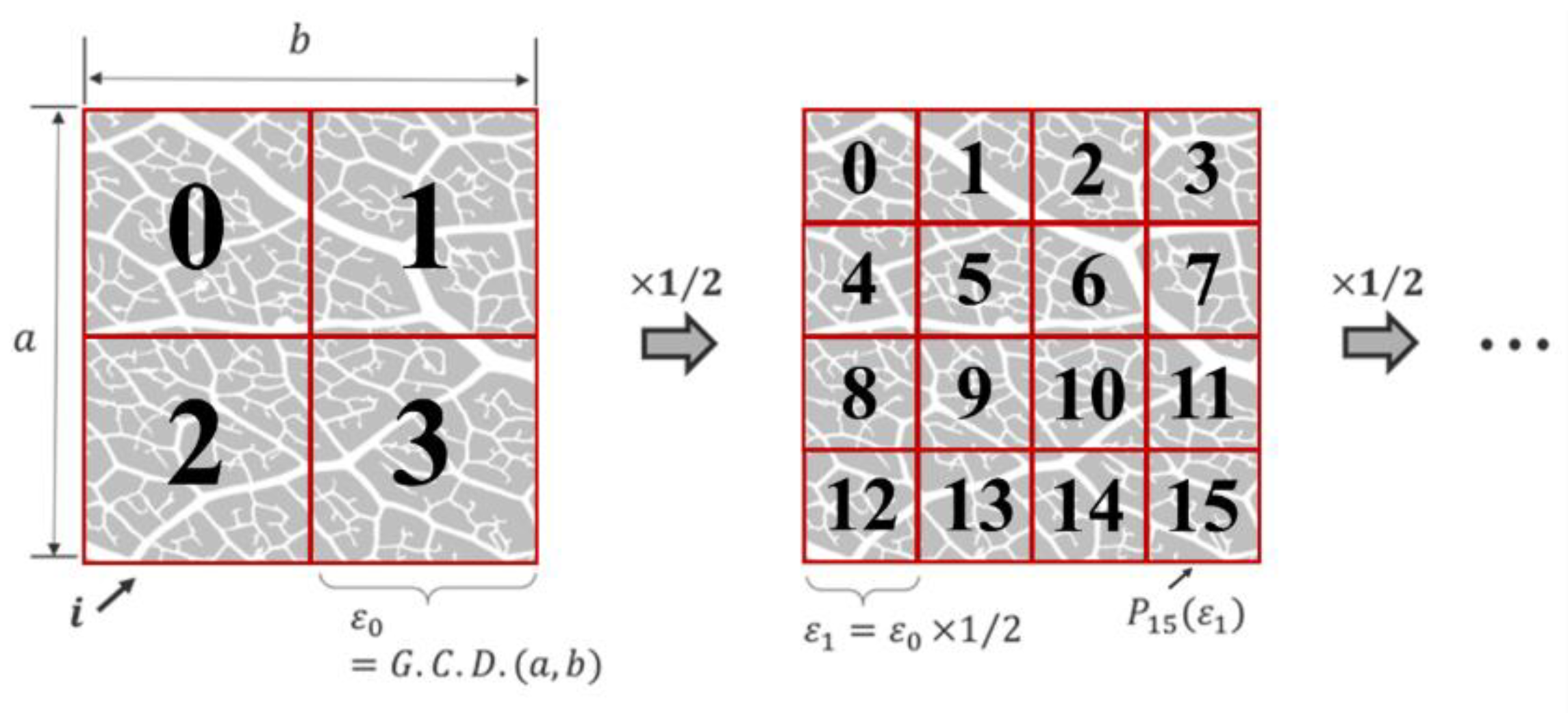
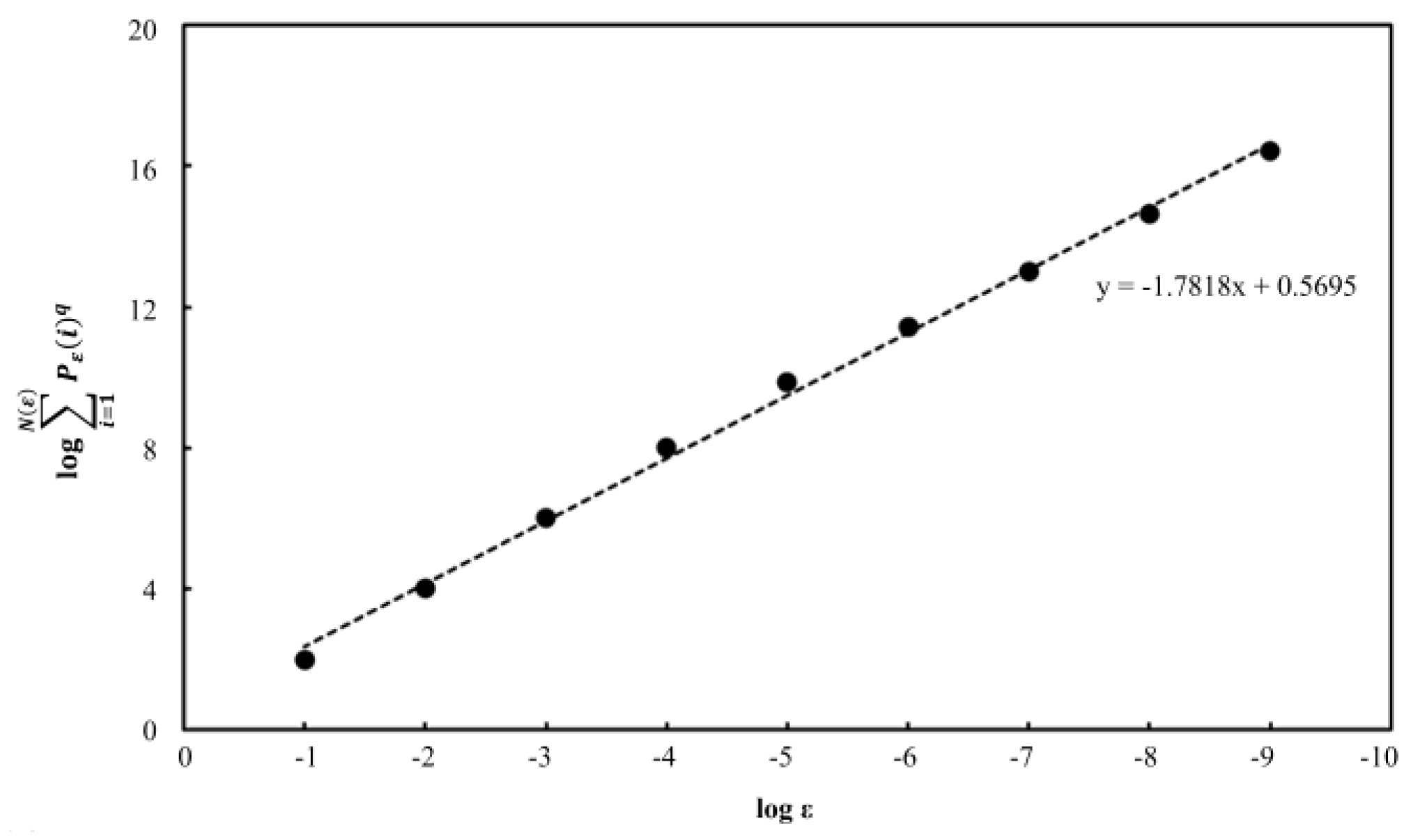
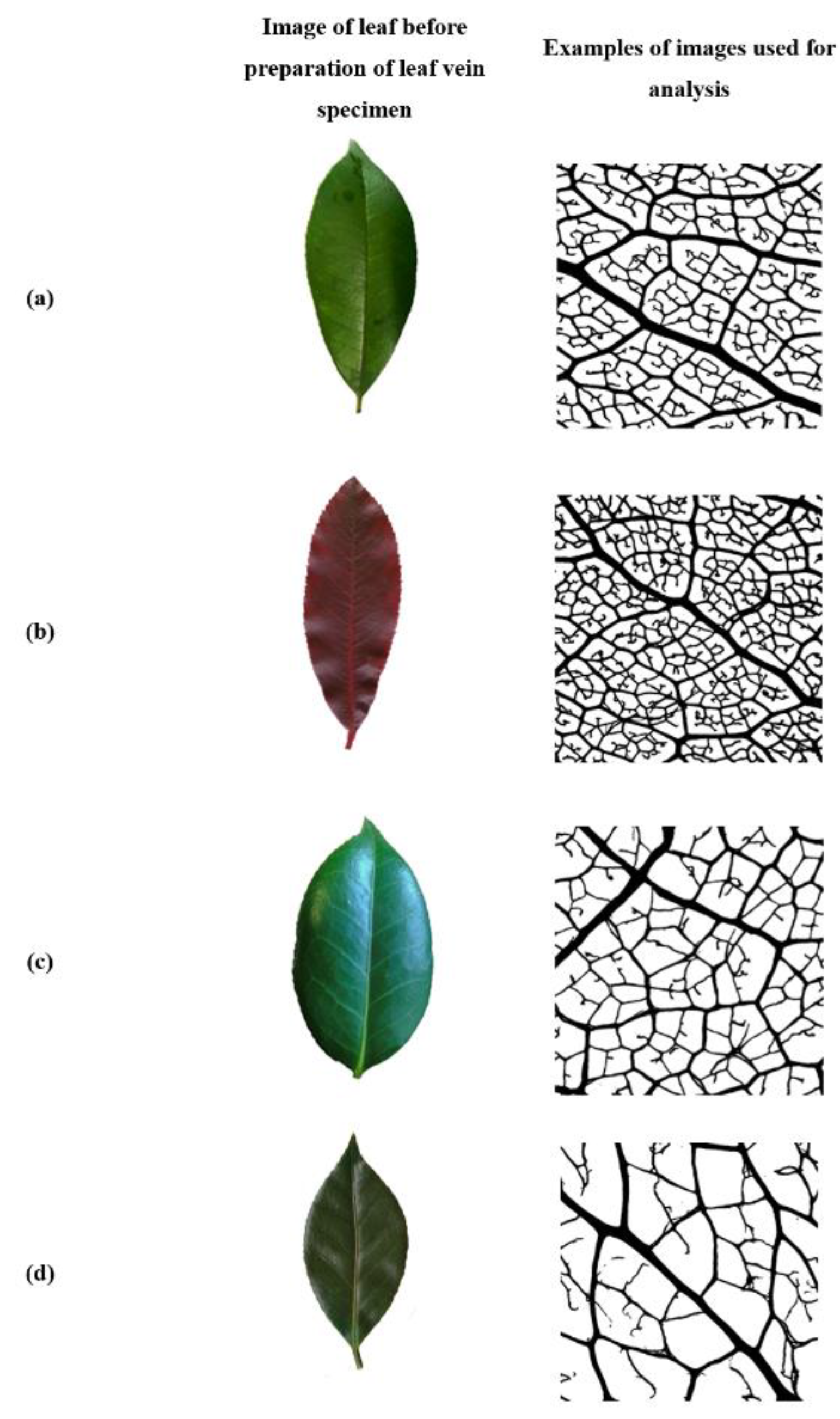
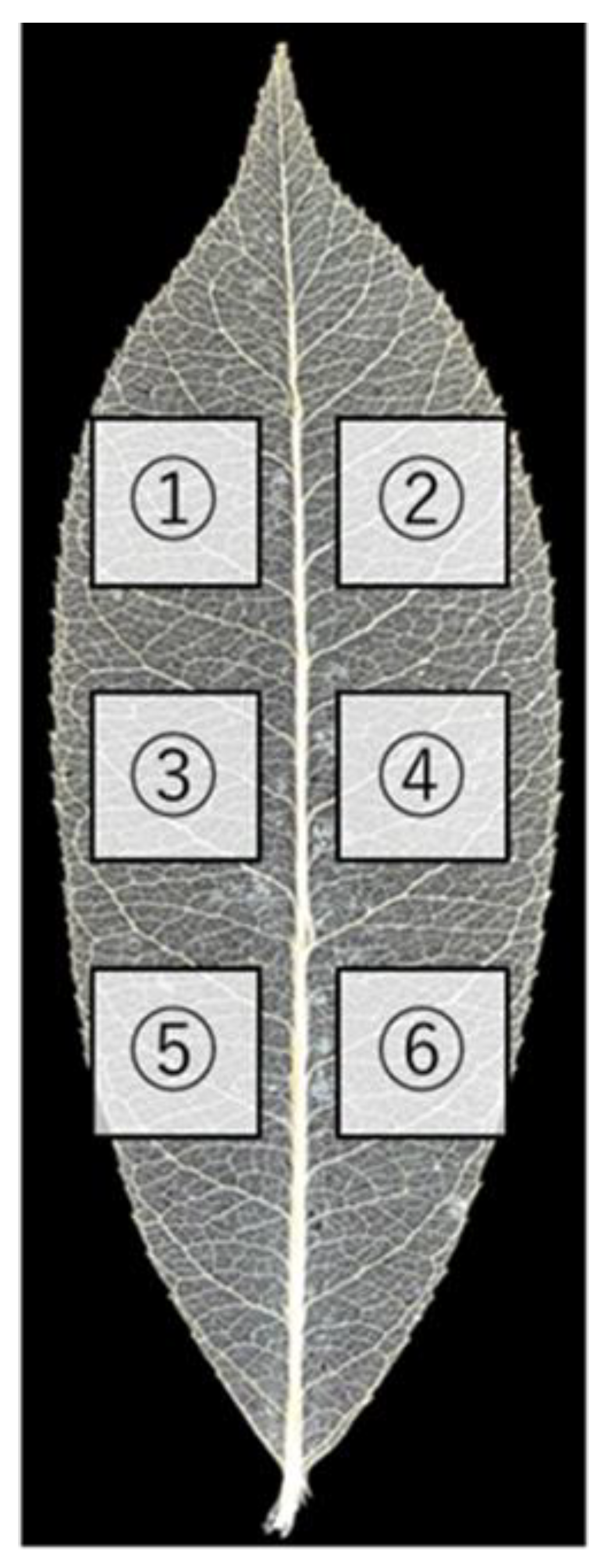
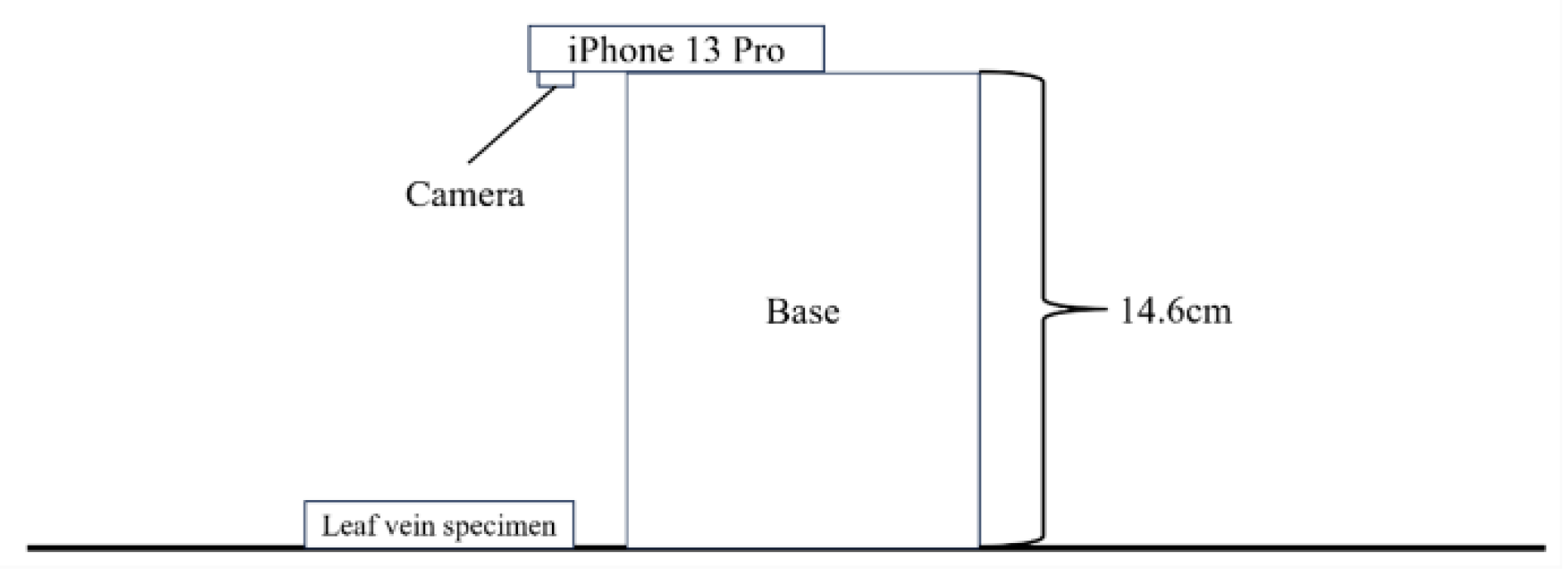
1. Introduction
There are so many plant species on the planet that as of 2020, an estimated 350,000 species of vascular plants have been identified [
1]. Because botany defines taxonomic groups, such as families and genera, and assigns individual plants to those taxonomic groups, experts determine which plant species they fall into, despite the diversity of plants. This process of assigning an unknown plant to a taxon is called plant identification [
2].
Accurate plant identification is an essential task when considering biodiversity assessment and environmental conservation [
3]. The plant species that inhabit a given area vary greatly depending on the sunlight conditions, human management conditions, and the ecosystem of the land. Because of the immobile nature of plants, vegetation can be used as an indicator to capture the local environment. Therefore, plant identification is the most fundamental step when considering biodiversity assessment and environmental conservation [
4]. In addition, with continued biodiversity loss, the demand for plant identification is expected to increase further [
5].
Plant identification is usually done manually using botanical illustrations. Assign plants to taxa based on their identifying and morphological characteristics until finally reaching a species. There are two types of characteristics: quantitative, such as plant height, flower width, and number of petals, and qualitative, such as leaf shape and flower color. Since plants of the same species cannot have exactly the same characteristics, some generalization is necessary for classification [
6]. Therefore, it is difficult for a layman to identify plants from illustrated books [
7]. Even if one is an expert, the task of identifying a single species from a large number of candidates listed in illustrated books is labor intensive and very costly in terms of time, due to the diversity of plant species [
8]. Against this background, the system of automatic plant species recognition is in demand not only by experts but also by laymen.
With the demand for automated species identification growing from many directions, in 2004 Gaston et al [
9] argued that with the development of artificial intelligence and image processing, automated species identification based on digital images will become concrete in the near future.
In fact, such research has been actively conducted in recent years, including studies attempting to identify plants from flower and leaf images [
10,
11], applying Minkowski's multiscale fractal dimension method (Multiscale Minkowski-Sausage) to images of leaf outlines and veins to attempt automatic [
12], and machine learning of leaf vein images and leaf shape images [
13].
Plants are composed of various organs such as flowers, leaves, and roots, and as mentioned above, various characteristics such as plant height, flower width, number of petals, leaf shape, and flower color can be obtained. These characteristics have been used in the past for manual identification, but many characteristics have also been studied as information for automatic recognition, and a number of methods for describing the characteristics have been investigated [
6].
Saito et al. [
10] proposed a flower recognition system that uses flower images taken in their natural state as input. Finally, 20 samples from each of 15 families and 30 species were tested using the leave-one-out method, resulting in a recognition rate of 98.6%. Although this method has been able to obtain a high recognition rate, there are some problems: flowers can often be collected only for a limited period of time during the year, there are large differences in morphological and color characteristics depending on the flowering period, and some flowers change their flowering status during the daytime. Among the characteristics that can be obtained from plants, flowers are the most difficult to handle [
6].
On the other hand, leaves are readily available as information because they can be easily collected year-round from almost any plant, including fossils and rare plants. Furthermore, leaf structures are planar, making them easy to collect, store, and image. Because these aspects simplify the data collection process, leaf morphology is the most studied feature in plant identification [
6,
14].
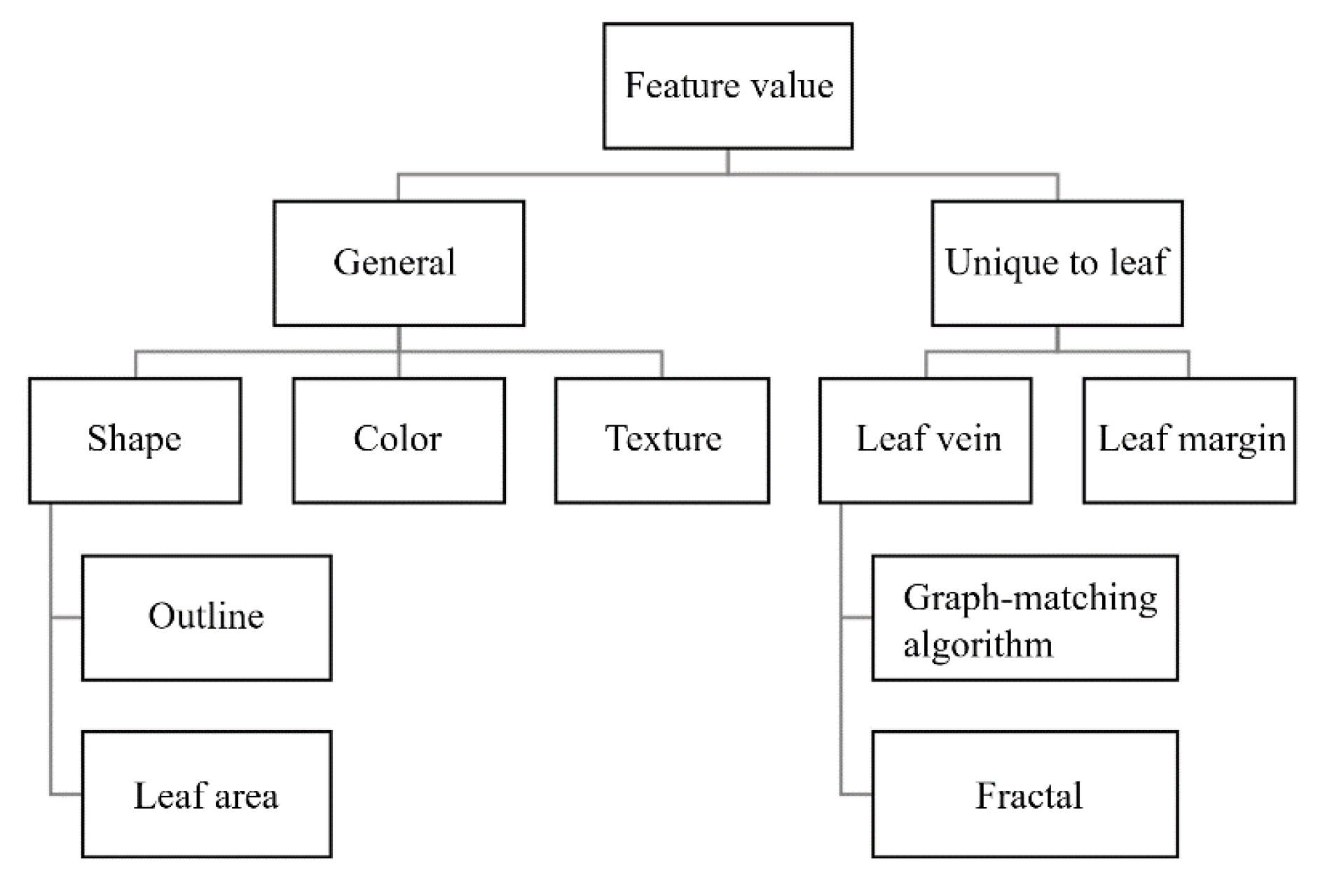
Figure 1 shows the main leaf features and their descriptors proposed for the automatic classification of plant species.
Leaf characteristics can be divided into two main categories: general characteristics such as shape, color, and texture, and characteristic quantities specific to leaves such as veins and leaf margins. Of these properties, shape is the one that has been studied most extensively [
14]. Research on automatic shape classification can be divided into two main categories: methods that quantify the outline of the leaf and methods that quantify the leaf region (area, axial length, etc.). While shape characteristics are simple and readily available and therefore easy to use as information, they often vary considerably even among leaves of the same species, making automatic classification by shape alone difficult. Therefore, shape features are usually used in combination with other features.
For color features, descriptors such as color moments (CM), color histograms (CH), color coherence spectra, and color correlograms have been proposed [
15]. The main challenge in describing color characteristics is that there are considerable differences due to the intensity and angle of the light at the time of capture, as well as different color temperatures [
14]. In addition, Yanikoglu et al [
16] reported that color information does not contribute at all to classification accuracy when combined with shape and texture descriptors. For these reasons, there are few studies on color characteristics.
For texture features, the main descriptors proposed are the Gabor filter (GF), fractal dimension (FD), and gray-level co-occurrence matrix (GLCM). GF has been widely employed to extract texture features from images, and Casanova et al [
17] applied GF to textured images without leaf margin and reported higher accuracy than traditional texture analysis methods such as FD and GLCM. On the other hand, some literature claims that fractal analysis is the most suitable method for texture analysis because objects in nature [
18], such as the surface of a leaf, have random and persistent patterns [
19]. Jarbas et al [
20] proposed a method that combines FD and lacunarity in a gravity model and found it to be superior to GF, FD, and GLCM. None of the texture analysis methods rely on complete leaf shape, as they can classify plants based on only one part of the leaf. Thus, it is very useful for the purpose of identifying plants that have been damaged by insect bites or other problems.
The leaf veins and leaf margins are characteristic quantities that are unique to the leaf. Since hand vein patterns are used in ecological authentication [
21], leaf vein features can be a powerful source of information in plant identification. Larese et al [
22] calculated 52 measurements (total number of edges, total number of nodes, network length, vein length, etc.) from the veins and examined the measurements needed for automatic identification. Nam et al [
23] reported applying a graph matching algorithm called Venation Matching (VM) to leaf veins and combining it with shape features to obtain high accuracy. Bruno et al [
12] applied Minkowski's multiscale fractal dimension method (Multiscale Minkowski-Sausage) to images of leaf outlines and veins to attempt automatic recognition, and reported a high recognition rate when the two methods were combined.
Bruno et al [
12] proposed an automatic plant identification method based on multiscale fractal dimension. The resulting average misclassification rate of 2.5% is good, indicating that multi-scaling fractal analysis is a useful method for feature extraction in leaf veins.
Bruno et al. extracted and analyzed only the central and lateral veins of the leaf veins and did not deal with information on the fine veins. In light of the aforementioned ecological authentication technology, it would be possible to improve accuracy by also analyzing veinlets. In fact, Wilf et al [
13] obtained excellent results in family and order classification by machine learning of vein images and leaf shape images, including veinlets.
However, Minkowski's multi-scaling fractal analysis used in this method uses the extrema of the resulting graph as features and does not interpret the features semantically. In contrast, in the other means of obtaining fractal dimension, called multifractal analysis, which extends the Hausdorff-Besicovitch dimension, the features obtained are values related to shape, entropy, etc. Leaf features can be used not only for plant identification, but also as indicators for other basic research, such as the study of potential leaf diseases. In the future, if breeding attempts are made to create disease-resistant plants based on these values, features that can be associated with shape and entropy could be more informative than features that have no semantic interpretation. Minkowski's multiscaling fractal analysis is also based on the assumption that objects in nature are unlikely to be mathematically perfect fractals, and multifractal analysis is based on the same assumption. Since multi-scaling fractal analysis is useful as a feature extraction method for leaf veins, we believe that multi-fractal analysis is also useful as a feature extraction method for leaf veins.
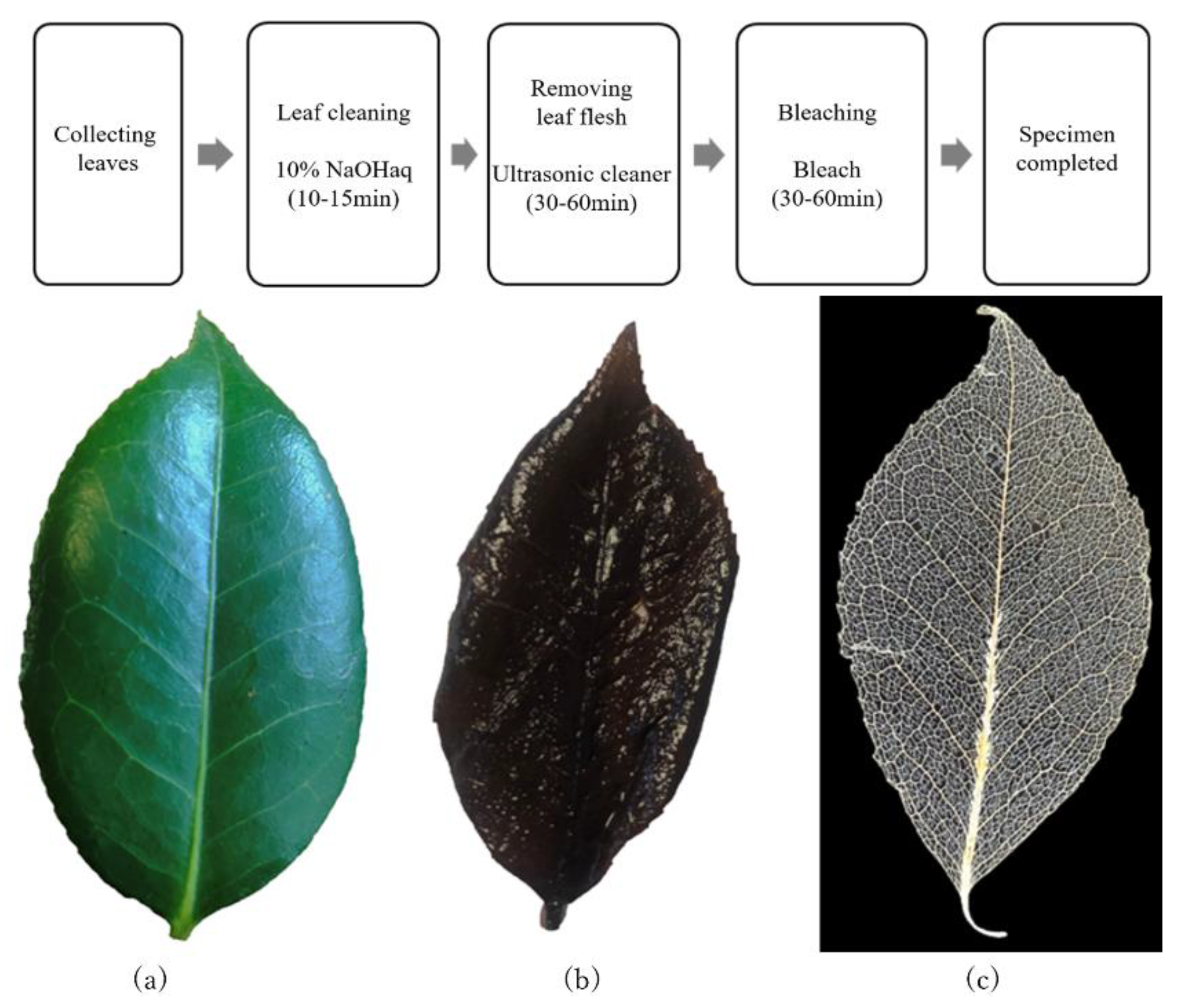
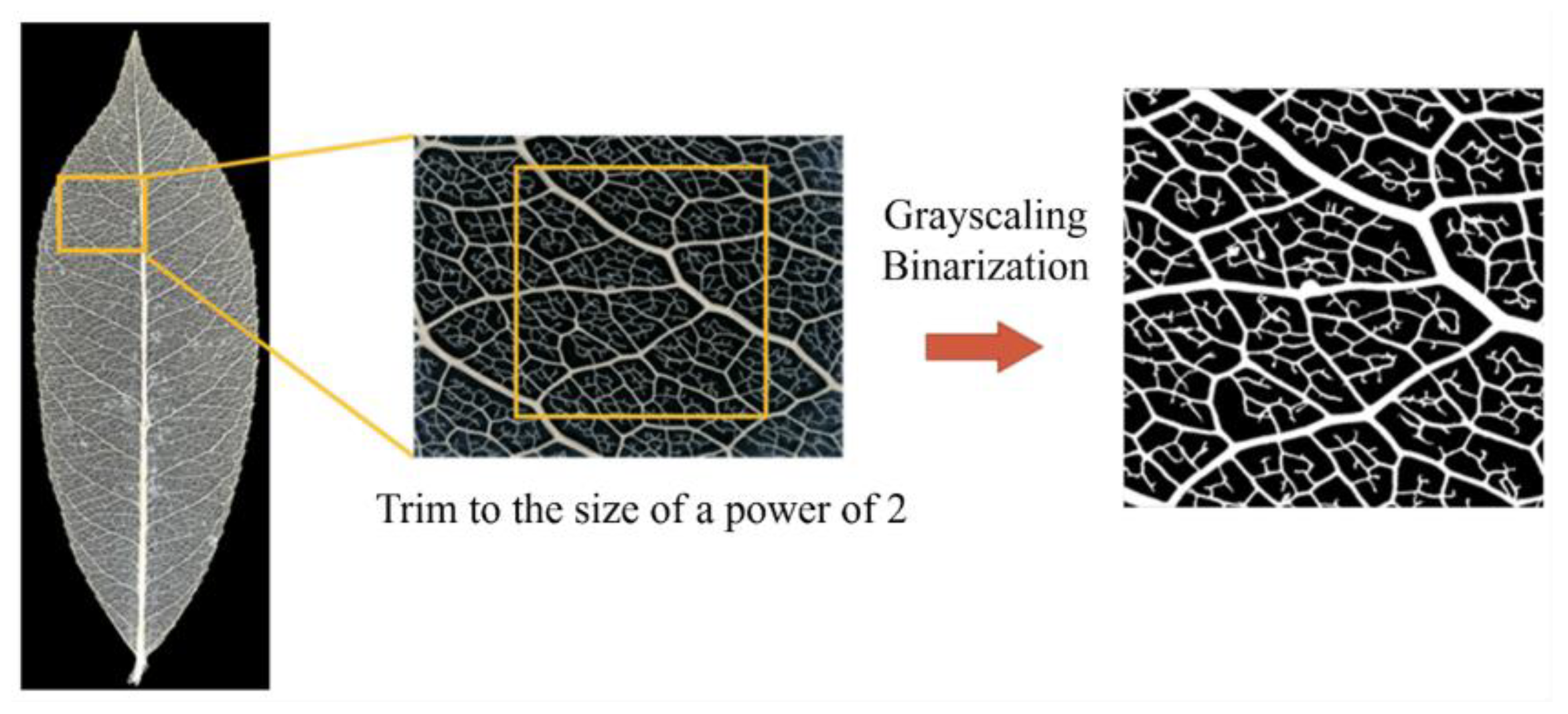
For these reasons, we propose a method of applying multifractal analysis to images of leaf veins, including veinlets.
4. Conclusions
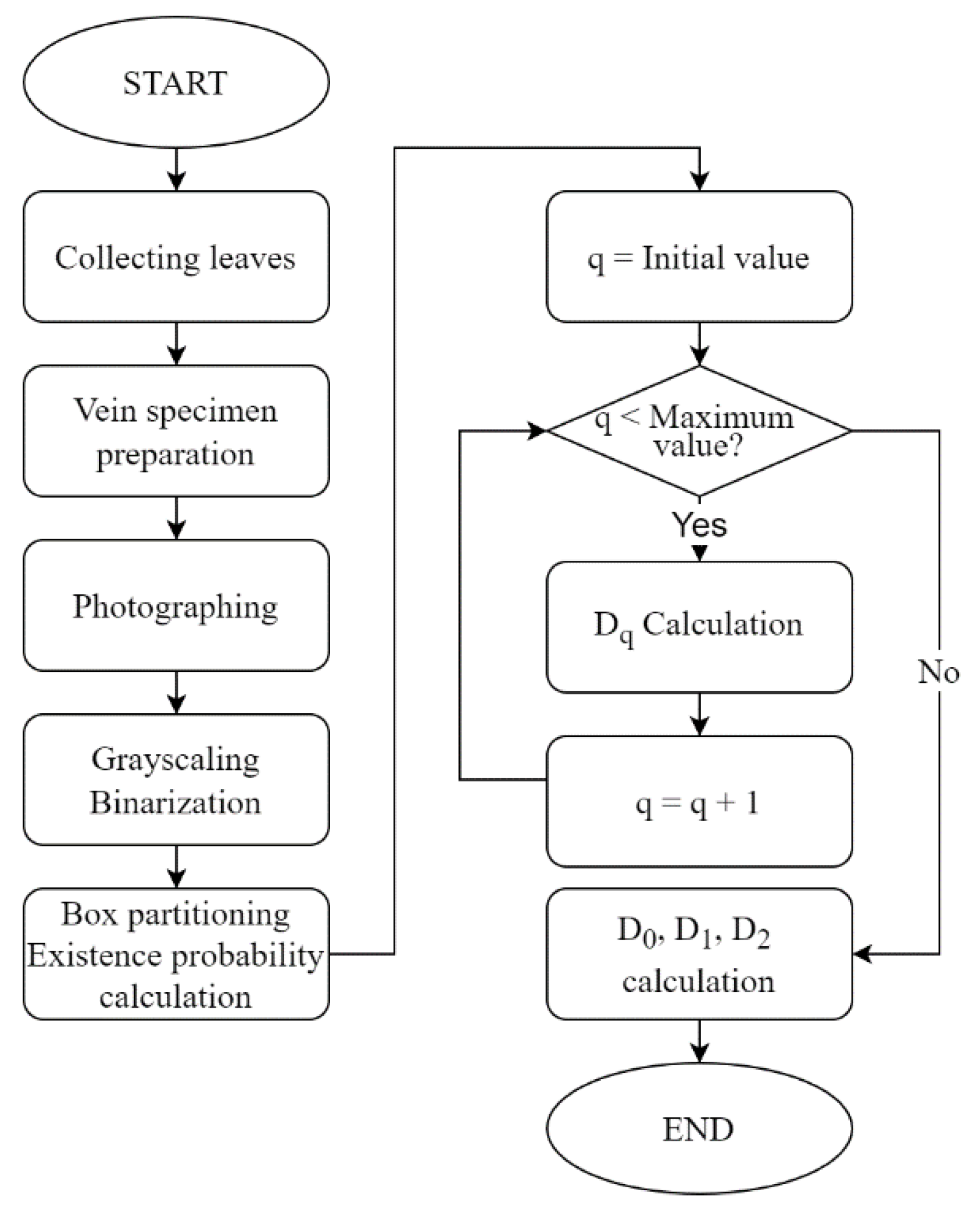
In this study, we proposed a method for automatic plant identification by applying multifractal analysis to leaf vein images that include veinlets. Two experiments were conducted to verify the effectiveness of the proposed method.
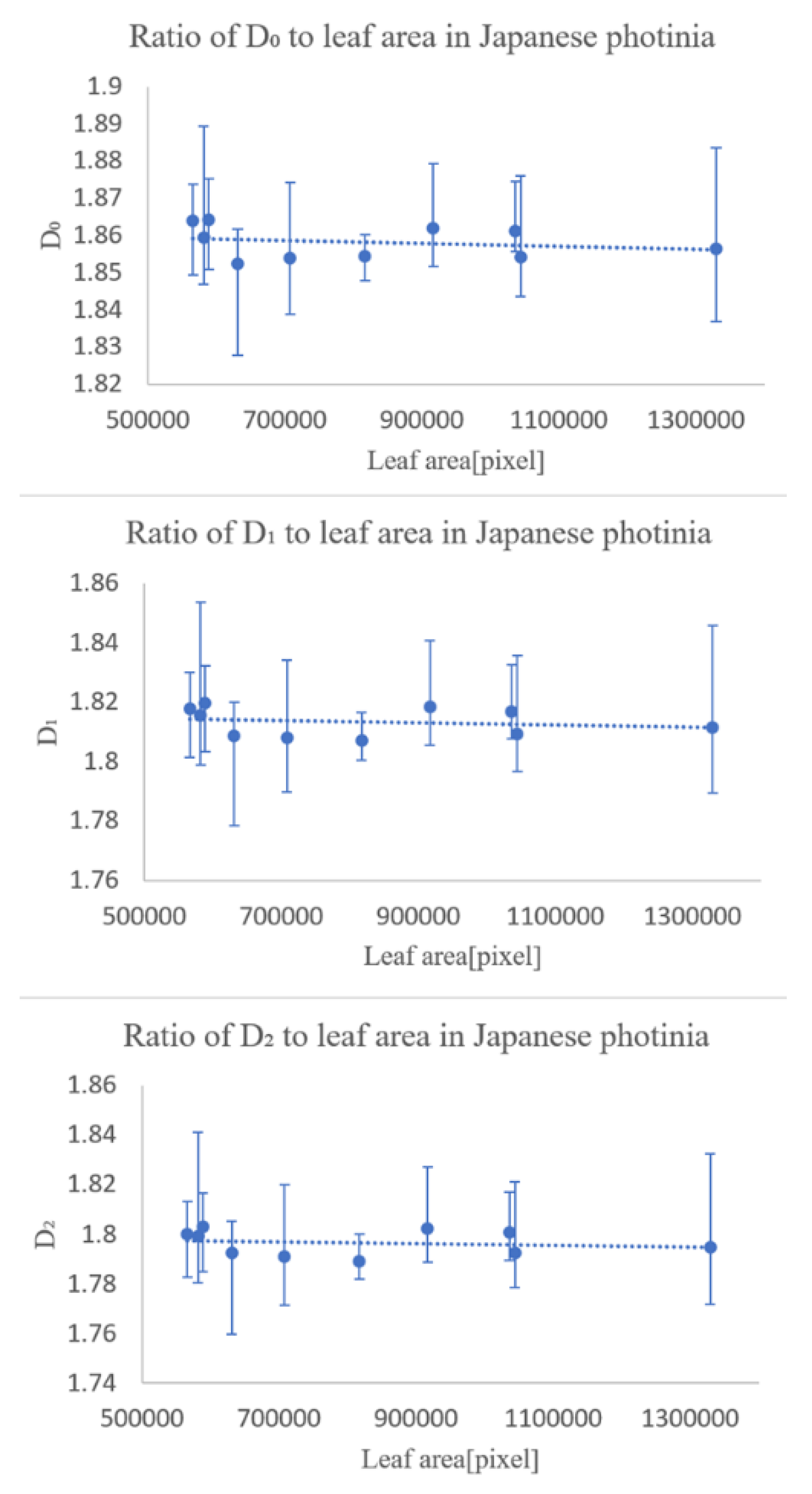
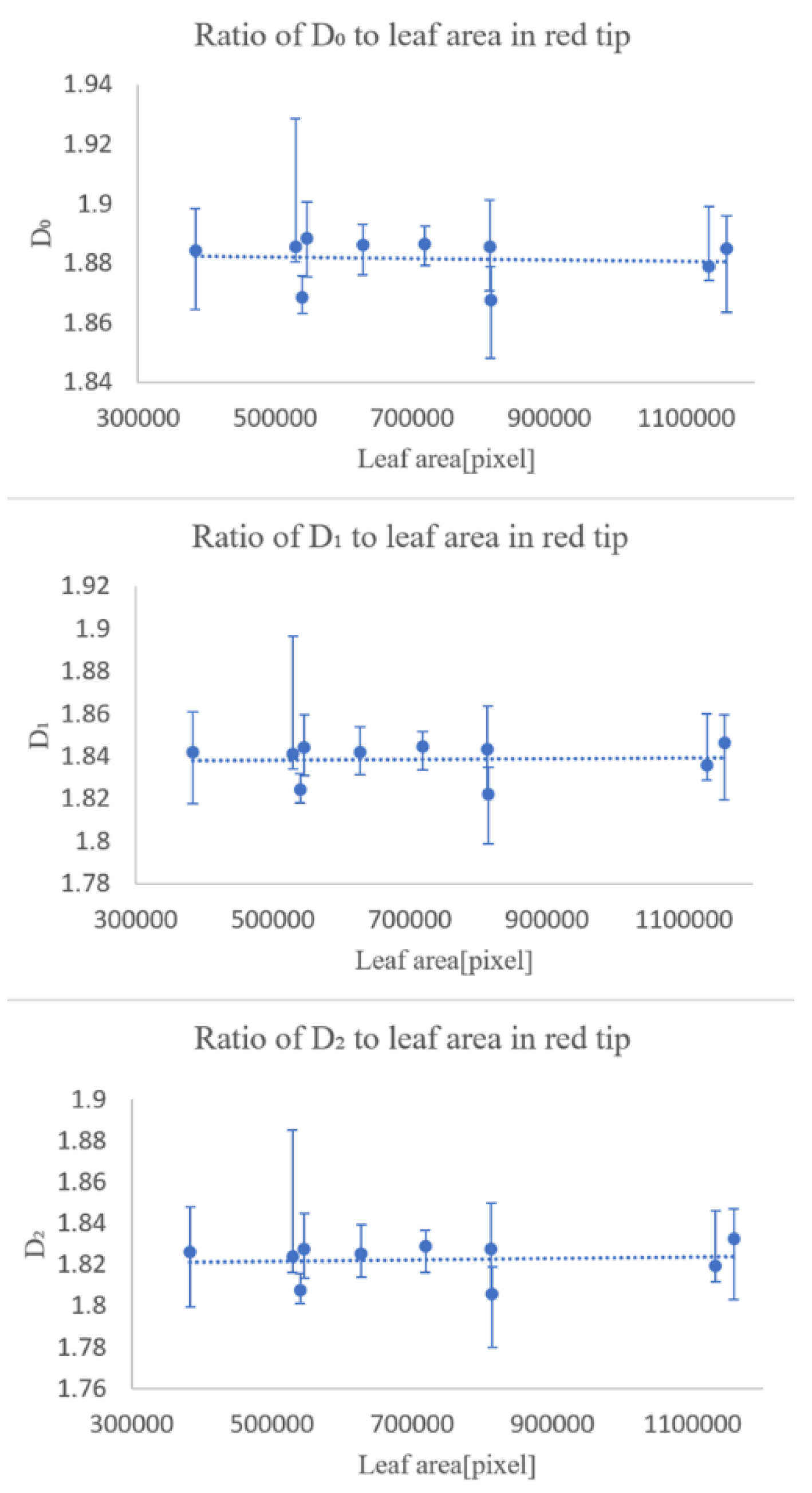
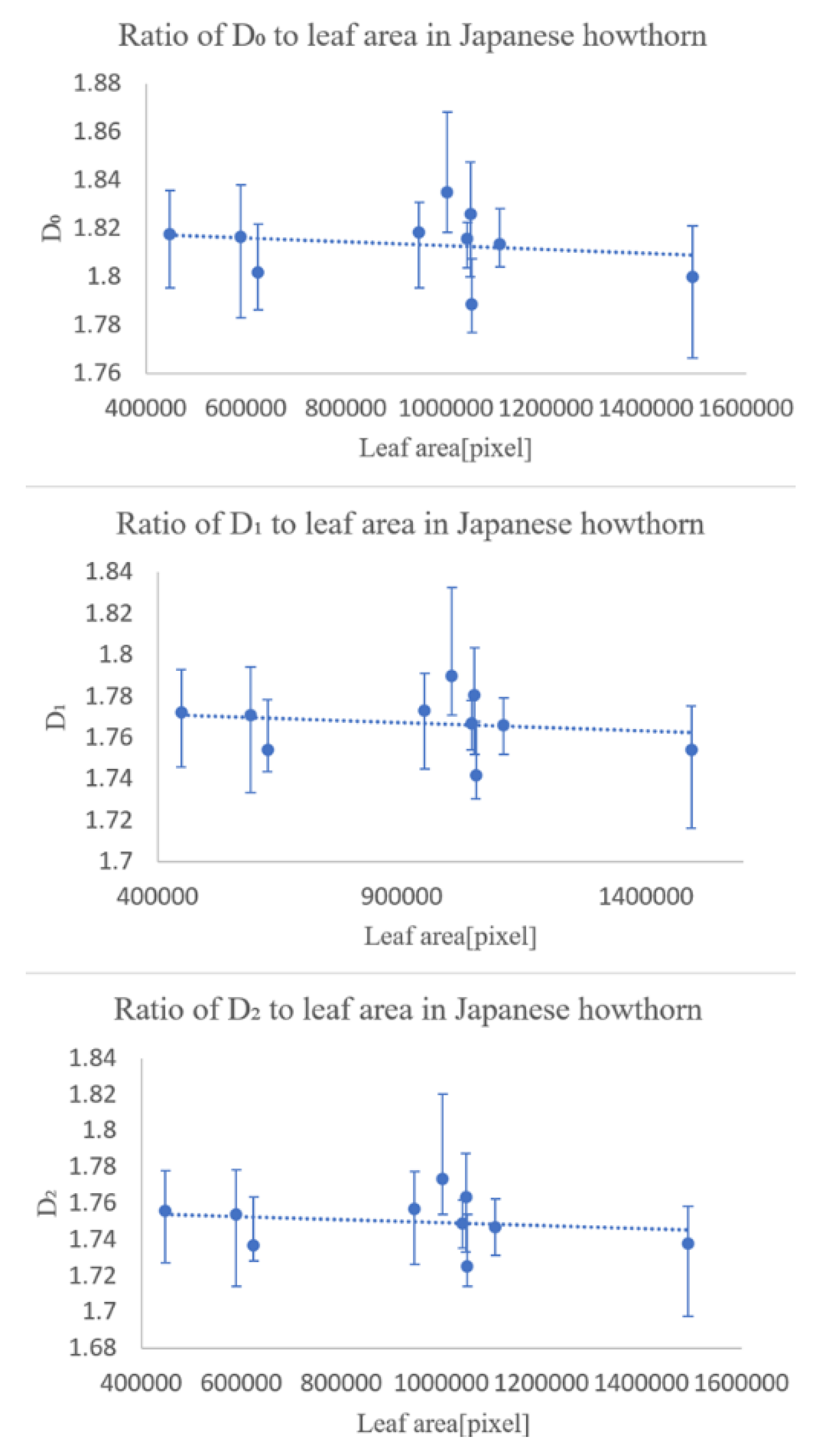
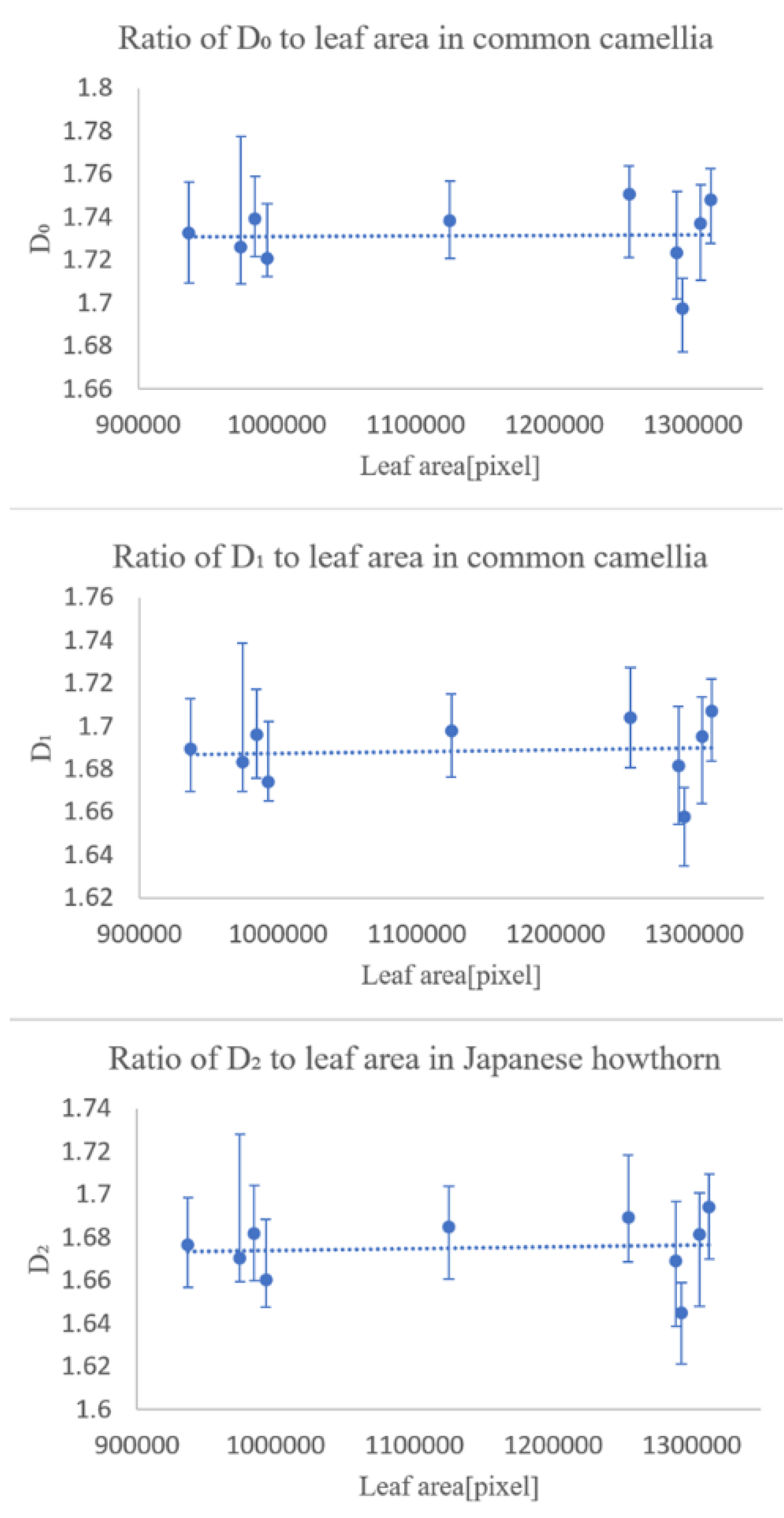
In the first experiment, we checked whether the fractal dimensionality varied with the degree of leaf growth. The results showed that the degree of leaf growth had little effect on the fractal dimension number of the leaf veins.
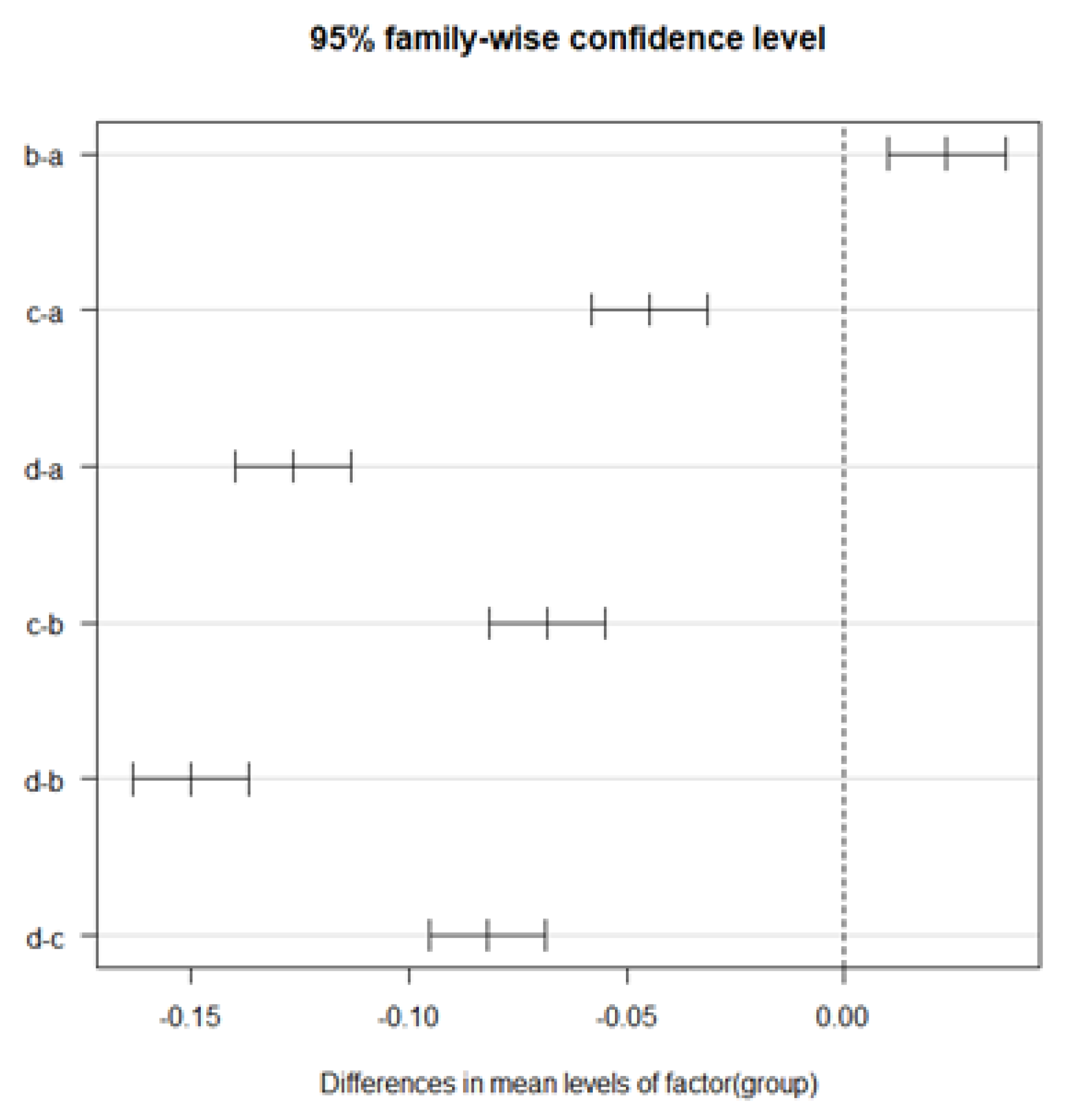
The second experiment was conducted to see how the fractal dimensionality varied among the different classification groups, and the results suggested that classification was possible for all of same family and genus, differing only in species, same family, differing genus, differing family, and differing species.
The experimental results suggest that multifractal analysis can be applied to images of leaf veins, including veinlets, to enable automatic plant identification. However, we would like to collect more data in the future, because the magnitude of the effect of errors caused by the shooting position in the sample is unclear, and we have only been able to conduct experiments on four types of trees.