Submitted:
15 October 2024
Posted:
15 October 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Sampling Location and Collection
2.2. Sediment Chemical Analysis
2.3. DNA Extraction and Illumina Sequencing
2.4. Data Analysis
3. Results
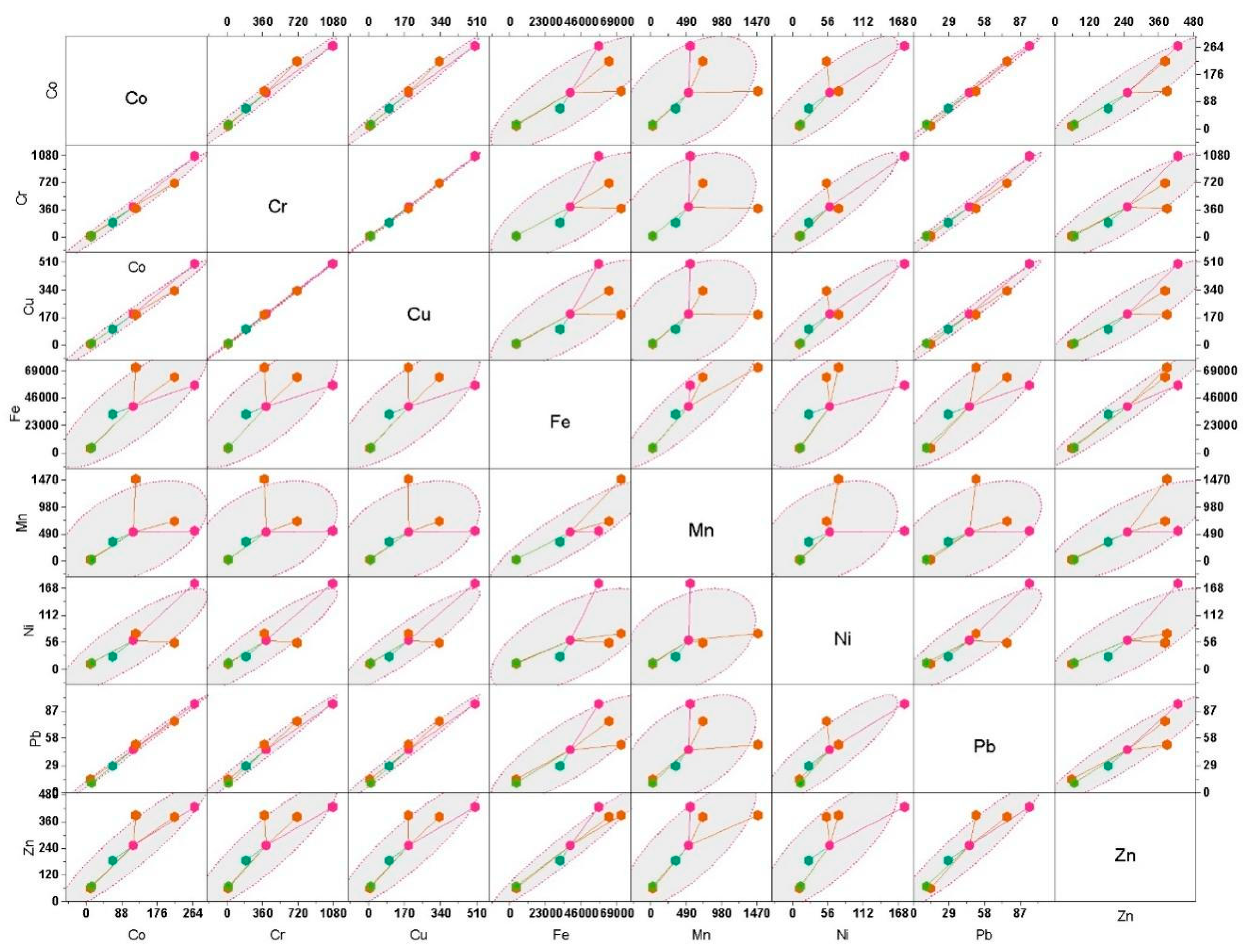
3.1. Sediment Chemical Properties
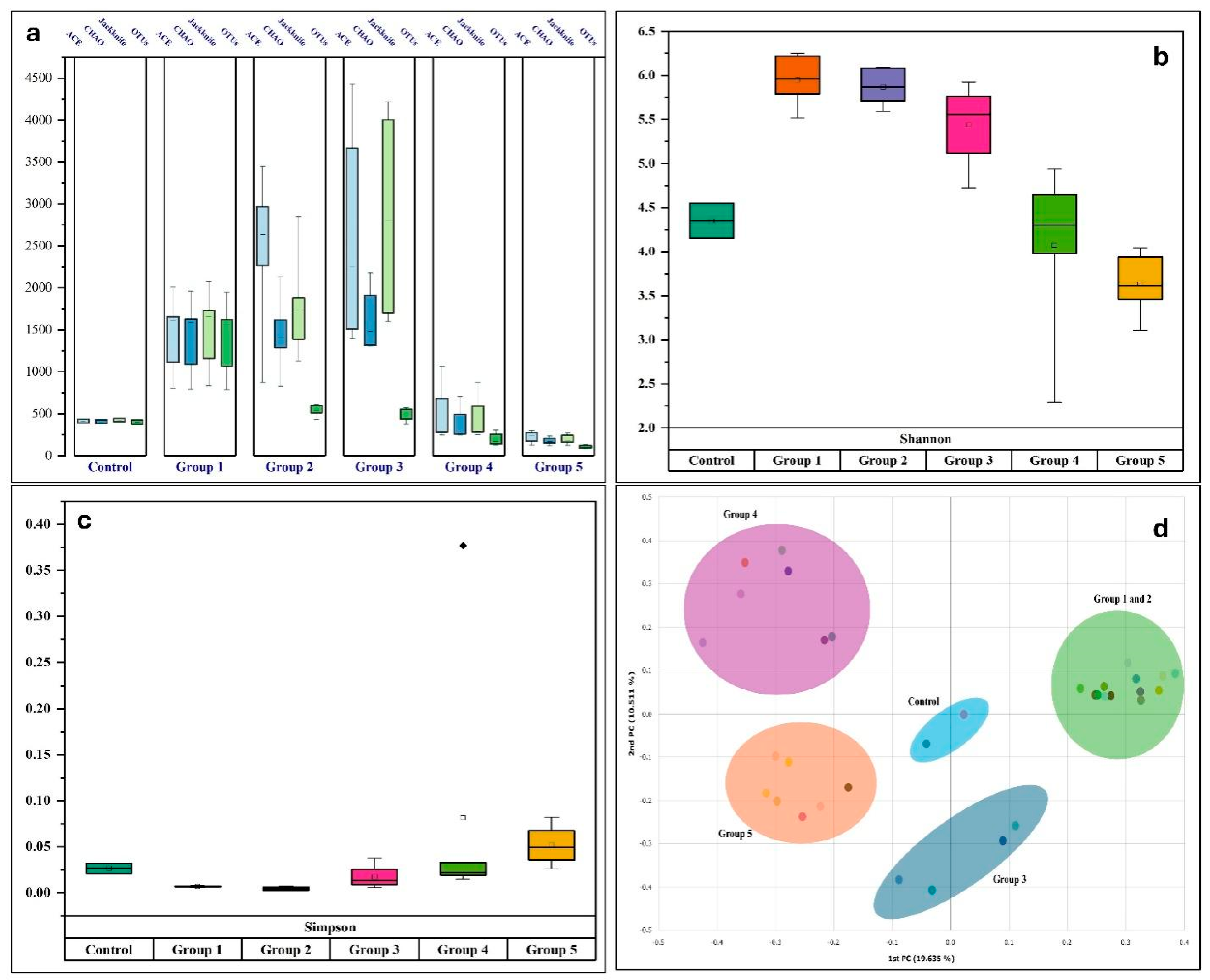
3.2. Diversity Indices of Bacterial Communities
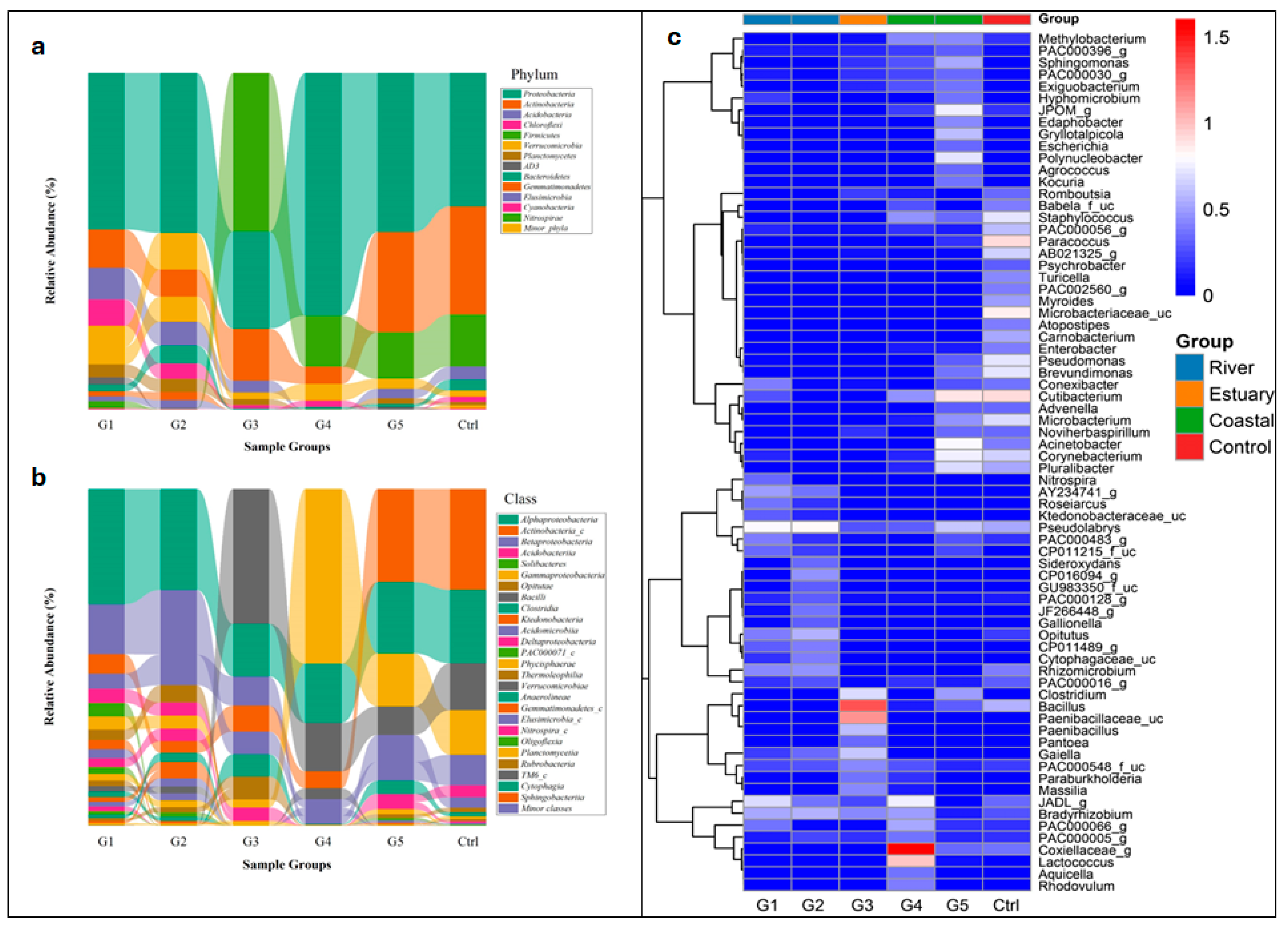
3.3. Bacterial Community Composition in Sediment Samples
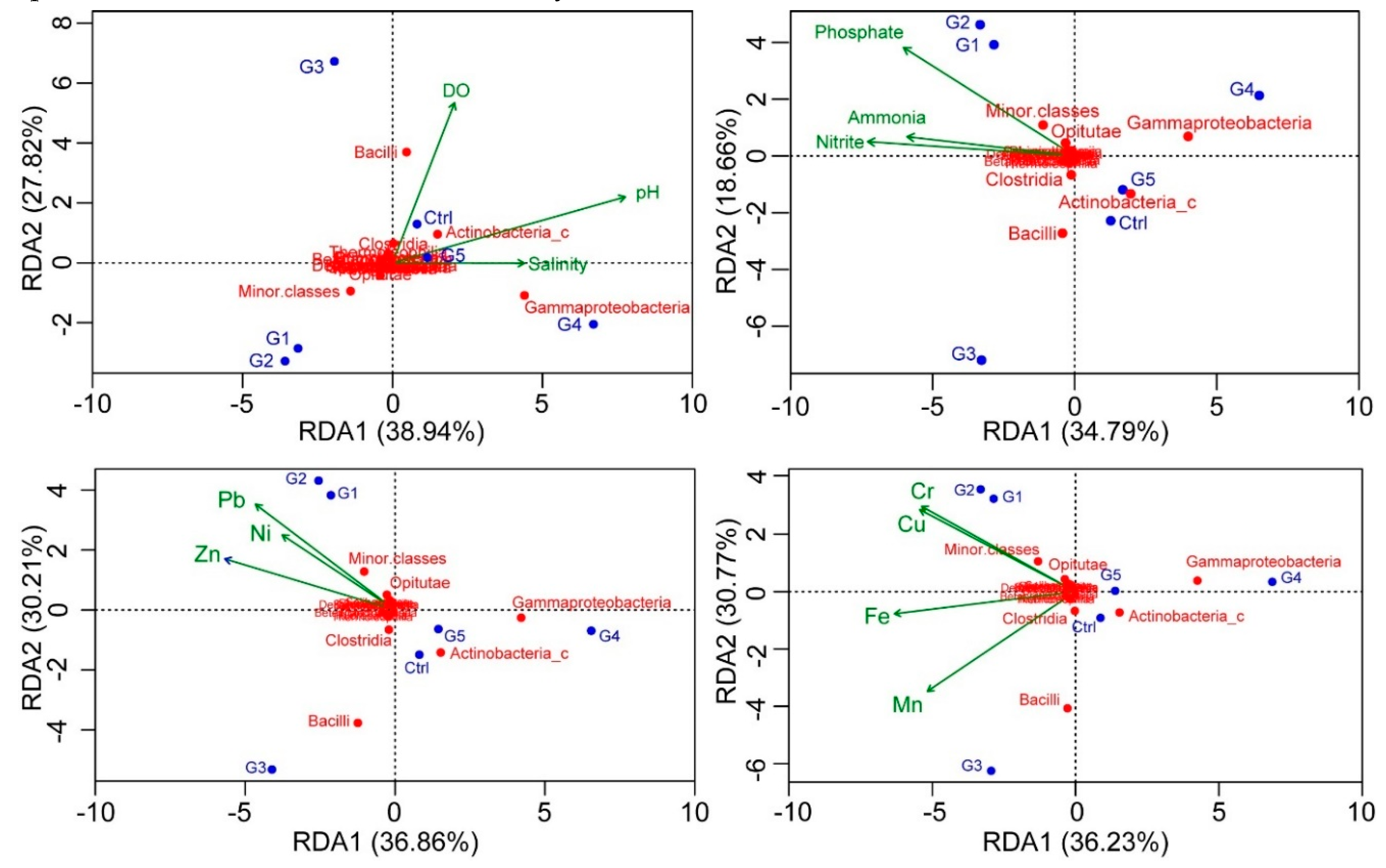
3.4. Redundancy Analysis
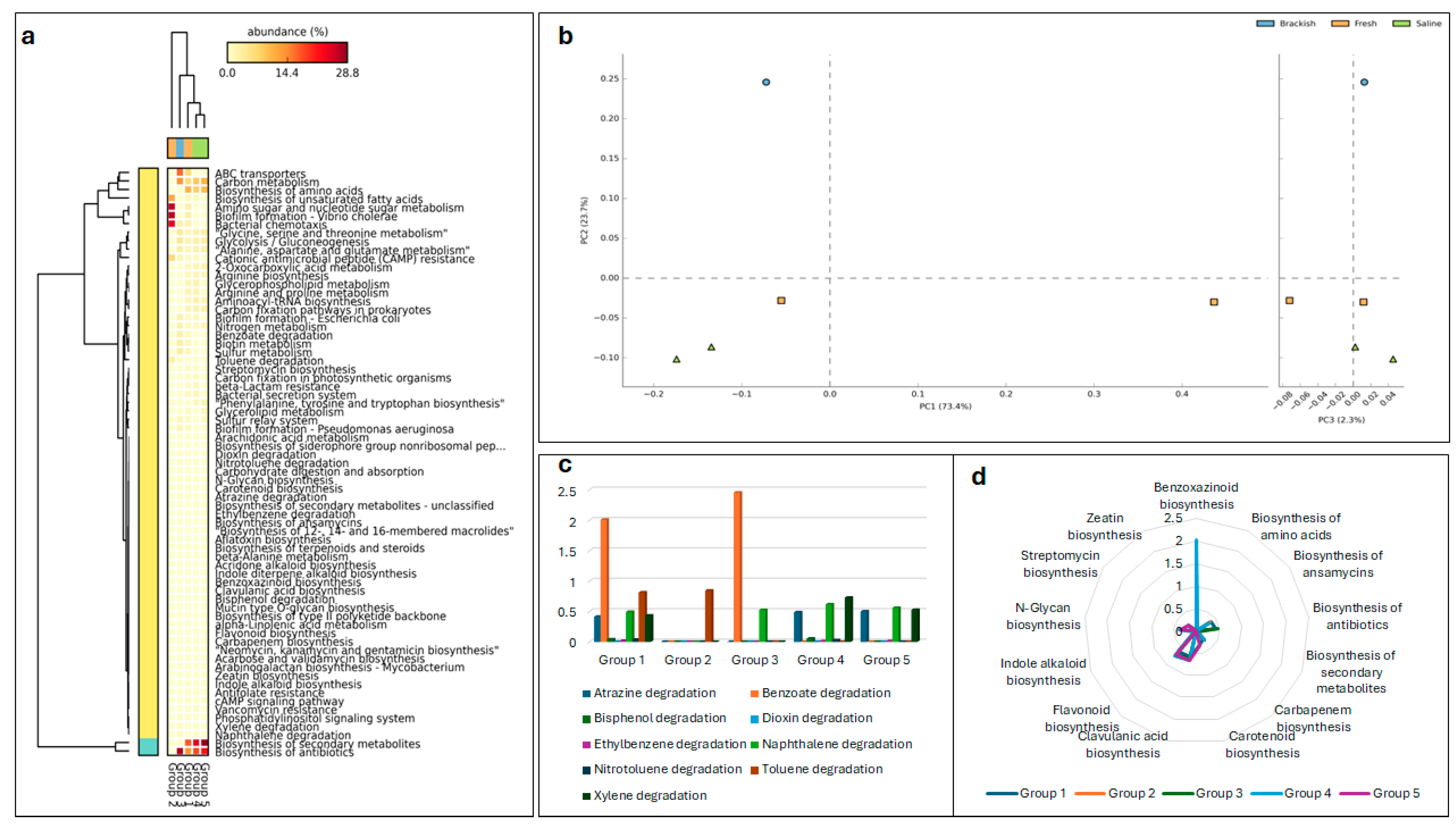
3.5. Functional Prediction Analysis
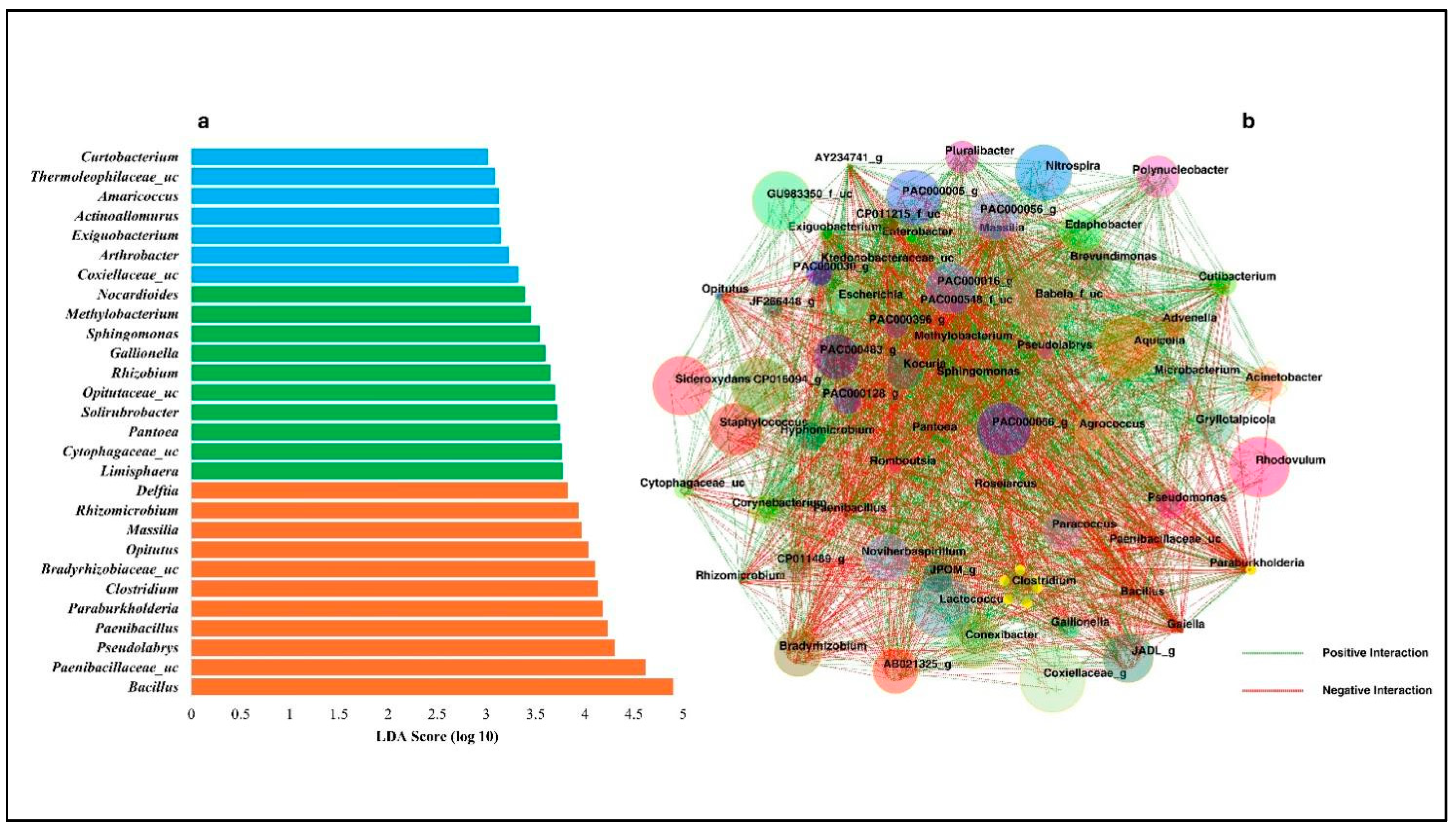
3.6. LEfSe and Co-Occurrence Network Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Vijayan, J.; Nathan, V.K.; Ammini, P.; Ammanamveetil, A.M.H. Bacterial Diversity in the Aquatic System in India Based on Metagenome Analysis—A Critical Review. Environ. Sci. Pollut. Res. 2023, 30, 28383–28406. [Google Scholar] [CrossRef] [PubMed]
- Nybakken, J.W. Seagrass Communities. Mar. Biol. An Ecol. Approach, fifth ed. Benjamin Cummings, an Impr. Addison-Wesley Longman, San Fr. 2001, 210–218. [Google Scholar]
- Karthigeyan, K.; Kumaraswamy, I.; Arisdason, W. An Assessment of Angiosperm Diversity of Adyar Estuary, Chennai - A Highly Degraded Estuarian Ecosystem, Tamil Nadu, India. Check List 2013, 9, 920–940. [Google Scholar] [CrossRef]
- Dupont, C.L.; Larsson, J.; Yooseph, S.; Ininbergs, K.; Goll, J.; Asplund-Samuelsson, J.; McCrow, J.P.; Celepli, N.; Allen, L.Z.; Ekman, M.; et al. Functional Tradeoffs Underpin Salinity-Driven Divergence in Microbial Community Composition. PLoS ONE 2014, 9. [Google Scholar] [CrossRef]
- Parvathi, A.; Catena, M.; Jasna, V.; Phadke, N.; Gogate, N. Influence of Hydrological Factors on Bacterial Community Structure in a Tropical Monsoonal Estuary in India. Environ. Sci. Pollut. Res. 2021, 28, 50579–50592. [Google Scholar] [CrossRef]
- Vajravelu, M.; Martin, Y.; Ayyappan, S.; Mayakrishnan, M. Seasonal Influence of Physico-Chemical Parameters on Phytoplankton Diversity, Community Structure and Abundance at Parangipettai Coastal Waters, Bay of Bengal, South East Coast of India. Oceanologia 2018, 60, 114–127. [Google Scholar] [CrossRef]
- Kisand, V.; Valente, A.; Lahm, A.; Tanet, G.; Lettieri, T. Phylogenetic and Functional Metagenomic Profiling for Assessing Microbial Biodiversity in Environmental Monitoring. PLoS ONE 2012. [Google Scholar] [CrossRef]
- DeLong, E.F.; Karl, D.M. Genomic Perspectives in Microbial Oceanography. Nature 2005, 437, 336–342. [Google Scholar] [CrossRef]
- Raina, V.; Panda, A.N.; Mishra, S.R.; Nayak, T.; Suar, M. Microbial Biodiversity Study of a Brackish Water Ecosystem in Eastern India; Elsevier Inc., 2019; ISBN 9780128148495. [Google Scholar]
- Rosa, S.; Palmeirim, J.M.; Moreira, F. Factors Affecting Waterbird Abundance and Species Richness in an Increasingly Urbanized Area of the Tagus Estuary in Portugal. Waterbirds 2003, 26, 226–232. [Google Scholar] [CrossRef]
- Rubalingeswari, N.; Thulasimala, D.; Giridharan, L.; Gopal, V.; Magesh, N.S.; Jayaprakash, M. Bioaccumulation of Heavy Metals in Water, Sediment, and Tissues of Major Fisheries from Adyar Estuary, Southeast Coast of India: An Ecotoxicological Impact of a Metropolitan City. Mar. Pollut. Bull. 2021, 163, 111964. [Google Scholar] [CrossRef]
- Barletta, M.; Lima, A.R.A.; Costa, M.F. Distribution, Sources and Consequences of Nutrients, Persistent Organic Pollutants, Metals and Microplastics in South American Estuaries. Sci. Total Environ. 2019, 651, 1199–1218. [Google Scholar] [CrossRef] [PubMed]
- Arrigo, K.R. Marine Microorganisms and Global Nutrient Cycles. Nature 2005, 437, 349–355. [Google Scholar] [CrossRef]
- Selvan, C.T.; Milton, J. Physicochemical Analysis of Coastal Water of East Coast of Tamil Nadu (Adyar Estuary). J. Zool. Stud. 2016, 3, 20–29. [Google Scholar]
- Janakiraman, A.; Naveed, M.S.; Asrar Sheriff, M.; Altaff, K. Ecological Restoration Assessment of Adyar Creek and Estuary Using Meiofaunal Communities as Ecological Indicators for Aquatic Pollution. Reg. Stud. Mar. Sci. 2017, 9, 135–144. [Google Scholar] [CrossRef]
- Altaff, K.; Janakiraman, A.; Naveed, M.S.; Asrar Sheriff, M.; War, M.; Sugumaran, J.; Mantha, G. Post-Restoration Ecological Assessment on the Zooplankton Dynamics of the Adyar Creek and Estuary. J. Coast. Conserv. 2019, 23, 473–483. [Google Scholar] [CrossRef]
- Veerasingam, S.; Venkatachalapathy, R.; Ramkumar, T. Historical Environmental Pollution Trend and Ecological Risk Assessment of Trace Metals in Marine Sediments off Adyar Estuary, Bay of Bengal, India. Environ. Earth Sci. 2014, 71, 3963–3975. [Google Scholar] [CrossRef]
- Rajkumar, A.N.; Barnes, J.; Ramesh, R.; Purvaja, R.; Upstill-Goddard, R.C. Methane and Nitrous Oxide Fluxes in the Polluted Adyar River and Estuary, SE India. Mar. Pollut. Bull. 2008, 56, 2043–2051. [Google Scholar] [CrossRef] [PubMed]
- Caruso, G.; La Ferla, R.; Azzaro, M.; Zoppini, A.; Marino, G.; Petochi, T.; Corinaldesi, C.; Leonardi, M.; Zaccone, R.; Fonda Umani, S. Microbial Assemblages for Environmental Quality Assessment: Knowledge, Gaps and Usefulness in the European Marine Strategy Framework Directive. Crit. Rev. Microbiol. 2016, 42, 883–904. [Google Scholar] [CrossRef] [PubMed]
- Astudillo-García, C.; Hermans, S.M.; Stevenson, B.; Buckley, H.L.; Lear, G. Microbial Assemblages and Bioindicators as Proxies for Ecosystem Health Status: Potential and Limitations. Appl. Microbiol. Biotechnol. 2019, 103, 6407–6421. [Google Scholar] [CrossRef]
- Eswaran, R.; Khandeparker, L. Seasonal Variation in β-Glucosidase-Producing Culturable Bacterial Diversity in a Monsoon-Influenced Tropical Estuary. Environ. Monit. Assess. 2019, 191, 1–11. [Google Scholar] [CrossRef]
- Nair, A. V; Vijayan, K.K.; Chakraborty, K.; Leo Antony, M. Diversity and Characterization of Antagonistic Bacteria from Tropical Estuarine Habitats of Cochin, India for Fish Health Management. World J. Microbiol. Biotechnol. 2012, 28, 2581–2592. [Google Scholar] [CrossRef] [PubMed]
- Ranjan Mishra, R.; Ranjan Swain, M.; Kanti Danga, T.; Thatoi, H. Diversity and Seasonal Fluctuation of Predominant Microbial Communities in Bhitarkanika, a Tropical Mangrove Ecosystem in India. Rev. Biol. Trop. 2012, 60, 909–924. [Google Scholar] [CrossRef]
- Thajudeen, J.; Yousuf, J.; Veetil, V.P.; Varghese, S.; Singh, A.; Abdulla, M.H. Nitrogen Fixing Bacterial Diversity in a Tropical Estuarine Sediments. World J. Microbiol. Biotechnol. 2017, 33, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Khandeparker, L.; Kuchi, N.; Kale, D.; Anil, A.C. Microbial Community Structure of Surface Sediments from a Tropical Estuarine Environment Using next Generation Sequencing. Ecol. Indic. 2017, 74, 172–181. [Google Scholar] [CrossRef]
- Chikere, C.B.; Mordi, I.J.; Chikere, B.O.; Selvarajan, R.; Ashafa, T.O.; Obieze, C.C. Comparative Metagenomics and Functional Profiling of Crude Oil-Polluted Soils in Bodo West Community, Ogoni, with Other Sites of Varying Pollution History. Ann. Microbiol. 2019, 69. [Google Scholar] [CrossRef]
- Okoye, A.U.; Selvarajan, R.; Chikere, C.B.; Okpokwasili, G.C.; Mearns, K. Characterization and Identification of Long-Chain Hydrocarbon-Degrading Bacterial Communities in Long-Term Chronically Polluted Soil in Ogoniland: An Integrated Approach Using Culture-Dependent and Independent Methods. Environ. Sci. Pollut. Res. 2024. [Google Scholar] [CrossRef] [PubMed]
- Osunmakinde, C.O.; Selvarajan, R.; Mamba, B.B.; Msagati, T.A.M. Profiling Bacterial Diversity and Potential Pathogens in Wastewater Treatment Plants Using High-Throughput Sequencing Analysis. Microorganisms 2019, 7. [Google Scholar] [CrossRef] [PubMed]
- Selvarajan, R.; Sibanda, T.; Venkatachalam, S.; Kamika, I.; Nel, W.A.J.W.A.J.; Ramganesh, S.; Timothy, S.; Venkatachalam, S.; Kamika, I.; Nel, W.A.J.W.A.J. Industrial Wastewaters Harbor a Unique Diversity of Bacterial Communities Revealed by High-Throughput Amplicon Analysis. Ann. Microbiol. 2018, 1–14. [Google Scholar] [CrossRef]
- Zheng, Y.; Hou, L.; Newell, S.; Liu, M.; Zhou, J.; Zhao, H.; You, L.; Cheng, X. Community Dynamics and Activity of Ammonia-Oxidizing Prokaryotes in Intertidal Sediments of the Yangtze Estuary. Appl. Environ. Microbiol. 2014, 80, 408–419. [Google Scholar] [CrossRef]
- Iloms, E.; Ololade, O.O.; Ogola, H.J.O.; Selvarajan, R. Investigating Industrial Effluent Impact on Municipal Wastewater Treatment Plant in Vaal, South Africa. Int. J. Environ. Res. Public Health 2020, 17. [Google Scholar] [CrossRef]
- Selvarajan, R.; Sibanda, T.; Ullah, H.; Abia, A.L.K. Beach Sand Mycobiome: The Silent Threat of Pathogenic Fungi and Toxic Metal Contamination for Beachgoers. Mar. Pollut. Bull. 2024, 198, 115895. [Google Scholar] [CrossRef] [PubMed]
- Ogola, H.J.O.; Selvarajan, R.; Tekere, M. Local Geomorphological Gradients and Land Use Patterns Play Key Role on the Soil Bacterial Community Diversity and Dynamics in the Highly Endemic Indigenous Afrotemperate Coastal Scarp Forest Biome. Front. Microbiol. 2021, 12. [Google Scholar] [CrossRef]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data 2010.
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.; Ghalith, G.A. Al; Alexander, H.; Alm, E.J.; Arumugam, M.; Bai, Y.; et al. QIIME 2: Reproducible, Interactive, Scalable, and Extensible Microbiome Data Science. PeerJ Prepr. 2018, 9–10. [Google Scholar] [CrossRef] [PubMed]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-Resolution Sample Inference from Illumina Amplicon Data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Crump, B.C.; Bowen, J.L. The Microbial Ecology of Estuarine Ecosystems. Ann. Rev. Mar. Sci. 2024, 16, 335–360. [Google Scholar] [CrossRef] [PubMed]
- Colares, G.B.; Melo, V.M.M. Relating Microbial Community Structure and Environmental Variables in Mangrove Sediments inside Rhizophora Mangle L. Habitats. Appl. Soil Ecol. 2013, 64, 171–177. [Google Scholar] [CrossRef]
- Campbell, B.J.; Kirchman, D.L. Bacterial Diversity, Community Structure and Potential Growth Rates along an Estuarine Salinity Gradient. ISME J. 2013, 7, 210–220. [Google Scholar] [CrossRef] [PubMed]
- Marengo, J.A.; Oliveira, G.S.; Alves, L.M.; Bergier, I.; Assine, M.L. Dynamics of the Pantanal Wetland in South America 2016, 227–238.
- Chen, Y.; Xu, Y.; Ma, Y.; Lin, J.; Ruan, A. Microbial Community Structure and Its Driving Mechanisms in the Hangbu Estuary of Chaohu Lake under Different Sedimentary Areas. Environ. Res. 2023, 238, 117153. [Google Scholar] [CrossRef]
- Ramganesh, S.; Maredza, A.T.; Tekere, M. Microbial Exploration in Extreme Conditions: Metagenomic Analysis and Future Perspectives. In Metagenomics - Methods, Applications and Perspectives; BENEDETTI, C., Ed.; Nova Science Publishers, Inc.: New York, 2014; ISBN 9781631172557. [Google Scholar]
- Sibanda, T.; Selvarajan, R.; Msagati, T.; Venkatachalam, S.; Meddows-Taylor, S. Defunct Gold Mine Tailings Are Natural Reservoir for Unique Bacterial Communities Revealed by High-Throughput Sequencing Analysis. Sci. Total Environ. 2019, 650, 2199–2209. [Google Scholar] [CrossRef]
- Du, M.; Zheng, M.; Liu, A.; Wang, L.; Pan, X.; Liu, J.; Ran, X. Effects of Emerging Contaminants and Heavy Metals on Variation in Bacterial Communities in Estuarine Sediments. Sci. Total Environ. 2022, 832, 155118. [Google Scholar] [CrossRef]
- Fernandes, S.O.; Gonsalves, M.J.; Nazareth, D.R.; Wong, S.K.; Haider, M.N.; Ijichi, M.; Kogure, K. Seasonal Variability in Environmental Parameters Influence Bacterial Communities in Mangrove Sediments along an Estuarine Gradient. Estuar. Coast. Shelf Sci. 2022, 270. [Google Scholar] [CrossRef]
- Lu, X.M.; Chen, C.; Zheng, T.L.; Chen, J.J. Temporal–Spatial Variation of Bacterial Diversity in Estuary Sediments in the South of Zhejiang Province, China. Appl. Microbiol. Biotechnol. 2016, 100, 2817–2828. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.; Chen, X.; Wang, K.; Chen, J.; Zheng, B.; Jiang, X. Comparison among the Microbial Communities in the Lake, Lake Wetland, and Estuary Sediments of a Plain River Network. Microbiologyopen 2019, 8, 1–13. [Google Scholar] [CrossRef]
- Anderson, C.R.; Peterson, M.E.; Frampton, R.A.; Bulman, S.R.; Keenan, S.; Curtin, D. Rapid Increases in Soil PH Solubilise Organic Matter, Dramatically Increase Denitrification Potential and Strongly Stimulate Microorganisms from the Firmicutes Phylum. PeerJ 2018, 6, e6090. [Google Scholar] [CrossRef]
- Lukhele, T.; Ogola, H.J.O.; Selvarajan, R.; Oruko, R.O.; Nyoni, H.; Mamba, B.B.; Msagati, T.A.M. Metagenomic Insights into Taxonomic Diversity and Metabolic Potential of Bacterial Communities Associated with Tannery Waste-Contaminated Soils. Int. J. Environ. Sci. Technol. 2022, 19, 2409–2424. [Google Scholar] [CrossRef]
- Das, M.; Royer, T. V; Leff, L.G. Diversity of Fungi, Bacteria, and Actinomycetes on Leaves Decomposing in a Stream. Appl. Environ. Microbiol. 2007, 73, 756–767. [Google Scholar] [CrossRef] [PubMed]
- Fenibo, E.O.; Selvarajan, R.; Abia, A.L.K.; Matambo, T. Medium-Chain Alkane Biodegradation and Its Link to Some Unifying Attributes of AlkB Genes Diversity. Sci. Total Environ. 2023, 877, 162951. [Google Scholar] [CrossRef]
- Jones, R.M.; Barrie Johnson, D. Acidithrix Ferrooxidans Gen. Nov., Sp. Nov.; a Filamentous and Obligately Heterotrophic, Acidophilic Member of the Actinobacteria That Catalyzes Dissimilatory Oxido-Reduction of Iron. Res. Microbiol. 2015, 166, 111–120. [Google Scholar] [CrossRef]
- Zhang, H.; Ma, B.; Huang, T.; Shi, Y. Nitrate Reduction by the Aerobic Denitrifying Actinomycete Streptomyces Sp. XD-11-6-2: Performance, Metabolic Activity, and Micro-Polluted Water Treatment. Bioresour. Technol. 2021, 326, 124779. [Google Scholar] [CrossRef]
- Sheng, P.; Yu, Y.; Zhang, G.; Huang, J.; He, L.; Ding, J. Bacterial Diversity and Distribution in Seven Different Estuarine Sediments of Poyang Lake, China. Environ. Earth Sci. 2016, 75, 479. [Google Scholar] [CrossRef]
- Dyksma, S.; Bischof, K.; Fuchs, B.M.; Hoffmann, K.; Meier, D.; Meyerdierks, A.; Pjevac, P.; Probandt, D.; Richter, M.; Stepanauskas, R. Ubiquitous Gammaproteobacteria Dominate Dark Carbon Fixation in Coastal Sediments. ISME J. 2016, 10, 1939–1953. [Google Scholar] [CrossRef] [PubMed]
- Gutierrez, T. Marine, Aerobic Hydrocarbon-Degrading Gammaproteobacteria: Overview. Taxon. genomics Ecophysiol. Hydrocarb. microbes 2019, 143–152. [Google Scholar]
- Gilbert, J.A.; Steele, J.A.; Caporaso, J.G.; Steinbrück, L.; Reeder, J.; Temperton, B.; Huse, S.; McHardy, A.C.; Knight, R.; Joint, I. Defining Seasonal Marine Microbial Community Dynamics. ISME J. 2012, 6, 298–308. [Google Scholar] [CrossRef] [PubMed]
- Gawade, L.; Sarma, V.; Rao, Y.V.; Hemalatha, K.P.J. Variation of Bacterial Metabolic Rates and Organic Matter in the Monsoon-Affected Tropical Estuary (Godavari, India). Geomicrobiol. J. 2017, 34, 628–640. [Google Scholar] [CrossRef]
- Douglas, A.E. How Multi-Partner Endosymbioses Function. Nat. Rev. Microbiol. 2016, 14, 731–743. [Google Scholar] [CrossRef]
- Tsao, H.F.; Scheikl, U.; Volland, J.M.; Köhsler, M.; Bright, M.; Walochnik, J.; Horn, M. “Candidatus Cochliophilus Cryoturris” (Coxiellaceae), a Symbiont of the Testate Amoeba Cochliopodium Minus. Sci. Rep. 2017, 7, 1–8. [Google Scholar] [CrossRef]
- Poretsky, R.S.; Sun, S.; Mou, X.; Moran, M.A. Transporter Genes Expressed by Coastal Bacterioplankton in Response to Dissolved Organic Carbon. Environ. Microbiol. 2010, 12, 616–627. [Google Scholar] [CrossRef]
- Gebhard, S. ABC Transporters of Antimicrobial Peptides in Firmicutes Bacteria - Phylogeny, Function and Regulation. Mol. Microbiol. 2012, 86, 1295–1317. [Google Scholar] [CrossRef]
- Huq, M.A.; Ma, J.; Srinivasan, S.; Parvez, M.A.K.; Rahman, M.M.; Naserkheil, M.; Abuhena, M.; Maitra, P.; Islam, F.; Nam, K. Massilia Agrisoli Sp. Nov., Isolated from Rhizospheric Soil of Banana. Int. J. Syst. Evol. Microbiol. 2023, 73, 5897. [Google Scholar] [CrossRef]
- Morel, M.A.; Iriarte, A.; Jara, E.; Musto, H.; Castro-Sowinski, S. Revealing the Biotechnological Potential of Delftia Sp. JD2 by a Genomic Approach.; AIMS, 2016.
- Xu, L.; Wang, G.; Zhang, S.; Li, T.; Xu, X.; Gong, G.; Zhou, W.; Pu, Y.; Jia, Y.; Li, Y. Inhibition of High Sulfur on Functional Microorganisms and Genes in Slightly Contaminated Soil by Cadmium and Chromium. Environ. Pollut. 2024, 344, 123421. [Google Scholar] [CrossRef]
- Wang, L.; Qiao, N.; Sun, F.; Shao, Z. Isolation, Gene Detection and Solvent Tolerance of Benzene, Toluene and Xylene Degrading Bacteria from Nearshore Surface Water and Pacific Ocean Sediment. Extremophiles 2008, 12, 335–342. [Google Scholar] [CrossRef] [PubMed]
- Plotnikova, E.G.; Yastrebova, O. V; Anan’ina, L.N.; Dorofeeva, L. V; Lysanskaya, V.Y.; Demakov, V.A. Halotolerant Bacteria of the Genus Arthrobacter Degrading Polycyclic Aromatic Hydrocarbons. Russ. J. Ecol. 2011, 42, 502–509. [Google Scholar] [CrossRef]
- Raes, J.; Bork, P. Molecular Eco-Systems Biology: Towards an Understanding of Community Function. Nat. Rev. Microbiol. 2008, 6, 693–699. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Zhang, H.; Huang, T.; Ma, B.; Miao, Y.; Shi, Y.; Xu, L.; Liu, K.; Huang, X. Aerobic Denitrifying Bacterial Communities Drive Nitrate Removal: Performance, Metabolic Activity, Dynamics and Interactions of Core Species. Bioresour. Technol. 2020, 316, 123922. [Google Scholar] [CrossRef] [PubMed]
- Xun, W.; Liu, Y.; Li, W.; Ren, Y.; Xiong, W.; Xu, Z.; Zhang, N.; Miao, Y.; Shen, Q.; Zhang, R. Specialized Metabolic Functions of Keystone Taxa Sustain Soil Microbiome Stability. Microbiome 2021, 9, 1–15. [Google Scholar] [CrossRef]
- Yang, P.; van Elsas, J.D. Mechanisms and Ecological Implications of the Movement of Bacteria in Soil. Appl. Soil Ecol. 2018, 129, 112–120. [Google Scholar] [CrossRef]
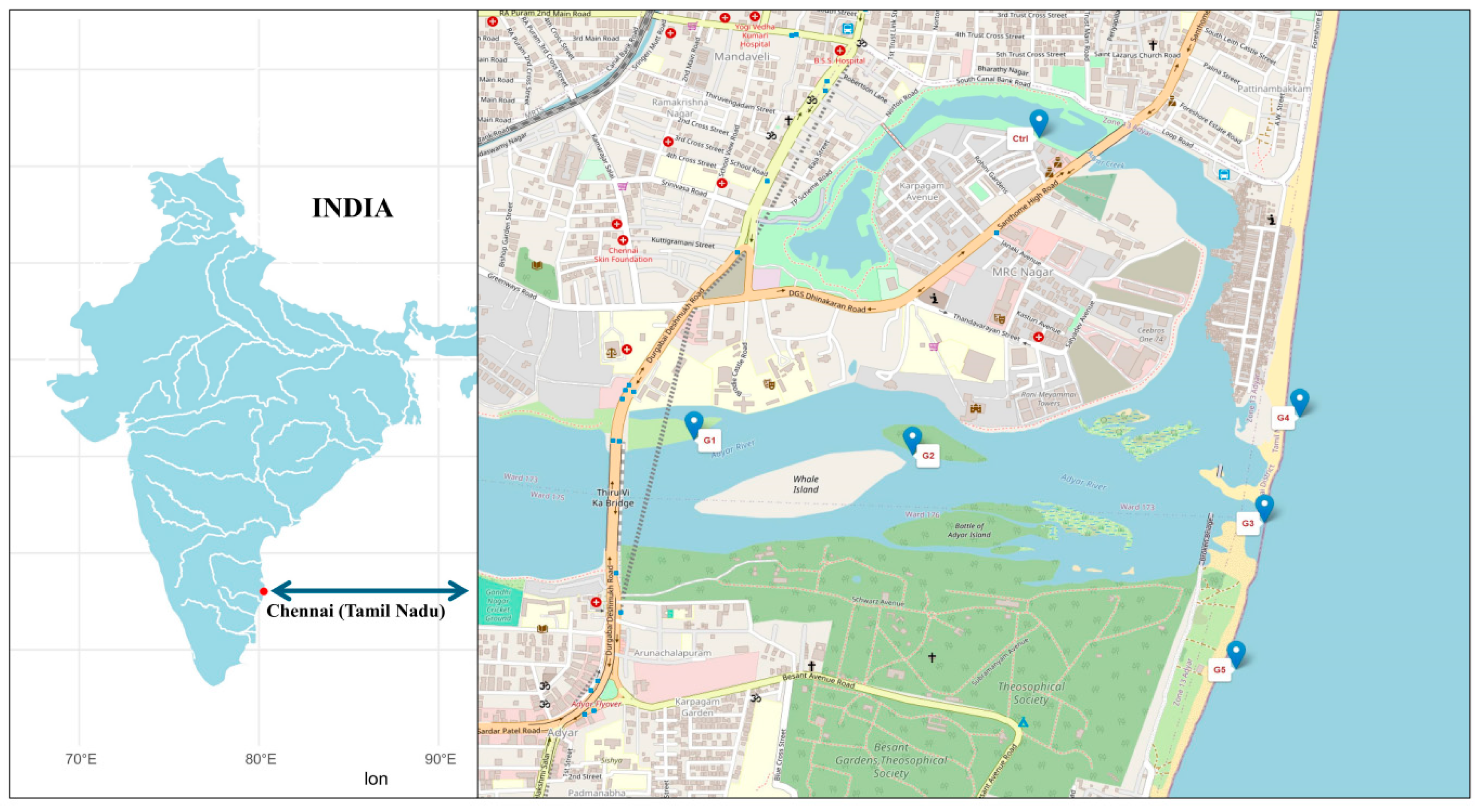
| Sampling points |
pH | Salinity | DO | NO3 | NH4 | PO4 | Co | Cr | Cu | Fe | Mn | Ni | Pb | Zn |
| G1 | 7.39 | 15.25 | 3.25 | 0.24 | 2.95 | 1.51 | 134.3 | 536.15 | 250.6 | 28550 | 273.05 | 89.1 | 47.3 | 213.2 |
| G2 | 7.44 | 17.29 | 2.97 | 0.22 | 4.39 | 1.46 | 109.45 | 357.1 | 167.25 | 31950 | 362.05 | 27.35 | 38.2 | 191.25 |
| G3 | 7.74 | 21.5 | 3.8 | 0.26 | 4.12 | 1.29 | 61.4 | 190.3 | 94.35 | 35900 | 742.8 | 36.9 | 25.6 | 194.3 |
| G4 | 8.03 | 28.17 | 3.43 | 0.11 | 1.72 | 1.13 | 5.25 | 6.6 | 3.695 | 2023.2 | 13.97 | 5.9 | 7.13 | 29.7 |
| G5 | 8.01 | 29.9 | 3.33 | 0.12 | 1.88 | 1.02 | 6.9 | 8.3 | 5.34 | 2108.1 | 15.35 | 6.65 | 5.23 | 33.55 |
| Ctrl | 7.57 | 6.25 | 4.56 | 0.14 | 0.51 | 1.24 | 32.9 | 95.75 | 49.35 | 16200 | 173.05 | 13.2 | 14.2 | 92.5 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





