Submitted:
10 October 2024
Posted:
11 October 2024
You are already at the latest version
Abstract
Keywords:
1. Executive Summary/Introduction
- COD1: Focuses on reductionist interpretation of collective omics data
- COD2: Involves the creation of curated dataset collections
- COD3: Trains participants in the re-analysis of omics data on a global scale
2. Historical Development and Overview of COD Training Modules
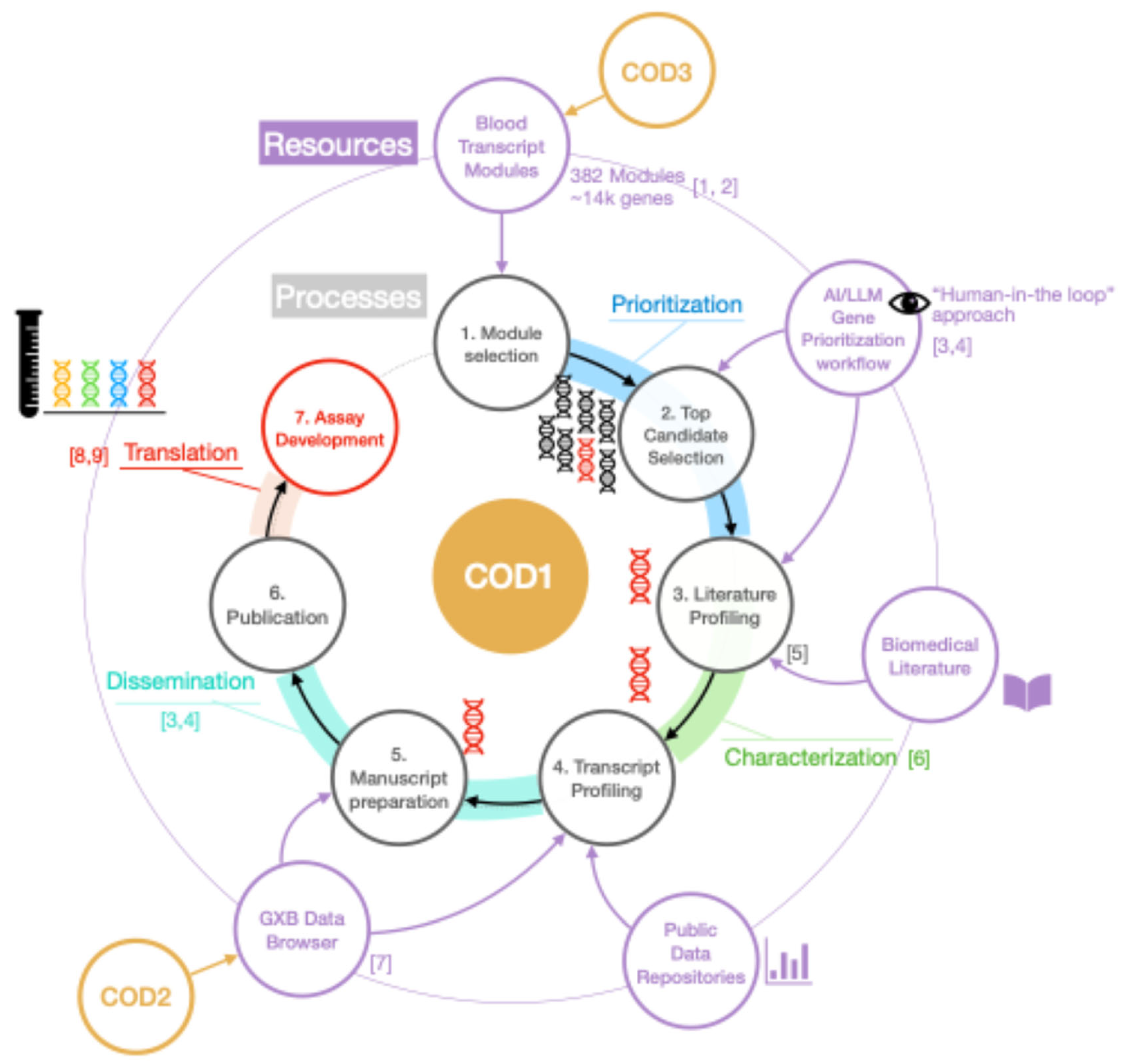
2.1. COD1: Reductionist Interpretation of Collective Omics Data
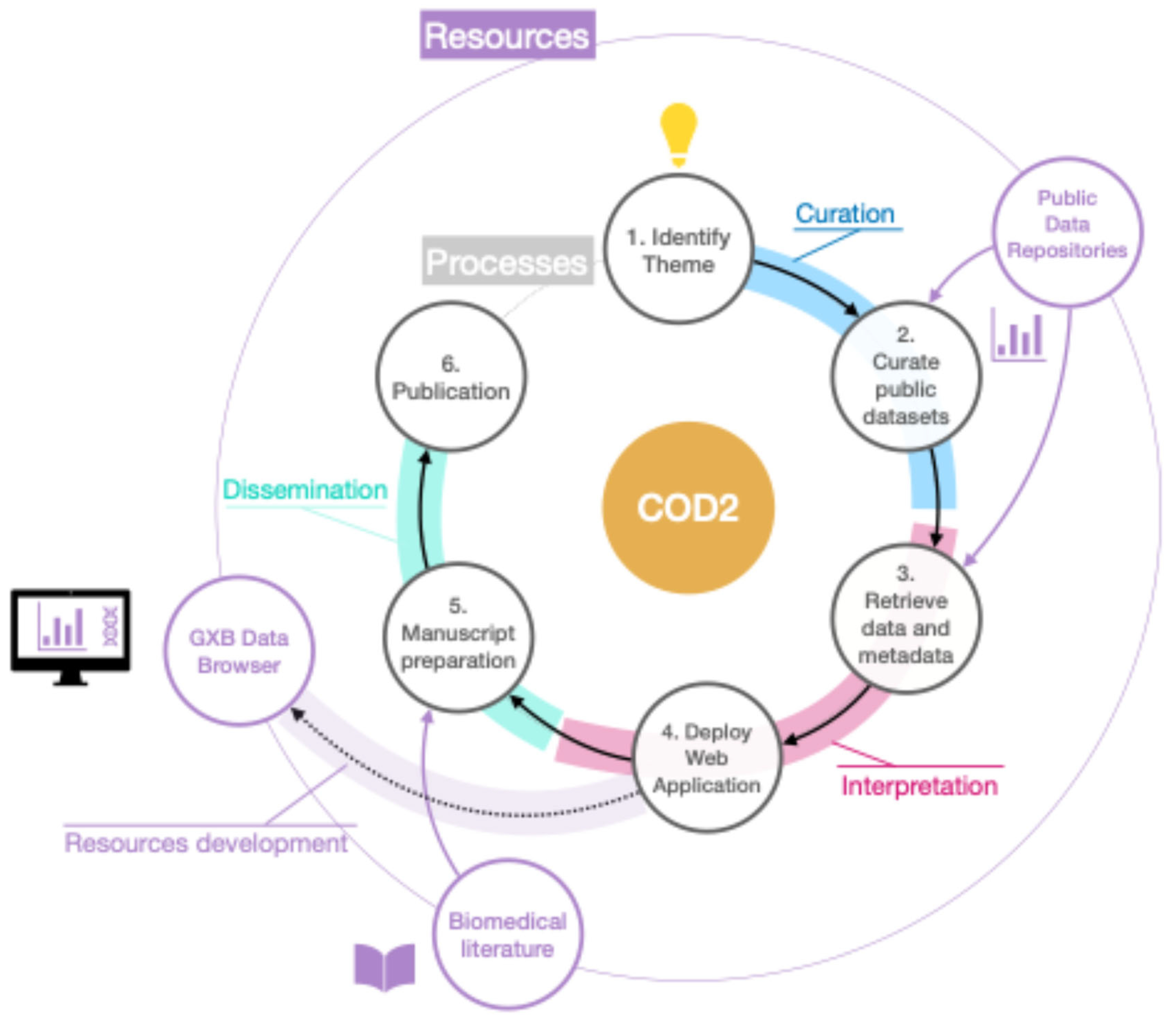
COD2: Creation of Curated Collective Omics Dataset Collections
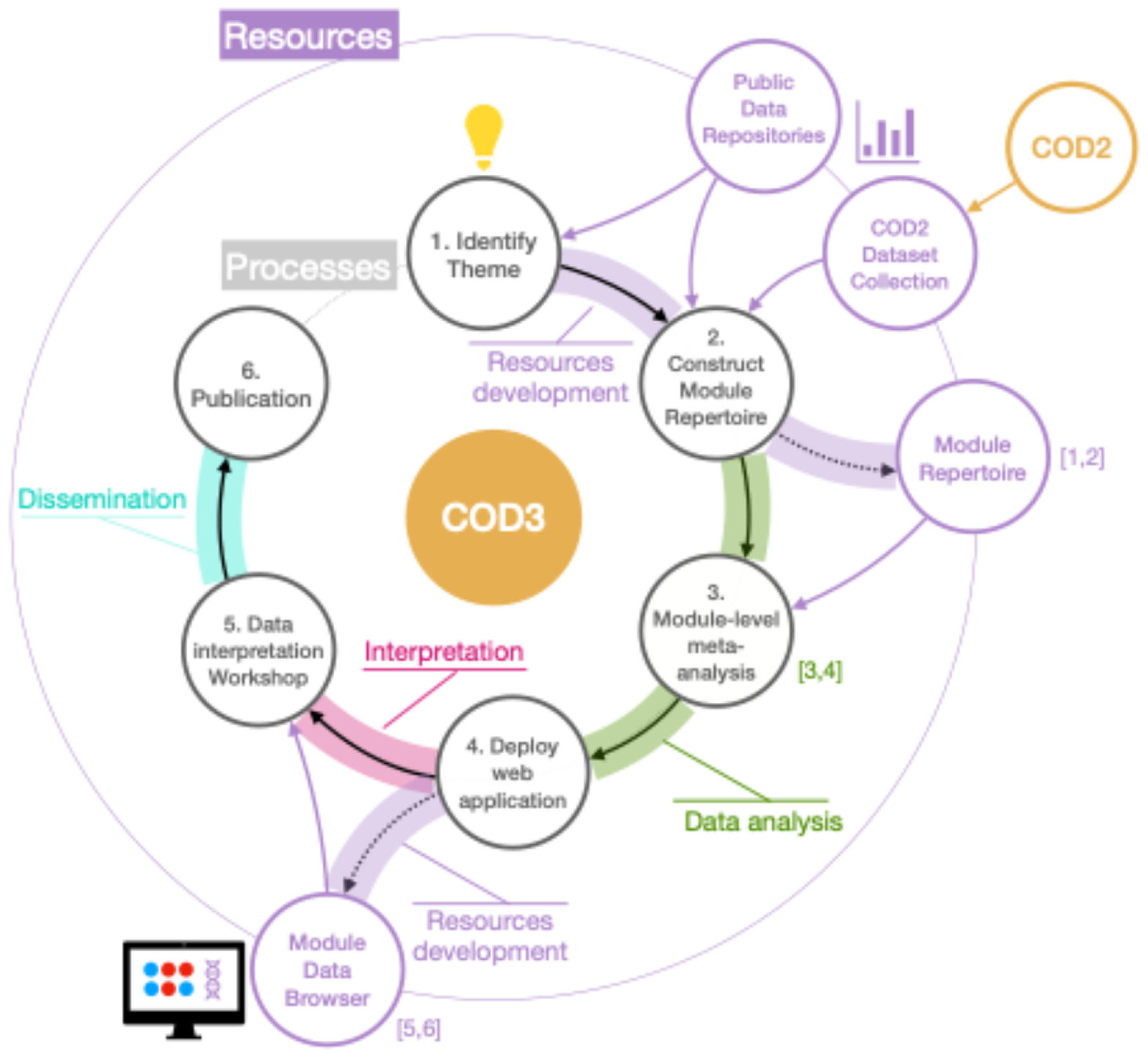
COD3: Re-Analysis of Collective Omics Data on a Global Scale
- Meta-analyses at the module level: These studies encompass multiple independent datasets, focusing either on investigating disease pathogenesis or biomarker discovery. For example, a study on psoriasis [31] utilized module-level meta-analysis to investigate the neutrophil-driven inflammatory signature characterizing the blood transcriptome fingerprint of the disease. Similarly, research on respiratory syncytial virus (RSV) infection [32] employed module-level meta-analysis for biomarker discovery, identifying erythroid cell-positive blood transcriptome phenotypes associated with severe RSV infection.
- Data and knowledge-driven biomarker discovery: Several studies have employed this approach, including work on pregnancy [33], COVID-19 [34], and most recently, the use of Large Language Models (LLMs) for developing a generic immune profiling assay [35]. This last study is particularly noteworthy as it demonstrates the potential of integrating artificial intelligence into the biomarker discovery process.
- Data interpretation workshops: These workshops, supported by BloodGen3 applications, involve participants with expertise in medicine and immunology who may not have extensive data science backgrounds. These activities emphasize the accessibility of COD3 workflows, even when focusing on the analysis of large-scale datasets. A manuscript describing this approach is currently in preparation.
Discussion
- Interdisciplinary Curriculum: The CD2K program integrates key concepts from genomics, bioinformatics, and immunology, as well as knowledge discovery and dissemination. This interdisciplinary approach provides trainees with a comprehensive understanding of how to translate omics data into actionable biomedical insights [45]. Unlike programs that focus solely on computational skills, CD2K emphasizes the biological context and translational potential of data analysis.
- Advanced Technology Integration: The recent incorporation of Large Language Models (LLMs) into the curriculum, particularly in the COD1 module, represents a significant advancement in biomedical data analysis training [35]. LLMs offer powerful tools for literature mining, hypothesis generation, and even candidate gene prioritization. By introducing trainees to these cutting-edge technologies, CD2K prepares them for the future of data-driven biomedical research.
- Publication as a Training Endpoint: A unique feature of CD2K is its emphasis on publication as a training outcome. This approach aligns practical training with tangible academic contributions, addressing a critical need for career advancement in research [46]. It provides trainees, especially those from LMICs, with opportunities to build their publication record and contribute to the global scientific discourse.
- Hands-On Training with Real-World Datasets: CD2K offers hands-on experience with actual research datasets, enhancing the practical learning experience. This approach bridges the gap between theoretical knowledge and real-world application, a challenge often faced in traditional bioinformatics training programs [47].
- Enhancing Data-to-Knowledge Translation: The program builds on traditional data science skills by teaching how to interpret complex datasets within broader scientific contexts. This focus on knowledge translation is crucial for developing actionable biomedical insights [48].
- Accessibility and Inclusivity: While the program includes advanced data analysis techniques, it is designed to be accessible to researchers without extensive computational backgrounds. This inclusivity is particularly valuable for engaging a broader range of biomedical researchers, including those from LMICs or with primarily wet-lab experience [49].
Limitations and Future Directions
References
- Chaussabel, D.; Rinchai, D. Using 'collective omics data' for biomedical research training. Immunology. 2018, 155, 18–23. [Google Scholar] [CrossRef]
- Toufiq, M.; Rinchai, D.; Bettacchioli, E.; et al. Harnessing large language models (LLMs) for candidate gene prioritization and selection. J Transl Med. 2023, 21, 728. [Google Scholar] [CrossRef]
- Preechanukul, A.; Yimthin, T.; Tandhavanant, S.; et al. Abundance of ACVR1B transcript is elevated during septic conditions: Perspectives obtained from a hands-on reductionist investigation. Front Immunol. 2023, 14, 1072732. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.S.Y.; Rinchai, D.; Toufiq, M.; et al. Transcriptomic profile investigations highlight a putative role for NUDT16 in sepsis. J Cell Mol Med. 2022, 26, 1714–1721. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.S.Y.; Toufiq, M.; Saraiva, L.R.; et al. Transcriptome and Literature Mining Highlight the Differential Expression of ERLIN1 in Immune Cells during Sepsis. Biology (Basel). 2021, 10, 755. [Google Scholar] [CrossRef] [PubMed]
- Toufiq, M.; Roelands, J.; Alfaki, M.; et al. Annexin A3 in sepsis: Novel perspectives from an exploration of public transcriptome data. Immunology. 2020, 161, 291–302. [Google Scholar] [CrossRef]
- Roelands, J.; Garand, M.; Hinchcliff, E.; et al. Long-Chain Acyl-CoA Synthetase 1 Role in Sepsis and Immunity: Perspectives From a Parallel Review of Public Transcriptome Datasets and of the Literature. Front Immunol. 2019, 10, 2410. [Google Scholar]
- Sawyer, A.J.; Garand, M.; Chaussabel, D.; Feng, C.G. Transcriptomic Profiling Identifies Neutrophil-Specific Upregulation of Cystatin F as a Marker of Acute Inflammation in Humans. Front Immunol. 2021, 12, 634119. [Google Scholar] [CrossRef]
- Le Berre, L.; Chesneau, M.; Danger, R.; et al. Connection of BANK1, Tolerance, Regulatory B cells, and Apoptosis: Perspectives of a Reductionist Investigation. Front Immunol. 2021, 12, 589786. [Google Scholar]
- Riyapa, D.; Rinchai, D.; Muangsombut, V.; et al. Transketolase and vitamin B1 influence on ROS-dependent neutrophil extracellular traps (NETs) formation. PLoS ONE. 2019, 14, e0221016. [Google Scholar] [CrossRef]
- Rinchai, D.; Kewcharoenwong, C.; Kessler, B.; et al. Increased abundance of ADAM9 transcripts in the blood is associated with tissue damage. F1000Res. 2015, 4, 89. [Google Scholar] [CrossRef] [PubMed]
- Chaussabel, D.; Baldwin, N. Democratizing systems immunology with modular transcriptional repertoire analyses. Nat Rev Immunol. 2014, 14, 271–280. [Google Scholar] [CrossRef]
- Barrett, T.; Wilhite, S.E.; Ledoux, P.; et al. NCBI GEO: Archive for functional genomics data sets--update. Nucleic Acids Res. 2013, 41, D991–D995. [Google Scholar] [CrossRef] [PubMed]
- Speake, C.; Presnell, S.; Domico, K.; et al. An interactive web application for the dissemination of human systems immunology data. J Transl Med. 2015, 13, 196. [Google Scholar] [CrossRef]
- Bettacchioli, E.; Chiche, L.; Chaussabel, D.; et al. An interactive web application for exploring systemic lupus erythematosus blood transcriptomic diversity. Database (Oxford). 2024, 2024, baae045. [Google Scholar] [CrossRef] [PubMed]
- Rinchai, D.; Brummaier, T.; Marr, A.A.; et al. A data browsing application for accessing gene and module-level blood transcriptome profiles of healthy pregnant women from high- and low-resource settings. Database (Oxford). 2024, 2024, baae021. [Google Scholar] [CrossRef]
- Bougarn, S.; Boughorbel, S.; Chaussabel, D.; Marr, N. A curated transcriptome dataset collection to investigate inborn errors of immunity. F1000Res. 2019, 8, 188. [Google Scholar] [CrossRef]
- Huang, S.S.Y.; Al Ali, F.; Boughorbel, S.; et al. A curated collection of transcriptome datasets to investigate the molecular mechanisms of immunoglobulin E-mediated atopic diseases. Database (Oxford). 2019, 2019, baz066. [Google Scholar] [CrossRef]
- Bougarn, S.; Boughorbel, S.; Chaussabel, D.; Marr, N. A curated transcriptome dataset collection to investigate the blood transcriptional response to viral respiratory tract infection and vaccination. F1000Res. 2019, 8, 284. [Google Scholar] [CrossRef]
- Roelands, J.; Decock, J.; Boughorbel, S.; et al. A collection of annotated and harmonized human breast cancer transcriptome datasets, including immunologic classification. F1000Res. 2017, 6, 296. [Google Scholar] [CrossRef]
- Mackeh, R.; Boughorbel, S.; Chaussabel, D.; Kino, T. A curated transcriptomic dataset collection relevant to embryonic development associated with in vitro fertilization in healthy individuals and patients with polycystic ovary syndrome. F1000Res. 2017, 6, 181. [Google Scholar] [CrossRef] [PubMed]
- Rahman, M.; Boughorbel, S.; Presnell, S.; et al. A curated transcriptome dataset collection to investigate the functional programming of human hematopoietic cells in early life. F1000Res. 2016, 5, 414. [Google Scholar] [CrossRef] [PubMed]
- Marr, A.K.; Boughorbel, S.; Presnell, S.; et al. A curated transcriptome dataset collection to investigate the development and differentiation of the human placenta and its associated pathologies. F1000Res. 2016, 5, 305. [Google Scholar] [CrossRef] [PubMed]
- Rinchai, D.; Boughorbel, S.; Presnell, S.; et al. A curated compendium of monocyte transcriptome datasets of relevance to human monocyte immunobiology research. F1000Res. 2016, 5, 291. [Google Scholar] [CrossRef] [PubMed]
- Blazkova, J.; Boughorbel, S.; Presnell, S.; et al. A curated transcriptome dataset collection to investigate the immunobiology of HIV infection. F1000Res. 2016, 5, 327. [Google Scholar] [CrossRef]
- Toufiq, M.; Huang, S.S.Y.; Boughorbel, S.; et al. SysInflam HuDB, a Web Resource for Mining Human Blood Cells Transcriptomic Data Associated with Systemic Inflammatory Responses to Sepsis. J Immunol. 2021, 207, 2195–2202. [Google Scholar] [CrossRef]
- Altman, M.C.; Rinchai, D.; Baldwin, N.; et al. Development of a fixed module repertoire for the analysis and interpretation of blood transcriptome data. Nat Commun. 2021, 12, 4385. [Google Scholar] [CrossRef]
- Rinchai, D.; Roelands, J.; Toufiq, M.; et al. BloodGen3Module: Blood transcriptional module repertoire analysis and visualization using R. Bioinformatics. 2021, 37, 2382–2389. [Google Scholar] [CrossRef]
- Bettacchioli, E.; Chiche, L.; Chaussabel, D.; et al. An interactive web application for exploring systemic lupus erythematosus blood transcriptomic diversity. Database (Oxford). 2024, 2024, baae045. [Google Scholar] [CrossRef]
- Rinchai, D.; Brummaier, T.; AMarr, A.; et al. A data browsing application for accessing gene and module-level blood transcriptome profiles of healthy pregnant women from high- and low-resource settings. Database (Oxford). 2024, 2024, baae021. [Google Scholar] [CrossRef]
- Rawat, A.; Rinchai, D.; Toufiq, M.; et al. A Neutrophil-Driven Inflammatory Signature Characterizes the Blood Transcriptome Fingerprint of Psoriasis. Front Immunol. 2020, 11, 587946. [Google Scholar] [CrossRef]
- Rinchai, D.; Altman, M.C.; Konza, O.; et al. Definition of erythroid cell-positive blood transcriptome phenotypes associated with severe respiratory syncytial virus infection. Clin Transl Med. 2020, 10, e244. [Google Scholar] [CrossRef] [PubMed]
- Brummaier, T.; Rinchai, D.; Toufiq, M.; et al. Design of a targeted blood transcriptional panel for monitoring immunological changes accompanying pregnancy. Front Immunol. 2024, 15, 1319949. [Google Scholar] [CrossRef]
- Rinchai, D.; Syed Ahamed Kabeer, B.; Toufiq, M.; et al. A modular framework for the development of targeted Covid-19 blood transcript profiling panels. J Transl Med. 2020, 18, 291. [Google Scholar] [CrossRef] [PubMed]
- Toufiq, M.; Rinchai, D.; Bettacchioli, E.; et al. Harnessing large language models (LLMs) for candidate gene prioritization and selection. J Transl Med. 2023, 21, 728. [Google Scholar] [CrossRef] [PubMed]
- Chaussabel, D.; Pulendran, B. A vision and a prescription for big data-enabled medicine. Nat Immunol. 2015, 16, 435–439. [Google Scholar] [CrossRef]
- Garmire, L.X.; Gliske, S.; Nguyen, Q.C.; et al. The training of next generation data scientists in biomedicine. Pac Symp Biocomput. 2017, 22, 640–645. [Google Scholar]
- Byrd, J.B.; Greene, A.C.; Prasad, D.V.; Jiang, X.; Greene, C.S. Responsible, practical genomic data sharing that accelerates research. Nat Rev Genet. 2020, 21, 615–629. [Google Scholar] [CrossRef] [PubMed]
- Bourne, P.E.; Bonazzi, V.; Dunn, M.; et al. The NIH Big Data to Knowledge (BD2K) initiative. J Am Med Inform Assoc. 2015, 22, 1114–1120. [Google Scholar] [CrossRef]
- Hancock, J.M.; Zvelebil, M.; Hollich, V.; et al. ELIXIR-UK role in bioinformatics training at the national level and across ELIXIR. F1000Res. 2016, 5, ELIXIR-952. [Google Scholar]
- Mulder, N.J.; Adebiyi, E.; Alami, R.; et al. H3ABioNet, a sustainable pan-African bioinformatics network for human heredity and health in Africa. Genome Res. 2016, 26, 271–277. [Google Scholar] [CrossRef] [PubMed]
- Welch, L.; Lewitter, F.; Schwartz, R.; et al. Bioinformatics Curriculum Guidelines: Toward a Definition of Core Competencies. PLoS Comput Biol. 2014, 10, e1003496. [Google Scholar] [CrossRef] [PubMed]
- Shaikh, A.R.; Butte, A.J.; Schully, S.D.; et al. Collaborative Biomedicine in the Age of Big Data: The case of cancer. J Med Internet Res. 2014, 16, e101. [Google Scholar] [CrossRef]
- Payne, P.R.O. Biomedical informatics meets data science: Current state and future directions for interaction. JAMIA Open 2018, 1, 136–141. [Google Scholar] [CrossRef]
- Chaussabel, D.; Baldwin, N. Democratizing systems immunology with modular transcriptional repertoire analyses. Nat Rev Immunol. 2014, 14, 271–280. [Google Scholar] [CrossRef]
- Bourne, P.E. Ten simple rules for making good oral presentations. PLoS Comput Biol. 2007, 3, e77. [Google Scholar] [CrossRef]
- Attwood, T.K.; Blackford, S.; Brazas, M.D.; et al. A global perspective on evolving bioinformatics and data science training needs. Brief Bioinform. 2019, 20, 398–404. [Google Scholar] [CrossRef]
- Strasser, B.J.; Edwards, P.M. Big data is the answer. .. but what is the question? Osiris. 2017, 32, 328–345. [Google Scholar]
- Mulder, N.; Schwartz, R.; Brazas, M.D.; et al. The development and application of bioinformatics core competencies to improve bioinformatics training and education. PLoS Comput Biol. 2018, 14, e1005772. [Google Scholar] [CrossRef]
- Al Ali, F.; Marr, A.K.; Tatari-Calderone, Z.; et al. Organizing training workshops on gene literature retrieval, profiling, and visualization for early career researchers. F1000Res. 2023, 10, 275. [Google Scholar] [CrossRef]
- Rinchai, D.; Chaussabel, D. A training curriculum for retrieving, structuring, and aggregating information derived from the biomedical literature and large-scale data repositories [version 1; peer review: 2 approved with reservations]. F1000Research. 2022, 11, 994. [Google Scholar] [CrossRef]
- Rinchai, D.; Chaussabel, D. Assessing the potential relevance of CEACAM6 as a blood transcriptional biomarker [version 2; peer review: 1 approved, 1 approved with reservations]. F1000Research. 2024, 11, 1294. [Google Scholar] [CrossRef] [PubMed]
| Gene | Number of Participants | Affiliations | Theme | Citation (PMID) |
| ALAS2 | 7 | The Jackson Laboratory, The Rockefeller University, INSERM, Sidra Medicine, University of Bretagne Occidentale, CHU de Brest | Erythroid cells | 37845713 |
| ACVR1B | 5 | Mahidol University, Sidra Medicine, University of Washington | Sepsis | 37020544 |
| NUDT16 | 6 | Sidra Medicine, Washington University School of Medicine | Sepsis | 35174610 |
| ERLIN1 | 6 | Sidra Medicine | Sepsis | 34439987 |
| CST7 | 4 | University of Sydney, Sidra Medicine | Acute inflammation | 33868254 |
| BANK1 | 6 | University of Nantes, Sidra Medicine | B cells, tolerance | 33815360 |
| ANXA3 | 10 | Sidra Medicine | Sepsis | 32682335 |
| ACSL1 | 11 | Sidra Medicine, The University of Texas MD Anderson Cancer Center | Sepsis | 31681299 |
| TKT | 9 | Mahidol University, Sidra Medicine, National Institute of Infectious Diseases (Japan), University of Western Australia | Inflammation | 31415630 |
| ADAM9 | 5 | Khon Kaen University, Sidra Medicine | Tissue damage | 27990250 |
| Participants | Theme | Datasets | Unique Profiles | PMID | Web Link |
|---|---|---|---|---|---|
| 6 | Systemic Lupus Erythematosus | 1 | 157 | 38805754 | https://immunology-research.shinyapps.io/LUPUCE/ |
| 11 | Pregnancy | 2+ | 15+ | 38564425 | https://thejacksonlaboratory.shinyapps.io/BloodGen3_Pregnancy/ |
| 4 | Primary Immunodeficiencies | 18 | Not specified | 31559014 | http://pid.gxbsidra.org/dm3/geneBrowser/list |
| 6 | IgE-mediated Atopic Diseases | 33 | 1860 | 31290545 | http://ige.gxbsidra.org/dm3/geneBrowser/list |
| 4 | Viral Respiratory Tract Infection and Vaccination | 31 | 6648 | 31231515 | http://vri1.gxbsidra.org/dm3/geneBrowser/list |
| 18 | Breast Cancer | 13 | 2142 | 29527288 | http://breastcancer.gxbsidra.org/dm3/geneBrowser/list |
| 4 | In Vitro Fertilization and Polycystic Ovary Syndrome | 12 | 85 | 28413616 | http://ivf.gxbsidra.org/dm3/landing.gsp |
| 7 | Hematopoietic Cells in Early Life | 32 | 2129 | 27347375 | http://developmentalimmunology.gxbsidra.org/dm3/geneBrowser/list |
| 6 | Placenta Development and Pathologies | 24 | 759 | 27303626 | http://placentalendocrinology.gxbsidra.org/dm3/landing.gsp |
| 5 | Human Monocyte Immunobiology | 93 | 4516 | 27158452 | http://monocyte.gxbsidra.org/dm3/landing.gsp |
| 5 | HIV Infection | 34 | 2717 | 27134731 | http://hiv.gxbsidra.org/dm3/geneBrowser/list |
| 8 | Systemic Inflammatory Responses to Sepsis | 62 | 5719 | 34663591 | http://sepsis.gxbsidra.org/dm3/geneBrowser/list |
| PMID | Title | Participants | Theme | Resource /Training |
Key Aspects |
|---|---|---|---|---|---|
| 34282143 | Development of a fixed module repertoire for the analysis and interpretation of blood transcriptome data | 37 | Development of BloodGen3 module repertoire | Resource | Fixed module repertoire for blood transcriptome analysis |
| 33624743 | BloodGen3Module: blood transcriptional module repertoire analysis and visualization using R | 7 | Development of R package for BloodGen3 | Resource | R package for module-level analysis and visualization |
| 33329570 | A Neutrophil-Driven Inflammatory Signature Characterizes the Blood Transcriptome Fingerprint of Psoriasis | 13 | Meta-analysis of psoriasis blood transcriptome | Training | Module-level meta-analysis for pathogenesis investigation |
| 33377660 | Definition of erythroid cell-positive blood transcriptome phenotypes associated with severe respiratory syncytial virus infection | 14 | Meta-analysis of RSV blood transcriptome | Training | Module-level meta-analysis for biomarker discovery |
| 38352867 | Design of a targeted blood transcriptional panel for monitoring immunological changes accompanying pregnancy | 14 | Development of pregnancy-specific blood transcriptome panel | Training | Data and knowledge-driven biomarker discovery |
| 32736569 | A modular framework for the development of targeted Covid-19 blood transcript profiling panels | 18 | Development of COVID-19 blood transcriptome panels | Training | Data and knowledge-driven biomarker discovery |
| 38805754 | An interactive web application for exploring systemic lupus erythematosus blood transcriptomic diversity | 6 | SLE blood transcriptome data browsing application | Resource | Web application for module-level data browsing |
| 38564425 | A data browsing application for accessing gene and module-level blood transcriptome profiles of healthy pregnant women from high- and low-resource settings | 11 | Pregnancy blood transcriptome data browsing application | Resource | Web application for module-level data browsing |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




