Submitted:
14 October 2024
Posted:
15 October 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
- This study aims to enhance transfer learning (TL) models to improve the detection of sugarcane leaf diseases. To achieve this, the TL models have been augmented with the incorporation of dense layers for regularization, batchnormalization layers, and dropout layers to prevent overfitting.
- A public dataset of sugarcane leaf diseases was used to compare five enhanced transfer learning (TL) models. The results showed a considerable improvement in each model’s test accuracy.
- A novel deep ensemble convolutional neural network (DECNN) model for the detection of sugarcane leaf diseases is proposed, utilizing a distinctive performance-based custom weighted ensemble method. The model achieves an accuracy of 99.17%, outperforming individual models in detection accuracy.
2. Related Work
3. Materials and Methods
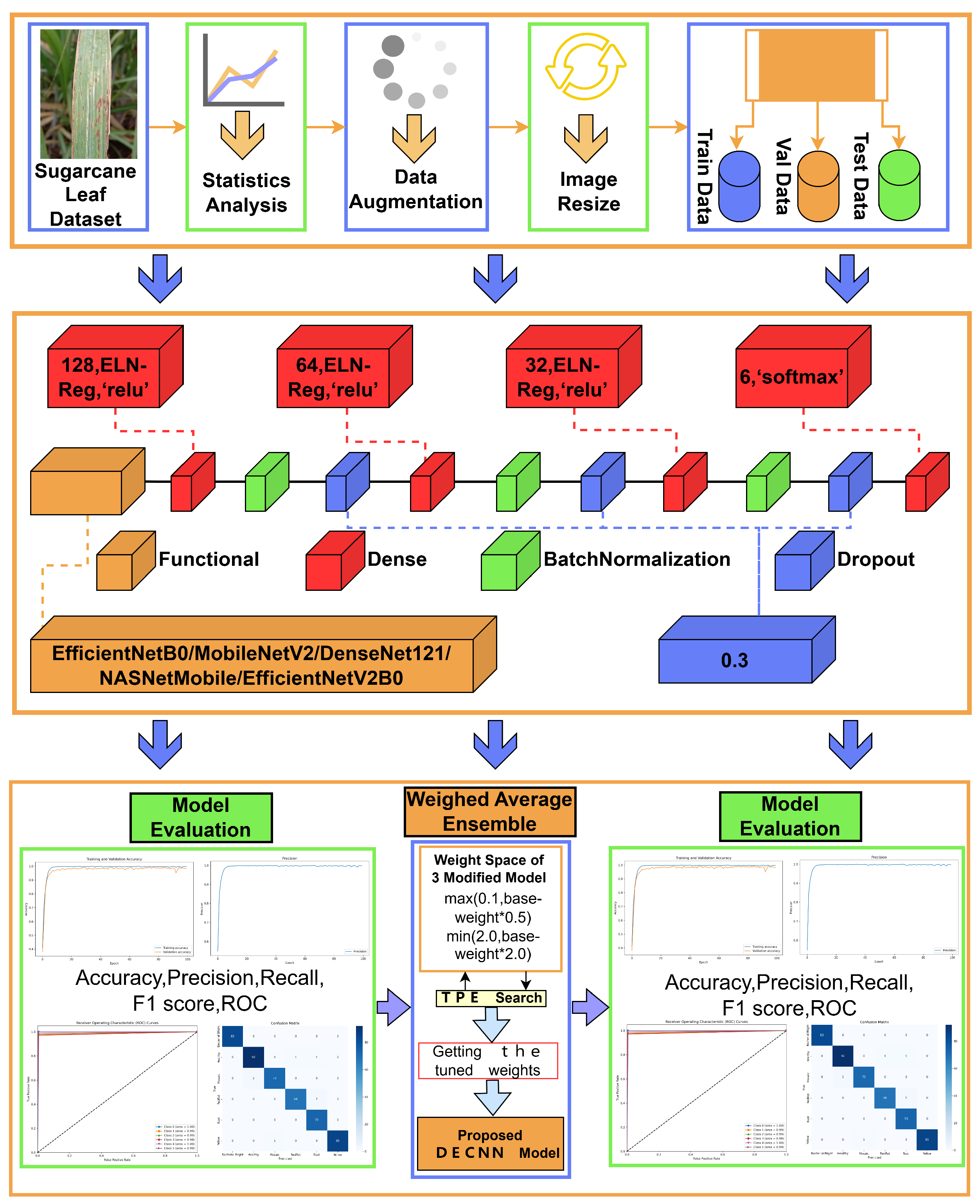
3.1. Dataset
3.2. Data Augmentation and Pre-Processing
3.3. Proposed DECNN Model
3.3.1. ELN-Reg Regularization
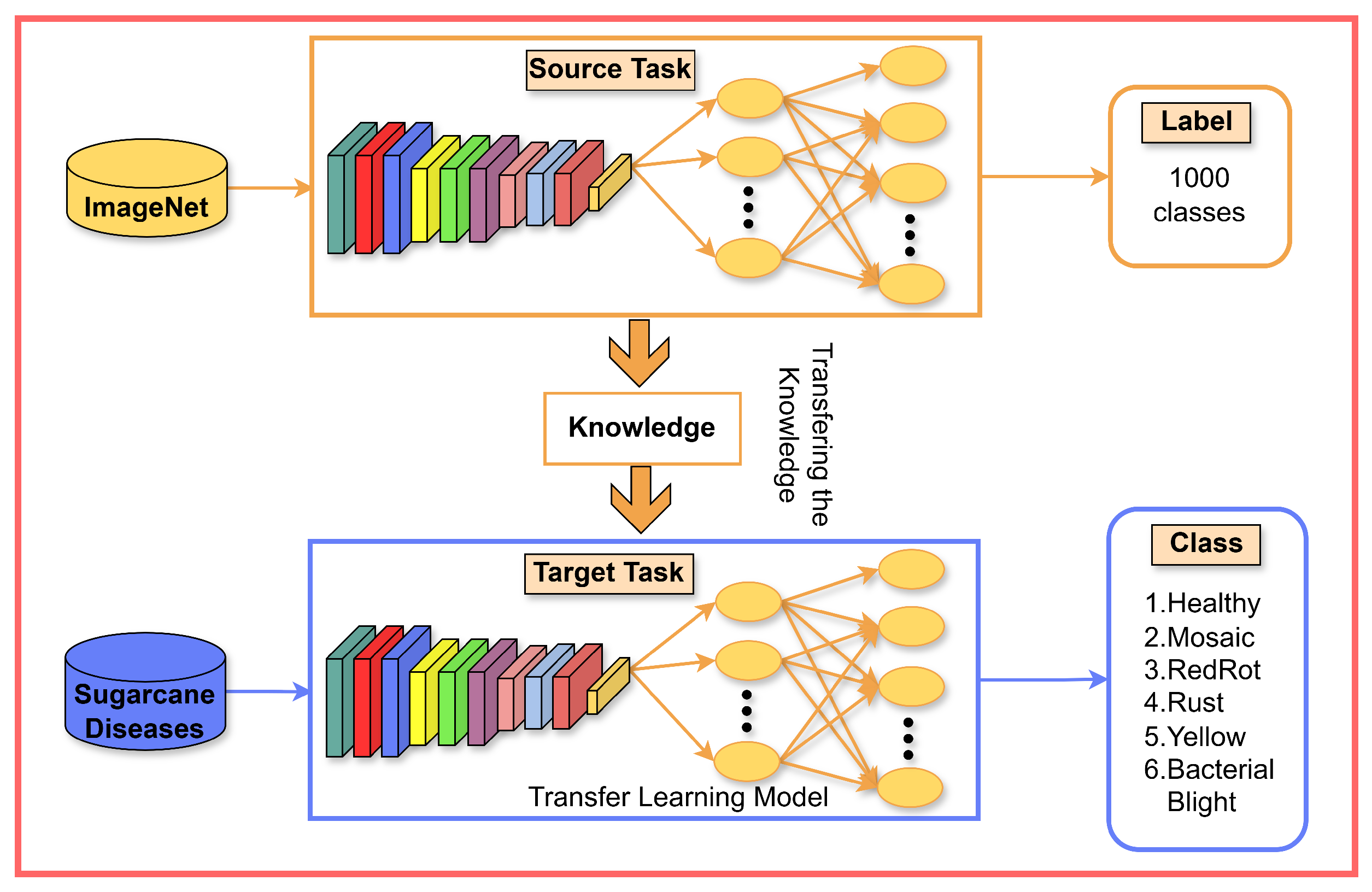
3.3.2. Modified Transfer Learning Models
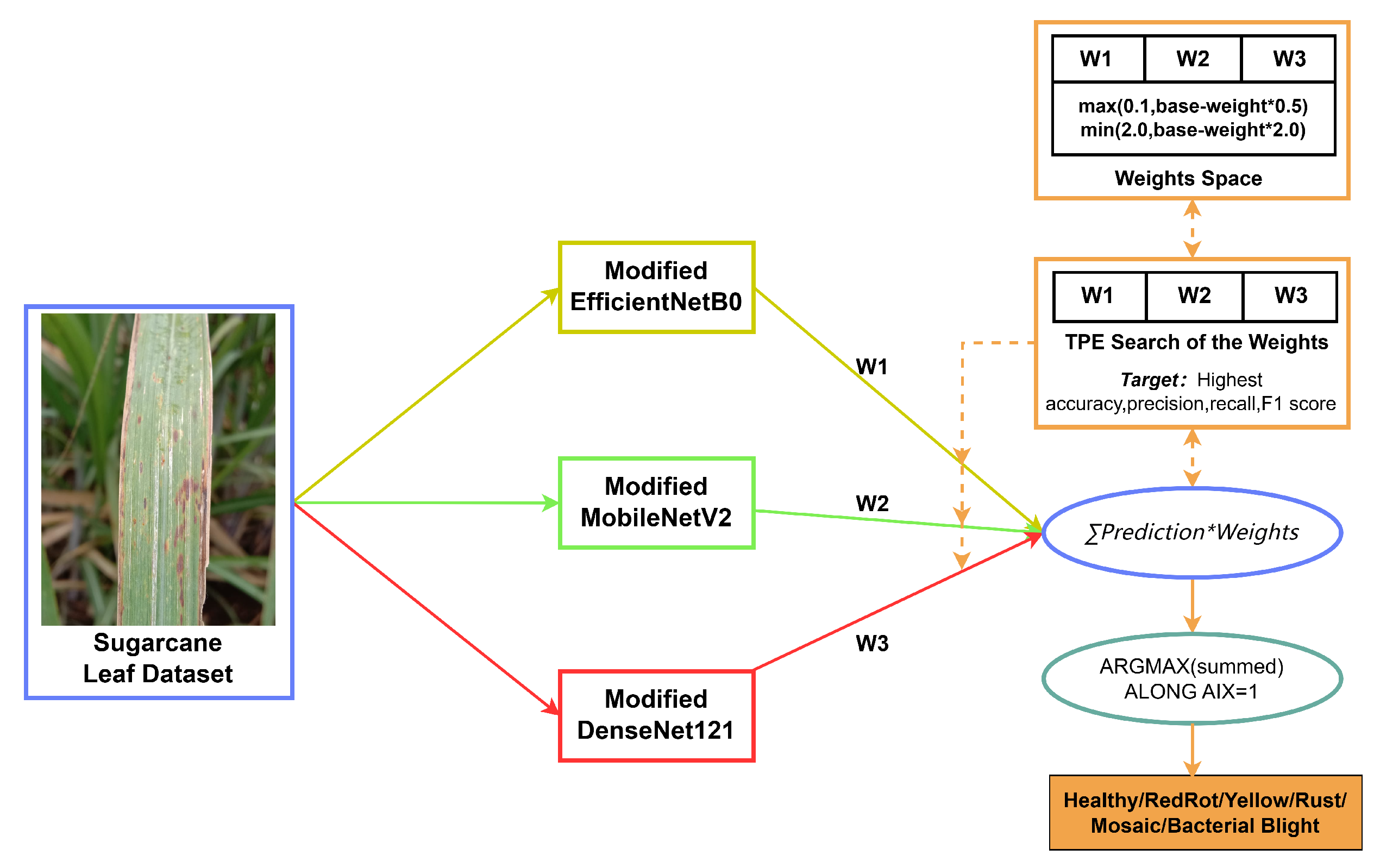
3.3.3. Ensemble Modified TL Model
| Algorithm 1: Weighted ensemble |
| Input: Test_set T, Models and Weight_set where k is the number of models |
| Output: |
| Ensemble_model |
| For do |
| Predict, |
| Confusion_matrix () |
| Classification_matrices () |
| End |
3.4. Model Performance Metrics
- Accuracy: the evaluation of a model heavily depends on the parameter of accuracy. The formula computes this ratio, which is the proportion of accurately anticipated data to all data:
- Precision: the proportion of correct predictions among the samples with positive predictions, as judged by the prediction results, calculated by the formula:
- Recall: the proportion of correctly predicted positive cases out of the total number of actual positive cases in the sample of actual positive cases, based on the judgment of the actual samples, which is calculated by the formula:
- F1 score: precision and recall are averaged together to get the F1 score. When comparing several models, it is computed as follows:
- Macro average: the arithmetic mean of every category linked to F1 score, precision, and recall is known as the macro average. It is determined by the following formula and is used to assess the multi-class classification’s overall effectiveness:
- Weighted average: a multi-category classification’s overall effectiveness can also be assessed using the weighted average. Using the following formula, it is determined as a weighted average for every category:
4. Results
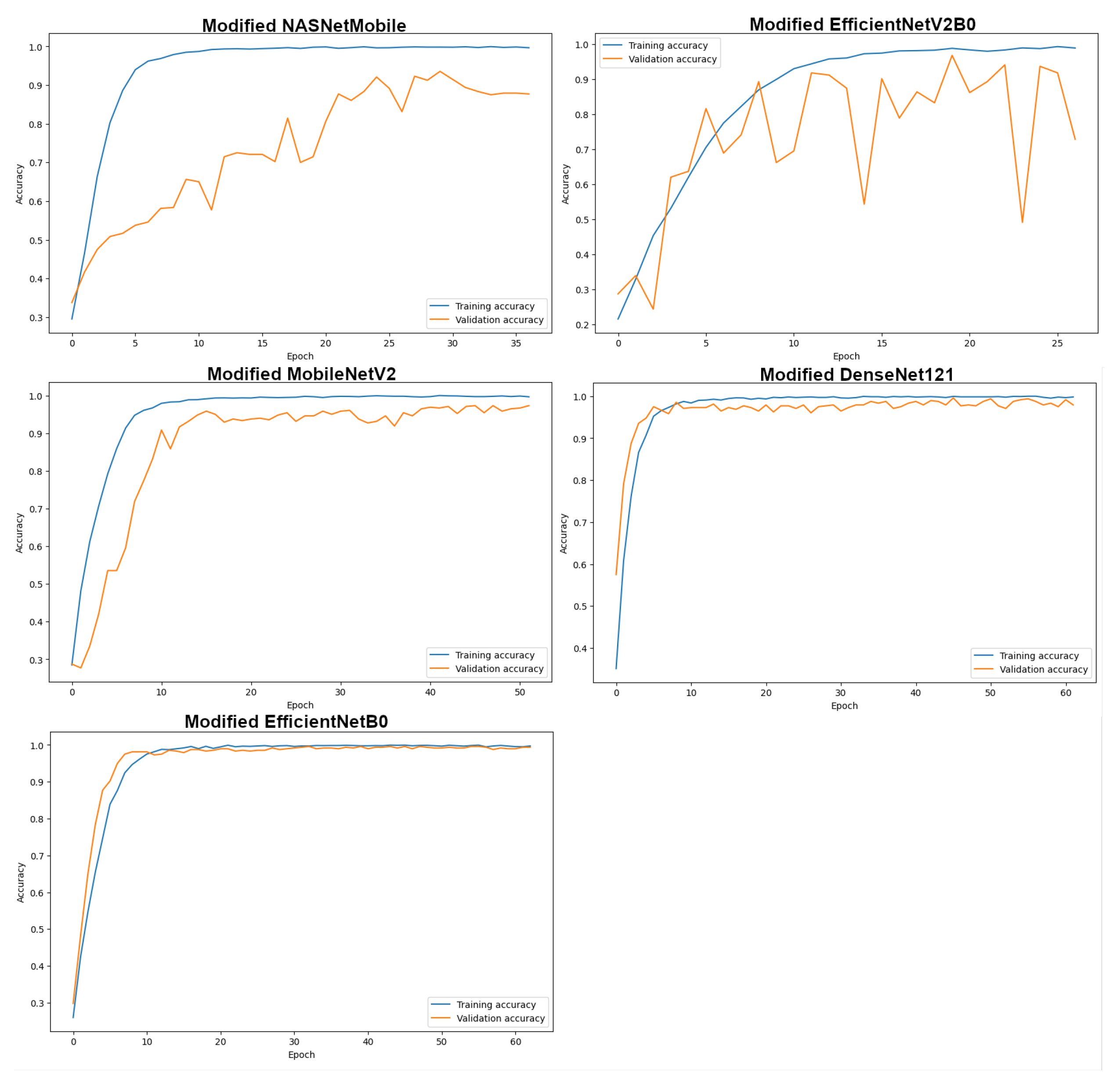
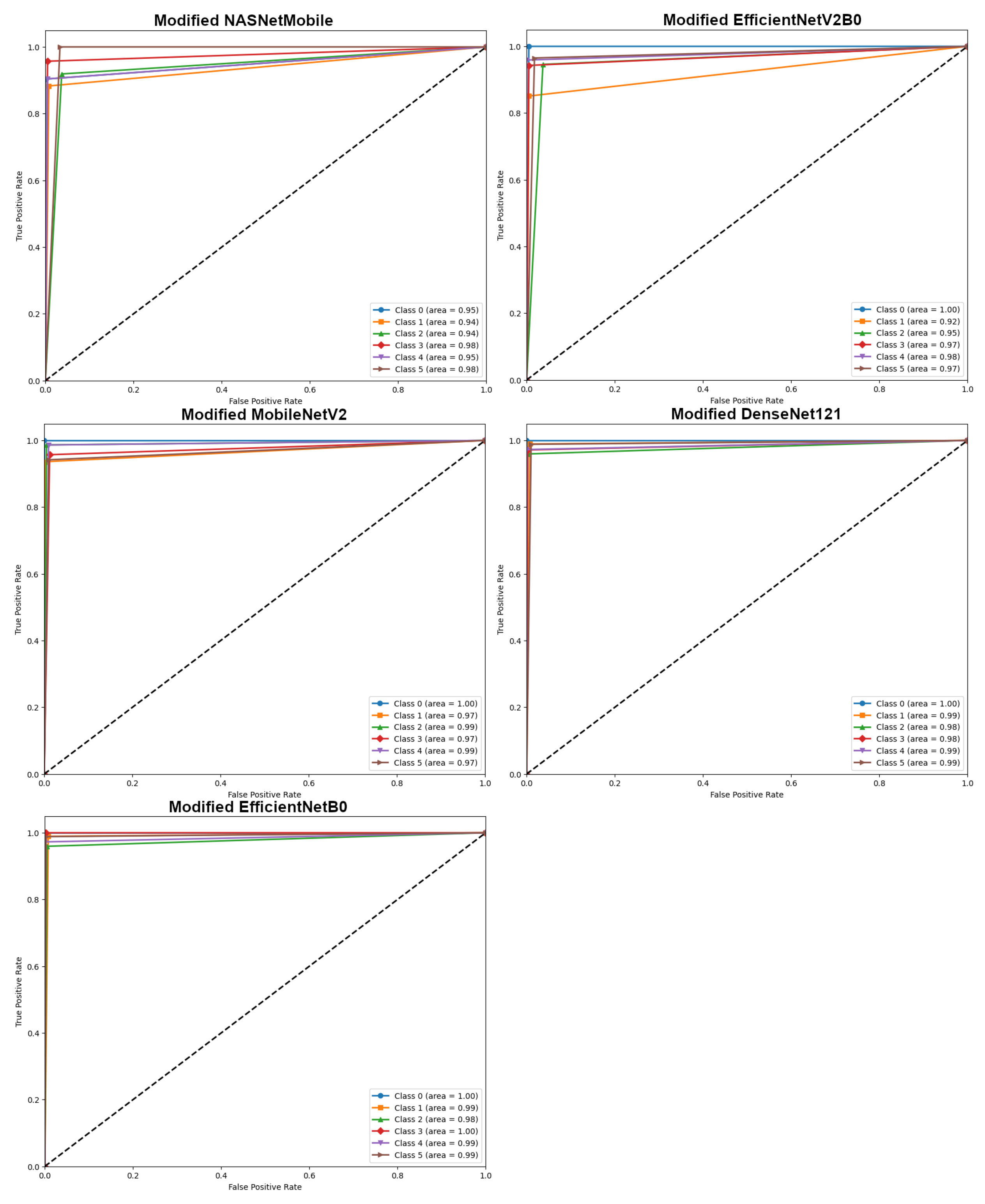
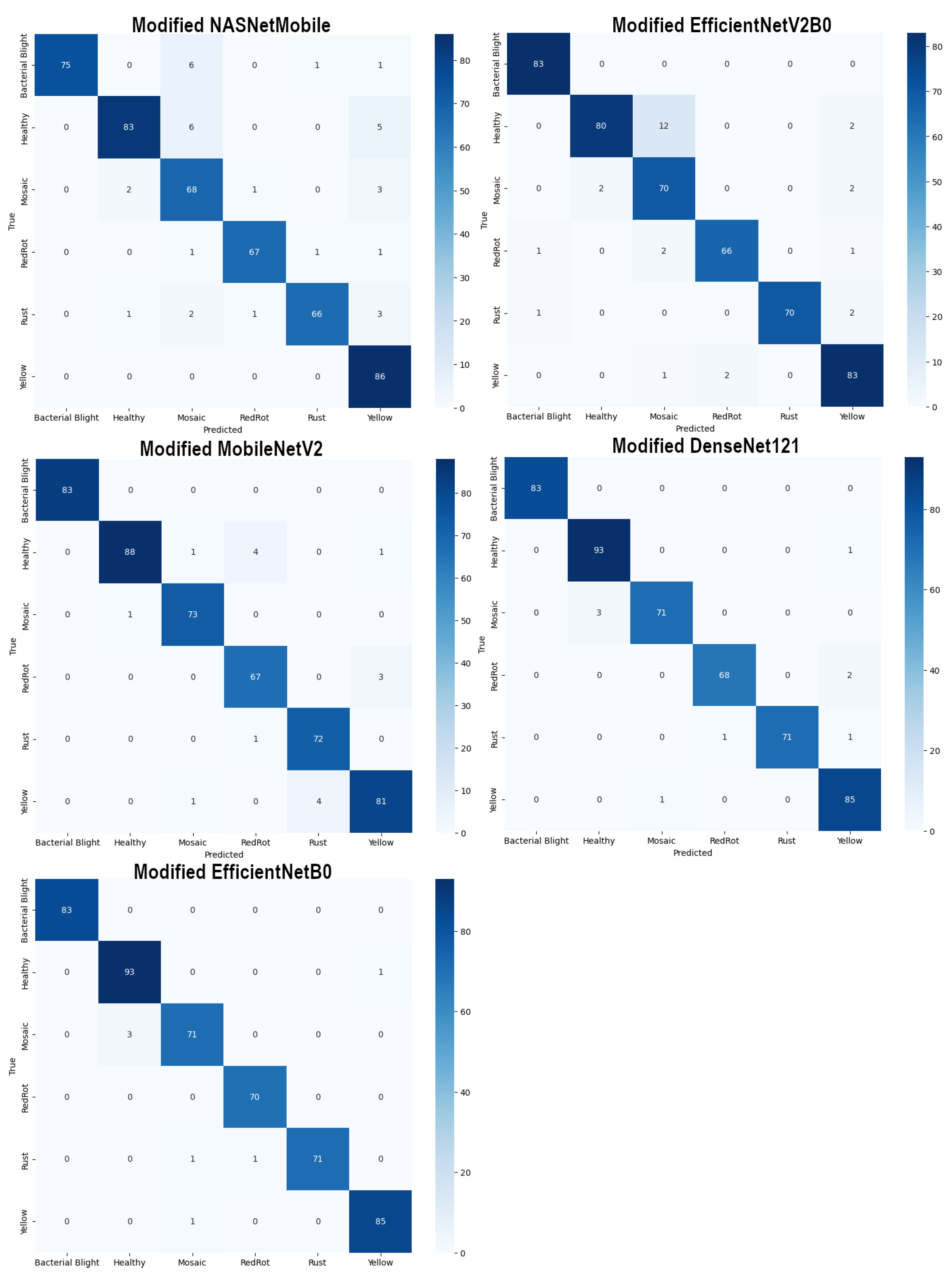
4.1. Results of Modified Transfer Learning models
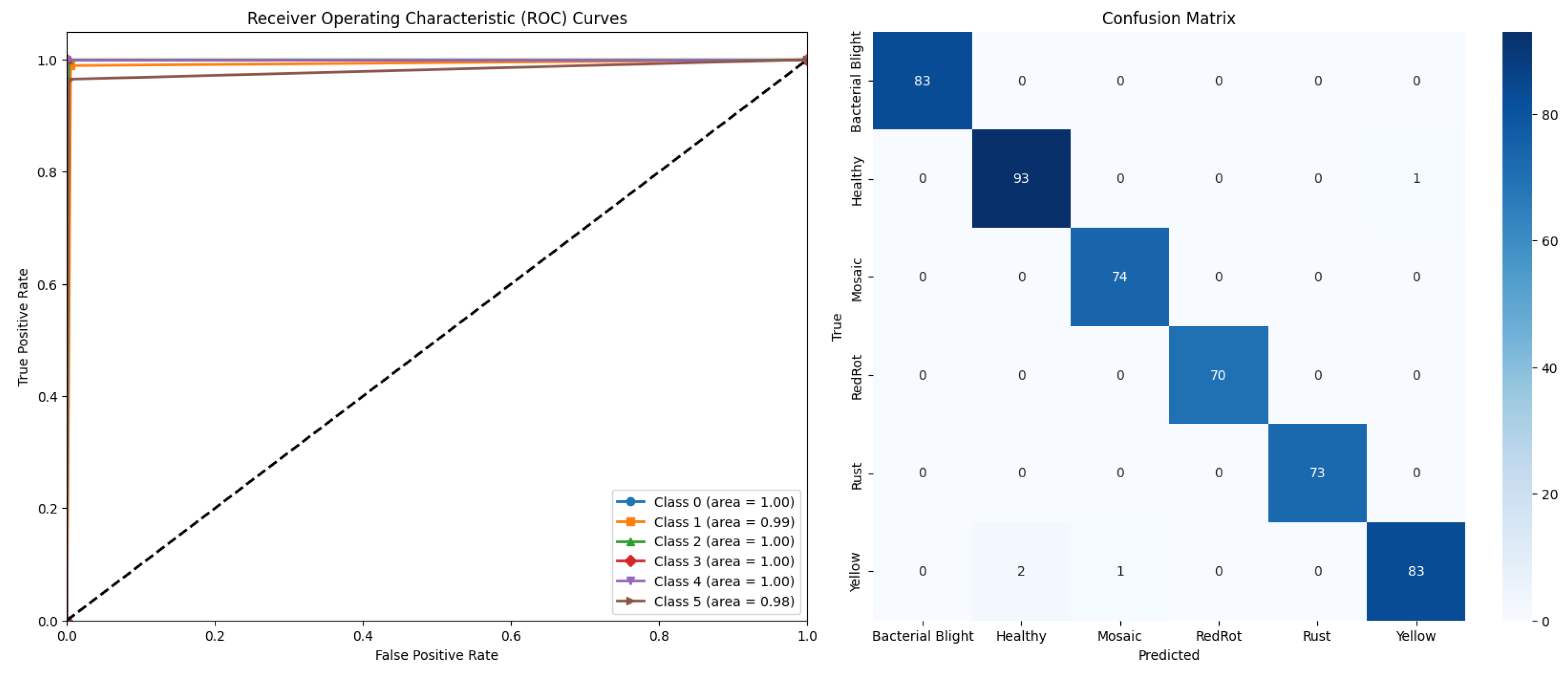
4.2. Results of Ensemble Modified TL Model DECNN
5. Discussion
6. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Rae, A.L.; Grof, C.P.L.; Casu, R.E.; Bonnett, G.D. Sucrose accumulation in the sugarcane stem: pathways and control points for transport and compartmentation. Field Crops Research 2005, 92, 159–168. [Google Scholar] [CrossRef]
- Ali, A.; Khan, M.; Sharif, R.; Mujtaba, M.; Gao, S.-J. Sugarcane Omics: An Update on the Current Status of Research and Crop Improvement. Plants 2019, 8, 344. [Google Scholar] [CrossRef]
- Hemalatha, N.K.; Brunda, R.N.; Prakruthi, G.S.; Prabhu, B.V.B.; Shukla, A.; Narasipura, O.S.J. Sugarcane leaf disease detection through deep learning. In Deep Learning for Sustainable Agriculture; Poonia, R.C., Singh, V., Nayak, S.R., Eds.; Elsevier Academic Press: London, United Kingdom, 2022; pp. 297–323. [Google Scholar]
- Li, A.-M.; Liao, F.; Wang, M.; Chen, Z.-L.; Qin, C.-X.; Huang, R.-Q.; Verma, K.K.; Li, Y.-R.; Que, Y.-X.; Pan, Y.-Q.; et al. Transcriptomic and Proteomic Landscape of Sugarcane Response to Biotic and Abiotic Stressors. Int. J. Mol. Sci. 2023, 24, 8913. [Google Scholar] [CrossRef]
- Yadav, S.; Singh, S.R.; Bahadur, L.; et al. Sugarcane Trash Ash Affects Degradation and Bioavailability of Pesticides in Soils. Sugar Tech 2023, 25, 77–85. [Google Scholar] [CrossRef]
- Ali, M.M.; Bachik, N.A.; Muhadi, N.A.; Yusof, T.N.T.; Gomes, C. Non-destructive techniques of detecting plant diseases: A review. Physiological and Molecular Plant Pathology 2019, 108, 101426. [Google Scholar] [CrossRef]
- Lu, T.; Han, B.; Chen, L.; Yu, F.; Xue, C. A generic intelligent tomato classification system for practical applications using DenseNet-201 with transfer learning. Sci. Rep. 2021, 11, 15824. [Google Scholar] [CrossRef]
- Kouadio, L.; El Jarroudi, M.; Belabess, Z.; Laasli, S.-E.; Roni, M.Z.K.; Amine, I.D.I.; Mokhtari, N.; Mokrini, F.; Junk, J.; Lahlali, R. A Review on UAV-Based Applications for Plant Disease Detection and Monitoring. Remote Sens. 2023, 15, 4273. [Google Scholar] [CrossRef]
- Krishnamoorthy, N.; Prasad, L.V.N.; Kumar, C.S.P.; Subedi, B.; Abraha, H.B.; Sathiskumar, V.E. Rice leaf diseases prediction using deep neural networks with transfer learning. Environ. Res. 2021, 198, 111275. [Google Scholar]
- Shorten, C.; Khoshgoftaar, T.M. A survey on Image Data Augmentation for Deep Learning. Journal of Big Data 2019, 6, 60. [Google Scholar] [CrossRef]
- Ayana, G.; Dese, K.; Choe, S.-w. Transfer Learning in Breast Cancer Diagnoses via Ultrasound Imaging. Cancers 2021, 13, 738. [Google Scholar] [CrossRef]
- Yigit, E.; Sabanci, K.; Toktas, A.; Kayabasi, A. A study on visual features of leaves in plant identification using artificial intelligence techniques. Comput. Electron. Agric. 2019, 156, 369–377. [Google Scholar] [CrossRef]
- Basavaiah, J.; Anthony, A.A. Tomato leaf disease classification using multiple feature extraction techniques. Wirel. Pers. Commun. 2020, 115, 633–651. [Google Scholar] [CrossRef]
- Chanda, M.; Biswas, M. Plant disease identification and classification using Back-Propagation Neural Network with Particle Swarm Optimization. In Proceedings of the 2019 3rd International Conference on Trends in Electronics and Informatics (ICOEI), Tirunelveli, India, 23-25 April 2019; pp. 1029–1036. [Google Scholar]
- Hossain, E.; Hossain, M.F.; Rahaman, M.A. A color and texture based approach for the detection and classification of plant leaf disease using KNN classifier. In Proceedings of the 2019 International Conference on Electrical, Computer and Communication Engineering (ECCE), Cox’s Bazar, Bangladesh, 7-9 February 2019; pp. 1–6. [Google Scholar]
- Wu, Y.; Feng, X.; Chen, G. Plant Leaf Diseases Fine-Grained Categorization Using Convolutional Neural Networks. IEEE Access 2022, 10, 41087–41096. [Google Scholar] [CrossRef]
- Patil, R.R.; Kumar, S. Rice-Fusion: A Multimodality Data Fusion Framework for Rice Disease Diagnosis. IEEE Access 2022, 10, 5207–5222. [Google Scholar] [CrossRef]
- Elfatimi, E.; Eryigit, R.; Elfatimi, L. Beans Leaf Diseases Classification Using MobileNet Models. IEEE Access 2022, 10, 9471–9482. [Google Scholar] [CrossRef]
- Rahaman Yead, M.A.; Rukhsara, L.; Rabeya, T.; Jahan, I. Deep Learning-Based Classification of Sugarcane Leaf Disease. In Proceedings of the 2024 6th International Conference on Electrical Engineering and Information & Communication Technology (ICEEICT), Dhaka, Bangladesh, 2–4 May 2024; pp. 818–823. [Google Scholar]
- Bagchi, A.K.; Haider Chowdhury, M.A.; Fattah, S.A. Sugarcane Disease Classification using Advanced Deep Learning Algorithms: A Comparative Study. In Proceedings of the 2024 6th International Conference on Electrical Engineering and Information & Communication Technology (ICEEICT), Dhaka, Bangladesh, 2–4 May 2024; pp. 887–892. [Google Scholar]
- Pham, T.N.; Tran, L.V.; Dao, S.V.T. Early Disease Classification of Mango Leaves Using Feed-Forward Neural Network and Hybrid Metaheuristic Feature Selection. IEEE Access 2020, 8, 189960–189973. [Google Scholar] [CrossRef]
- Sun, C.; Zhou, X.; Zhang, M.; Qin, A. SE-VisionTransformer: Hybrid Network for Diagnosing Sugarcane Leaf Diseases Based on Attention Mechanism. Sensors 2023, 23, 8529. [Google Scholar] [CrossRef]
- Daphal, S.D.; Koli, S.M. Enhancing sugarcane disease classification with ensemble deep learning: A comparative study with transfer learning techniques. Heliyon 2023, 9, e18261. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S. Five Crop Diseases Dataset. Available online: https://www.kaggle.com/datasets/shubham2703/five-crop-diseases-dataset (accessed on 10 August 2024).
- Sankalana, N. Sugarcane Leaf Disease Dataset. Available online: https://www.kaggle.com/datasets/nirmalsankalana/sugarcane-leaf-disease-dataset (accessed on 10 August 2024).
- Ali, A.; Ali, S.; Husnain, M.; Missen, M.M.S.; Samad, A.; Khan, M. Detection of Deficiency of Nutrients in Grape Leaves Using Deep Network. Mathematical Problems in Engineering 2022, 2022, 1–12. [Google Scholar] [CrossRef]
- Bansal, P.; Kumar, R.; Kumar, S. Disease detection in Apple leaves using deep convolutional neural network. Agriculture 2021, 11, 617. [Google Scholar] [CrossRef]
- Fan, C.; Chen, M.; Wang, X.; Wang, J.; Huang, B. A Review on Data Preprocessing Techniques Toward Efficient and Reliable Knowledge Discovery From Building Operational Data. Front. Energy Res. 2021, 9, 652801. [Google Scholar] [CrossRef]
- Zou, H.; Hastie, T. Regularization and Variable Selection Via the Elastic Net. Journal of the Royal Statistical Society Series B: Statistical Methodology 2005, 67, 301–320. [Google Scholar] [CrossRef]
- Dialameh, M.; Hamzeh, A.; Rahmani, H.; Dialameh, H.; Kwon, H.J. DL-Reg: A deep learning regularization technique using linear regression. Expert Systems with Applications 2024, 247, 123182. [Google Scholar] [CrossRef]
- Akiba, T.; Sano, S.; Yanase, T.; Ohta, T.; Koyama, M. Optuna: A Next-generation Hyperparameter Optimization Framework. In Proceedings of the 25th ACM SIGKDD International Conference on Knowledge Discovery & Data Mining (KDD ’19), Anchorage, AK, USA, 4-8 August 2019; pp. 2623–2631. [Google Scholar]
- Hosna, A.; Merry, E.; Gyalmo, J.; Alom, Z.; Aung, Z.; Azim, M.A. Transfer learning: a friendly introduction. Journal of Big Data 2022, 9, 102. [Google Scholar] [CrossRef] [PubMed]
- Neyshabur, B.; Sedghi, H.; Zhang, C. What is being transferred in transfer learning? In Proceedings of the 34th International Conference on Neural Information Processing Systems (NIPS ’20), Vancouver, BC, Canada, 6-12 December 2020; pp. 512–523. [Google Scholar]
- Liu, L.; Chen, J.; Fieguth, P.; Zhao, G.; Chellappa, R.; Pietikäinen, M. From BoW to CNN: Two Decades of Texture Representation for Texture Classification. Int. J. Comp. Vis. 2019, 127, 74–109. [Google Scholar] [CrossRef]
- Yan, Y.; Ren, W.; Cao, X. Recolored Image Detection via a Deep Discriminative Model. IEEE Transactions on Information Forensics and Security 2019, 14, 5–17. [Google Scholar] [CrossRef]
- Tan, M.; Le, Q.V. EfficientNet: Rethinking Model Scaling for Convolutional Neural Networks. arXiv 2019, arXiv:1905.11946. [Google Scholar]
- Sandler, M.; Howard, A.; Zhu, M.; Zhmoginov, A.; Chen, L.-C. MobileNetV2: Inverted Residuals and Linear Bottlenecks. In Proceedings of the 2018 IEEE/CVF Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18-23 June 2018; pp. 4510–4520. [Google Scholar]
- Huang, G.; Liu, Z.; Maaten, L.V.D.; Weinberger, K.Q. Densely Connected Convolutional Networks. In Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017; pp. 2261–2269. [Google Scholar]
- Zoph, B.; Vasudevan, V.; Shlens, J.; Le, Q.V. Learning Transferable Architectures for Scalable Image Recognition. In Proceedings of the 2018 IEEE/CVF Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18-23 June 2018; pp. 8697–8710. [Google Scholar]
- Tan, M.; Le, Q.V. EfficientNetV2: Smaller Models and Faster Training. arXiv 2021, arXiv:2104.00298. [Google Scholar]
- Sagi, O.; Rokach, L. Ensemble learning: A survey. WIREs Data Mining and Knowledge Discovery 2018, 8, e1249. [Google Scholar] [CrossRef]
- Dietterichl, T.G. Ensemble learning. In The Handbook of Brain Theory and Neural Networks; Arbib, M.A., Ed.; MIT Press: Cambridge, MA, USA, 2002; pp. 405–408. [Google Scholar]
- Bergstra, J.; Yamins, D.; Cox, D.D. Making a science of model search: hyperparameter optimization in hundreds of dimensions for vision architectures. In Proceedings of the 30th International Conference on International Conference on Machine Learning - Volume 28 (ICML’13), Atlanta, GA, USA, 16-21 June 2013; pp. I–115–I–123. [Google Scholar]


| References | Model Used | Dataset | Number of Images | Number of Classes | Transfer learning | Ensemble Learning | Data augmentation | Accuracy |
|---|---|---|---|---|---|---|---|---|
| [12] | SVM | Multi-plant (Folio) | 637 | 32 | No | No | No | 92.91% |
| [16] | CNN(FGIA) | Peach,Tomato (PlantVillage) | 2657,18162 | 2,10 | No | No | No | 95.48% |
| [17] | CNN, MLP | Rice(own) | 3200 | 4 | No | Yes | No | 95.31% |
| [18] | MobileNetV2 | Bean(ibean) | 1296 | 3 | Yes | No | No | 92.97% |
| [19] | ResNet50, VGG16,VGG19 DenseNet201, InceptionV3 | Sugarcane (Mendeley) | 2511 | 5 | Yes | No | No | 95.69% |
| [20] | EfficientNetB0, CSPDarknet53 | Sugarcane (Mendeley) | 2522 | 5 | Yes | Yes | Yes | 96.80% |
| [21] | ANN | Mango(own) | 450 | 4 | No | No | No | 89.41% |
| [22] | SE-VIT | Multi-plant (PlantVillage), Sugarcane(own) | 60343,1877 | 38,5 | Yes | No | Yes | 89.57% |
| [23] | CNN,VGG19, ResNet50, Xception, MobileNetV2, EfficientNetB7 | Sugarcane(own) | 2569 | 5 | Yes | Yes | No | 86.53% |
| Serial No. | Augmentation Technique | Parameter with Value |
|---|---|---|
| 1 | Rotation | rotation_range=20 |
| 2 | Width shift | width_shift_range=0.2 |
| 3 | Height shift | height_shift_range=0.2 |
| 4 | Shear | shear_range=0.2 |
| 5 | Zoom | zoom_range=0.2 |
| 6 | Horizontal flip | horizontal_flip=True |
| 7 | Brightness | brightness_range=[0.5, 1.5] |
| Classes | Original dataset | Data augmentation | ||||||
|---|---|---|---|---|---|---|---|---|
| Total | Training | Validation | Testing | Total | Training | Validation | Testing | |
| Healthy | 522 | 420 | 54 | 48 | 800 | 631 | 75 | 94 |
| Mosaic | 462 | 366 | 49 | 47 | 800 | 658 | 68 | 74 |
| RedRot | 518 | 413 | 49 | 56 | 800 | 653 | 77 | 70 |
| Rust | 514 | 416 | 45 | 53 | 800 | 644 | 83 | 73 |
| Yellow | 505 | 400 | 56 | 49 | 800 | 618 | 96 | 86 |
| BacterialBlight | 125 | 101 | 12 | 12 | 800 | 636 | 81 | 83 |
| Total | 2646 | 2116 | 265 | 265 | 4800 | 3840 | 480 | 480 |
| Method | Test Classification Accuracy |
|---|---|
| NULL | 96.39 |
| L1 | 97.92 |
| L1+Dropout | 98.12 |
| L2 | 97.50 |
| L2+Dropout | 98.12 |
| ELN-Reg | 98.12 |
| ELN-Reg+Dropout | 98.54 |
| Model | Total Parameter | Trainable Parameters | Non-Trainable Parameters |
|---|---|---|---|
| EfficientNetB0 | 5,330,571 | 5,288,548 | 42,023 |
| MobileNetV2 | 3,538,984 | 3,504,872 | 34,112 |
| DenseNet121 | 8,062,504 | 7,978,856 | 83,648 |
| NASNetMobile | 5,326,716 | 5,289,978 | 36,738 |
| EfficientNetV2B0 | 7,200,312 | 7,139,704 | 60,608 |
| Layer (Type) | Output Shape | Parameters |
|---|---|---|
| Input Layer | [(None,224,224,3)] | 0 |
| efficientnet-b0 | (None, 1280) | 4049564 |
| Dense | (None, 128) | 163968 |
| BatchNormalization | (None, 128) | 896 |
| Dropout | (None, 128) | 0 |
| Dense | (None, 64) | 8256 |
| BatchNormalization | (None, 64) | 448 |
| Dropout | (None, 64) | 0 |
| Dense | (None, 32) | 2080 |
| BatchNormalization | (None, 32) | 224 |
| Dropout | (None, 32) | 0 |
| Dense | (None, 6) | 198 |
| Model | Original | Modified | Improvement |
|---|---|---|---|
| NASNetMobile | 85.00 | 92.71 | +%7.71 |
| EfficientNetV2B0 | 90.21 | 94.17 | +%3.96 |
| MobileNetV2 | 92.50 | 96.67 | +%4.17 |
| DenseNet121 | 95.83 | 98.12 | +%2.29 |
| EfficientNetB0 | 97.08 | 98.54 | +%1.46 |
| Model (Modified) | Macro Average | Weighted Average | Accuracy | ||||
|---|---|---|---|---|---|---|---|
| Precision | Recall | F1 score | Precision | Recall | F1 score | ||
| NASNetMobile | 93.24 | 92.78 | 92.80 | 93.31 | 92.71 | 92.78 | 92.71 |
| EfficientNetV2B0 | 94.47 | 94.40 | 94.27 | 94.57 | 94.17 | 94.20 | 94.17 |
| MobileNetV2 | 96.60 | 96.76 | 96.64 | 96.75 | 96.67 | 96.67 | 96.67 |
| DenseNet121 | 98.22 | 98.11 | 98.16 | 98.14 | 98.12 | 98.12 | 98.12 |
| EfficientNetB0 | 98.59 | 98.50 | 98.53 | 98.58 | 98.54 | 98.54 | 98.54 |
| Proposed DECNN | 99.23 | 99.13 | 99.18 | 99.17 | 99.17 | 99.17 | 99.17 |
| Model (Modified) | Weight Values |
|---|---|
| EfficientNetB0 | 0.58 |
| MobileNetV2 | 0.17 |
| DenseNet121 | 0.21 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





