Submitted:
15 October 2024
Posted:
17 October 2024
You are already at the latest version
Abstract
Keywords:
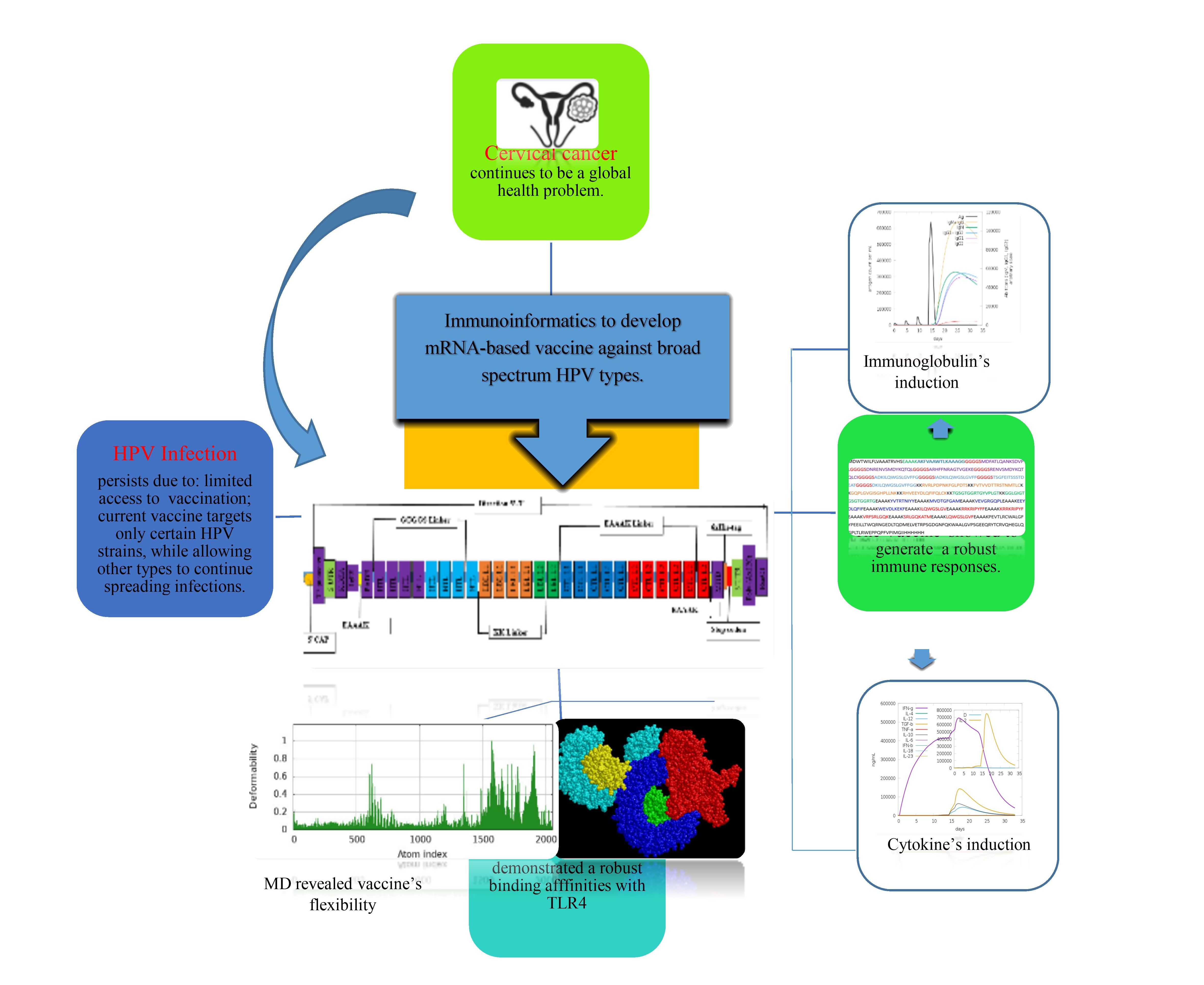
1. Introduction
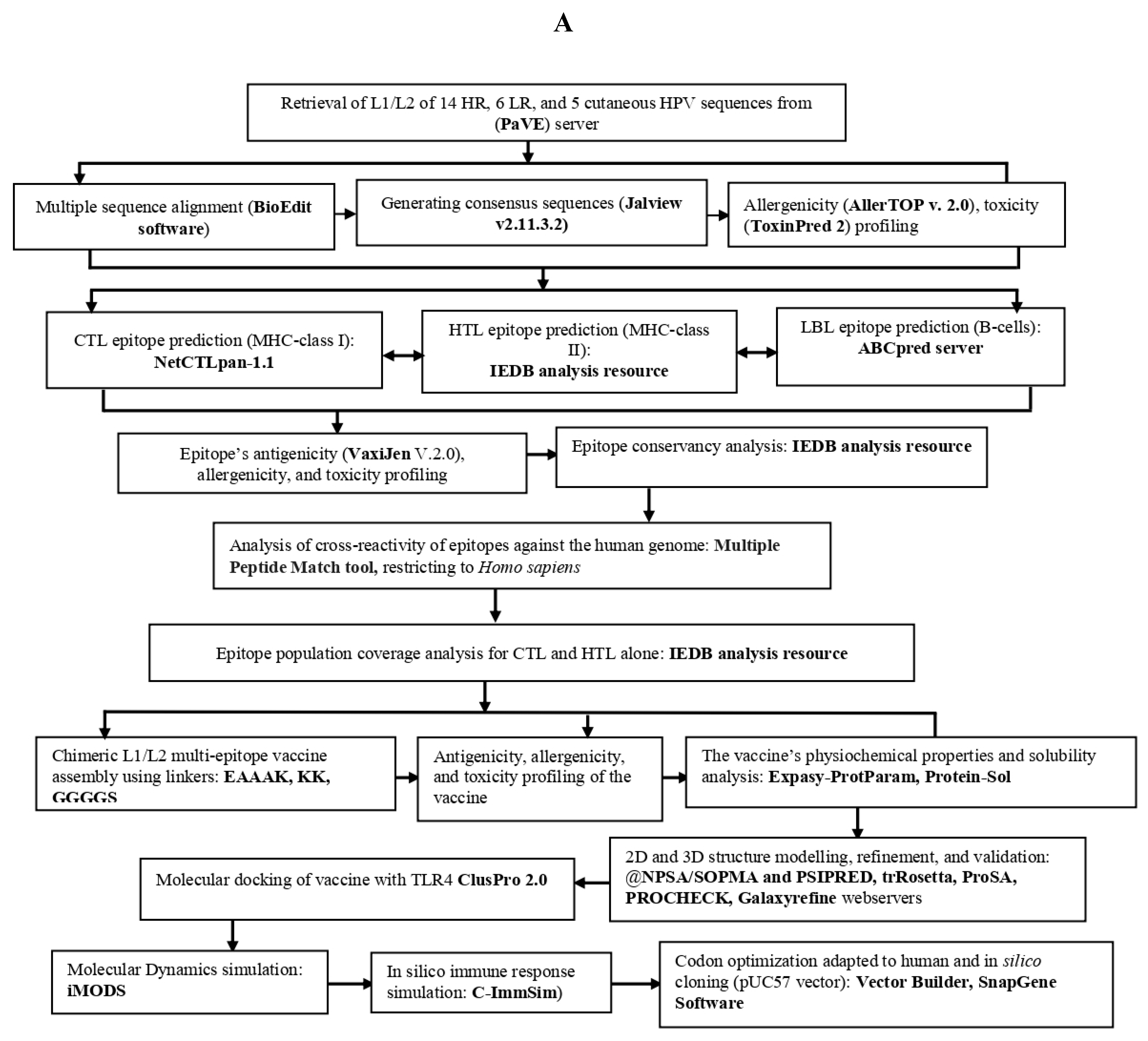
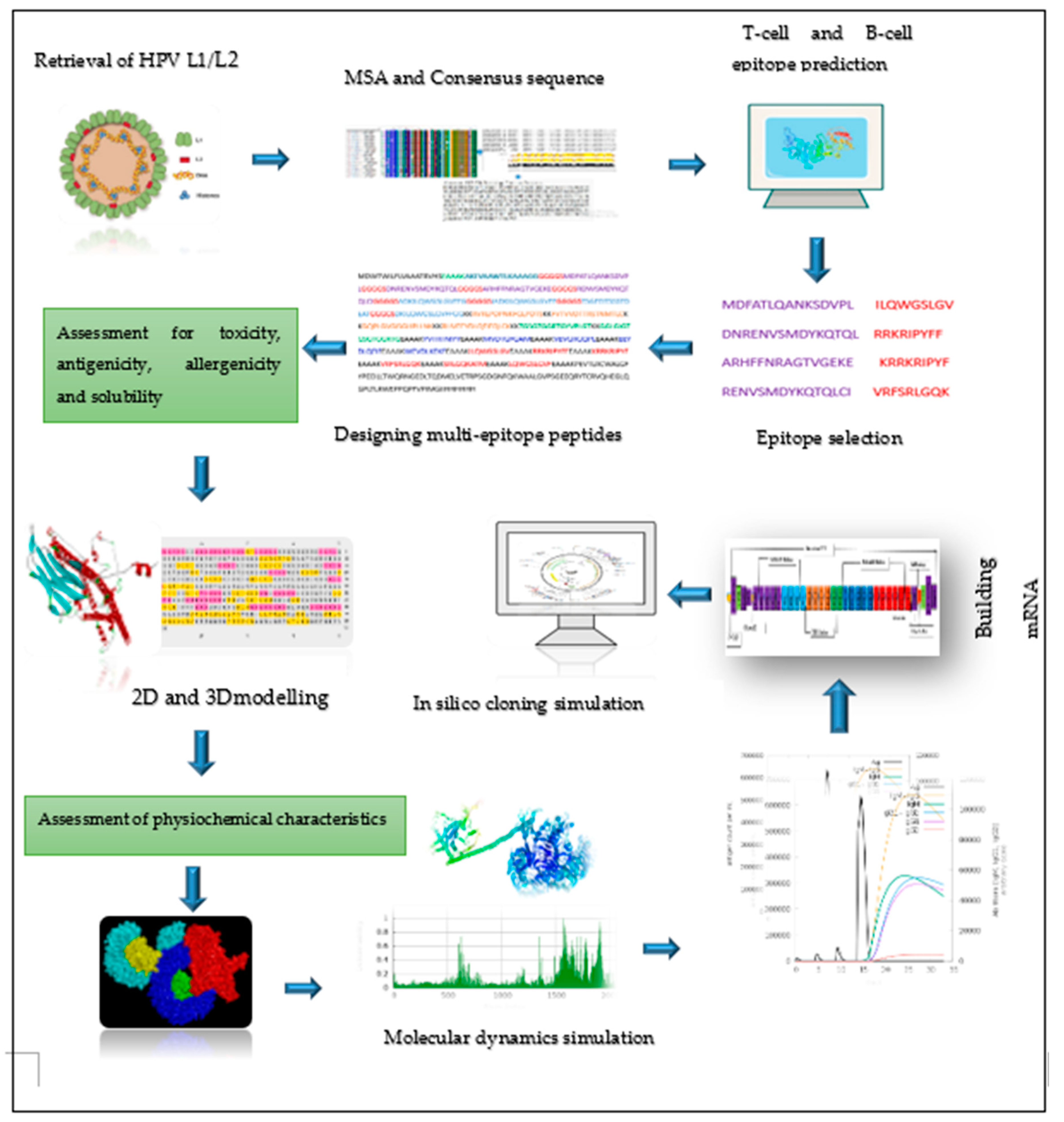
2. Materials and Methods
2.1. Retrieval of protein sequence, identification of conserved regions and generating consensus sequences
2.2. Prediction of immune cell epitopes
2.3. Screening the best epitopes for toxicity, antigenicity, allergenicity, and evaluation of vaccine physiochemical characteristics and solubility
2.4. Epitope human homology and conservancy analysis
2.5. Population coverage analysis of CTL and HTL epitopes
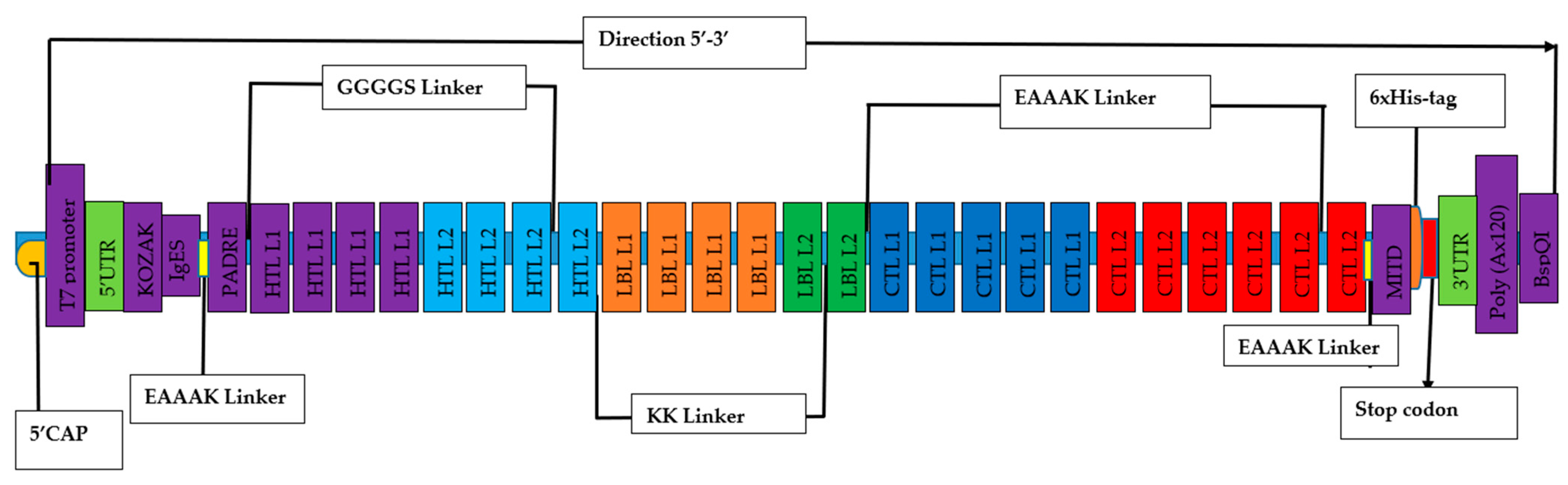
2.6. Designing of mRNA-based vaccine
2.7. Predicting 2D and 3D structures, refining 3D structure, and validating the vaccine candidates
2.8. Identification of discontinuous B-cells epitopes
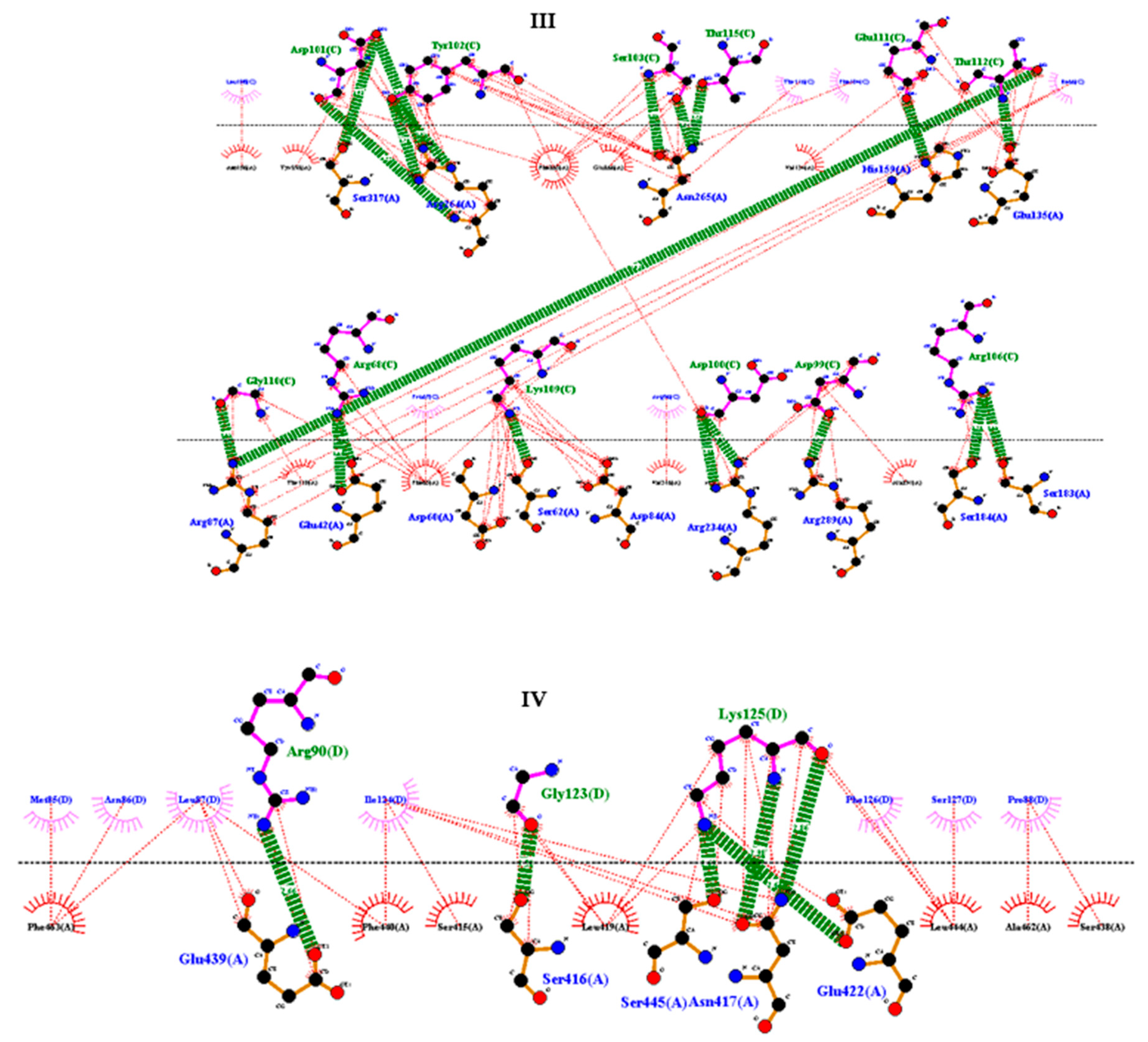
2.9. Molecular docking between the vaccine and TRL4
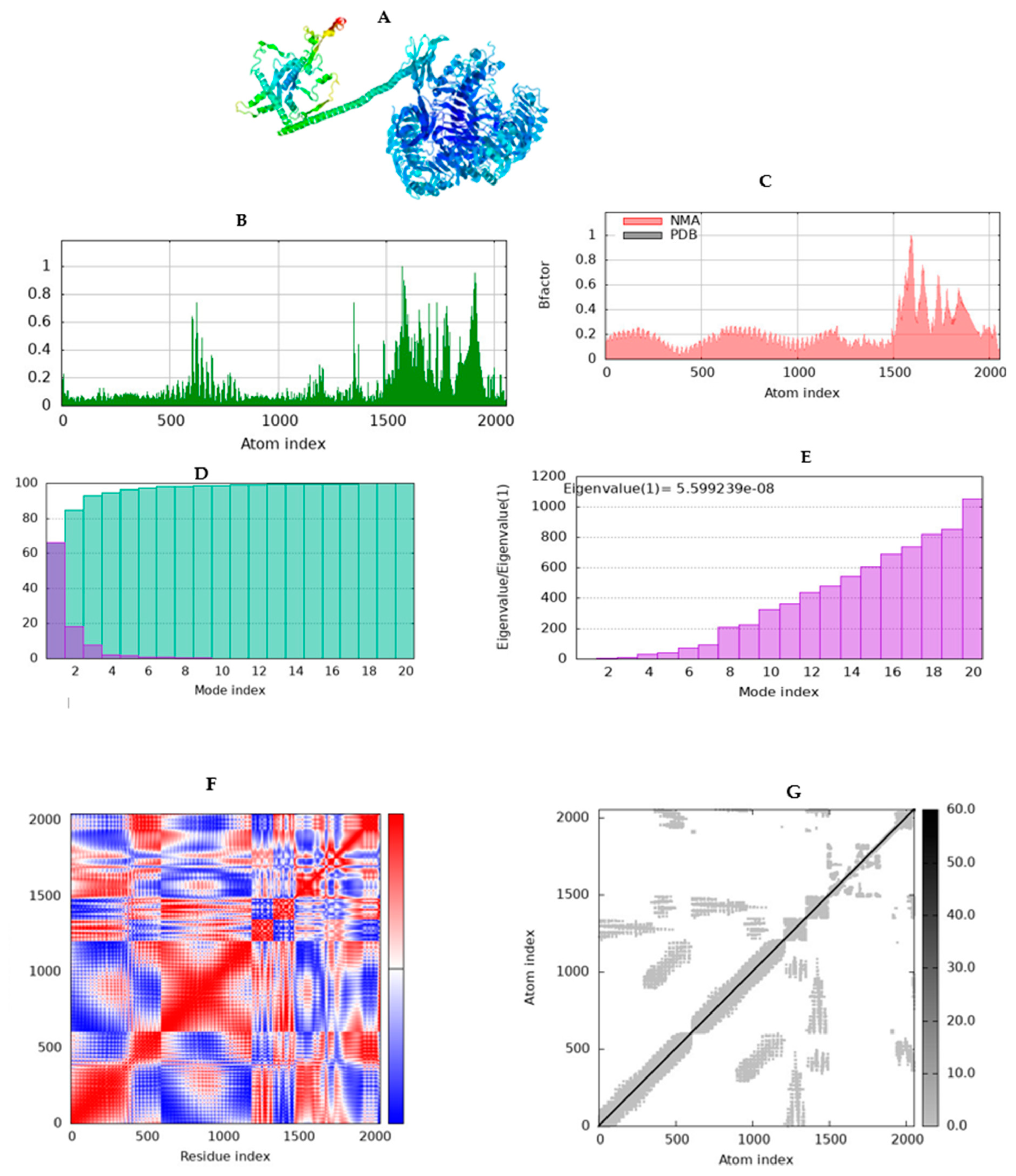
2.10. Simulating molecular dynamics of docking complex
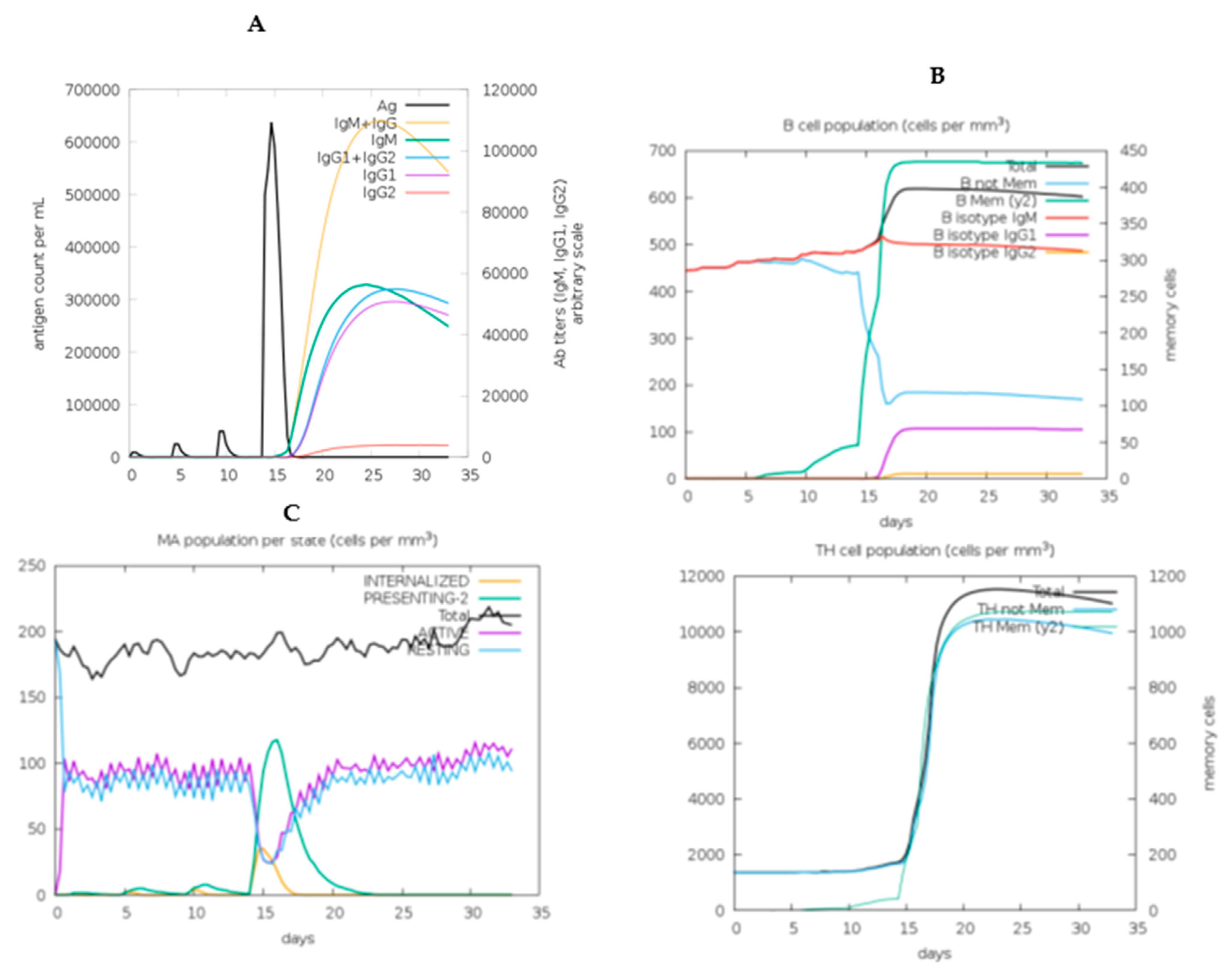
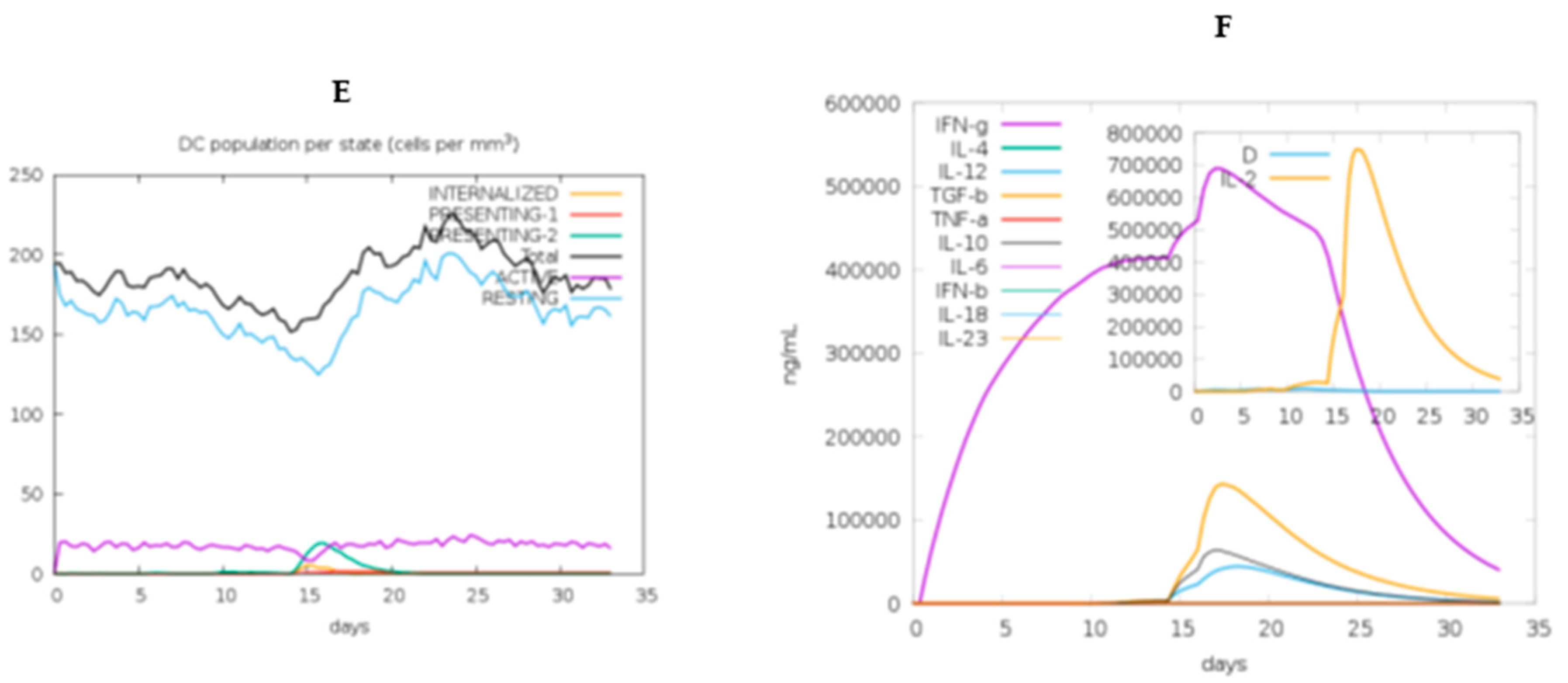
2.11. In Silico immune response simulation
2.12. Codon optimization, secondary structure prediction for mRNA vaccine, and cloning simulation
3. Results
3.1. Retrieval of protein sequences, multiple sequence alignment and generating consensus sequence
3.2. Prediction and evaluation of MHC class I and MHC class II and B CELL (LBL) epitopes based on L1 protein
3.3. Prediction and evaluation of CTL and HTL) and B CELL (LBL) based on L2 protein
3.4. Epitope human homology
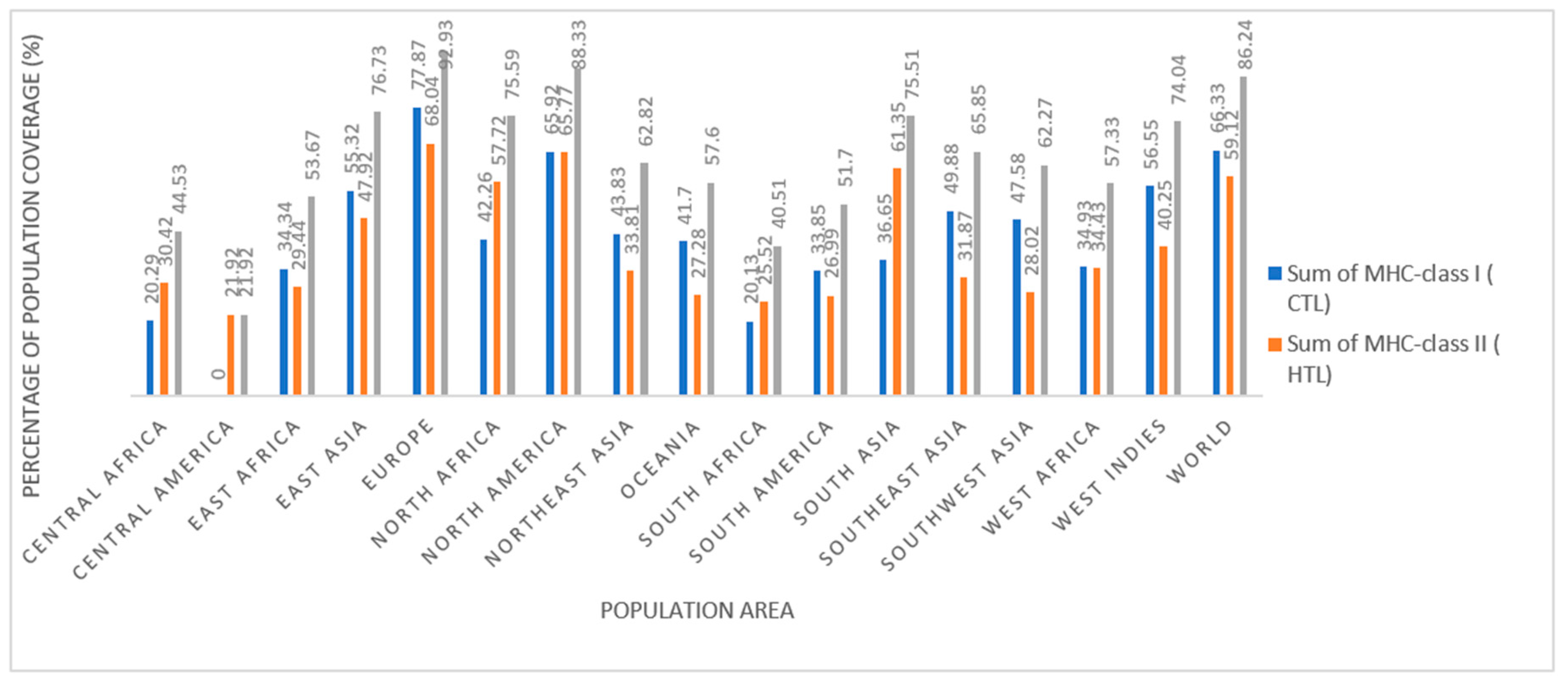
1. Population coverage analysis
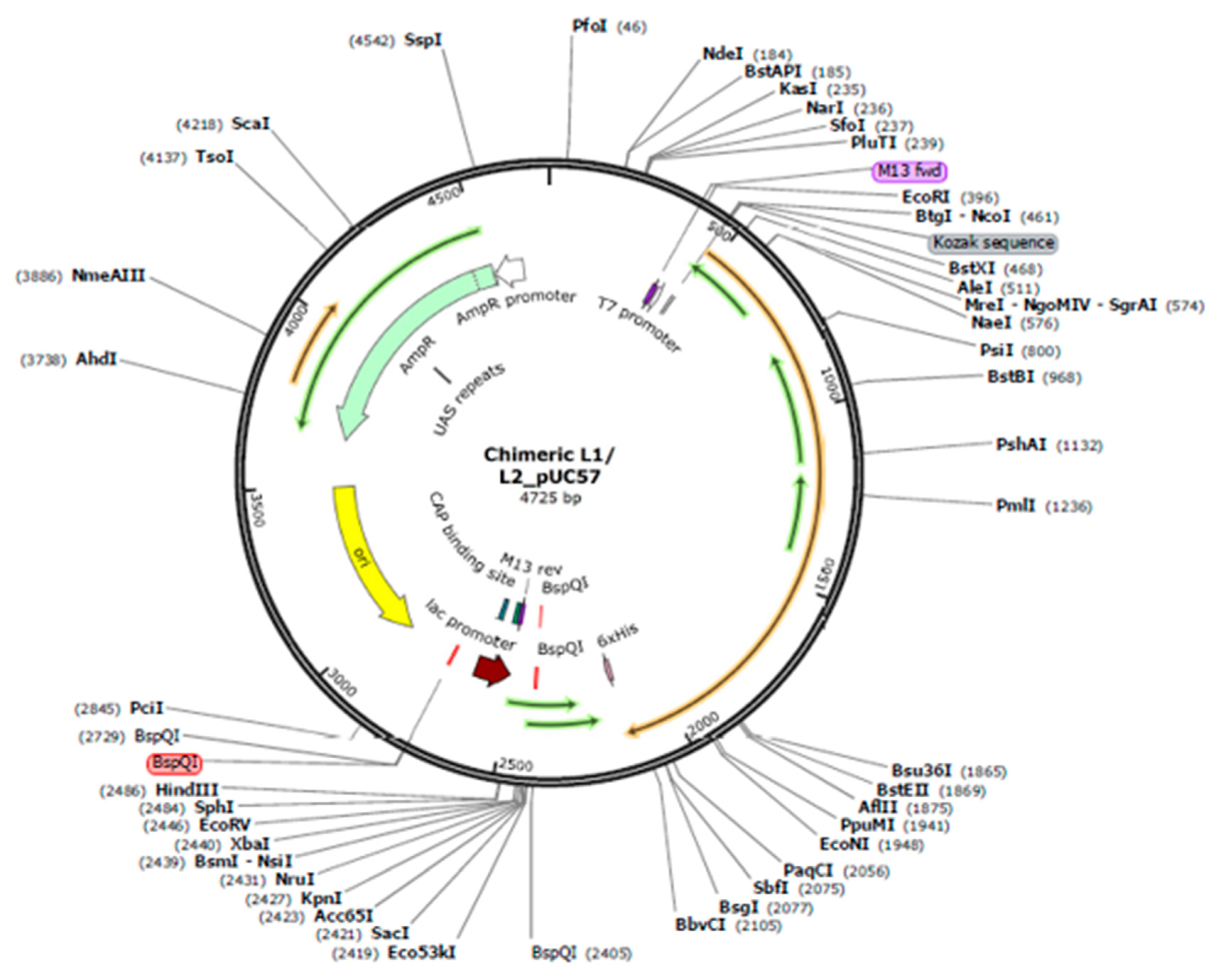
3.5. Chimeric mRNA–based vaccine assembly
3.5. Assessment of antigenicity, allergenicity, and solubility profiling and physiochemical properties of the vaccine design
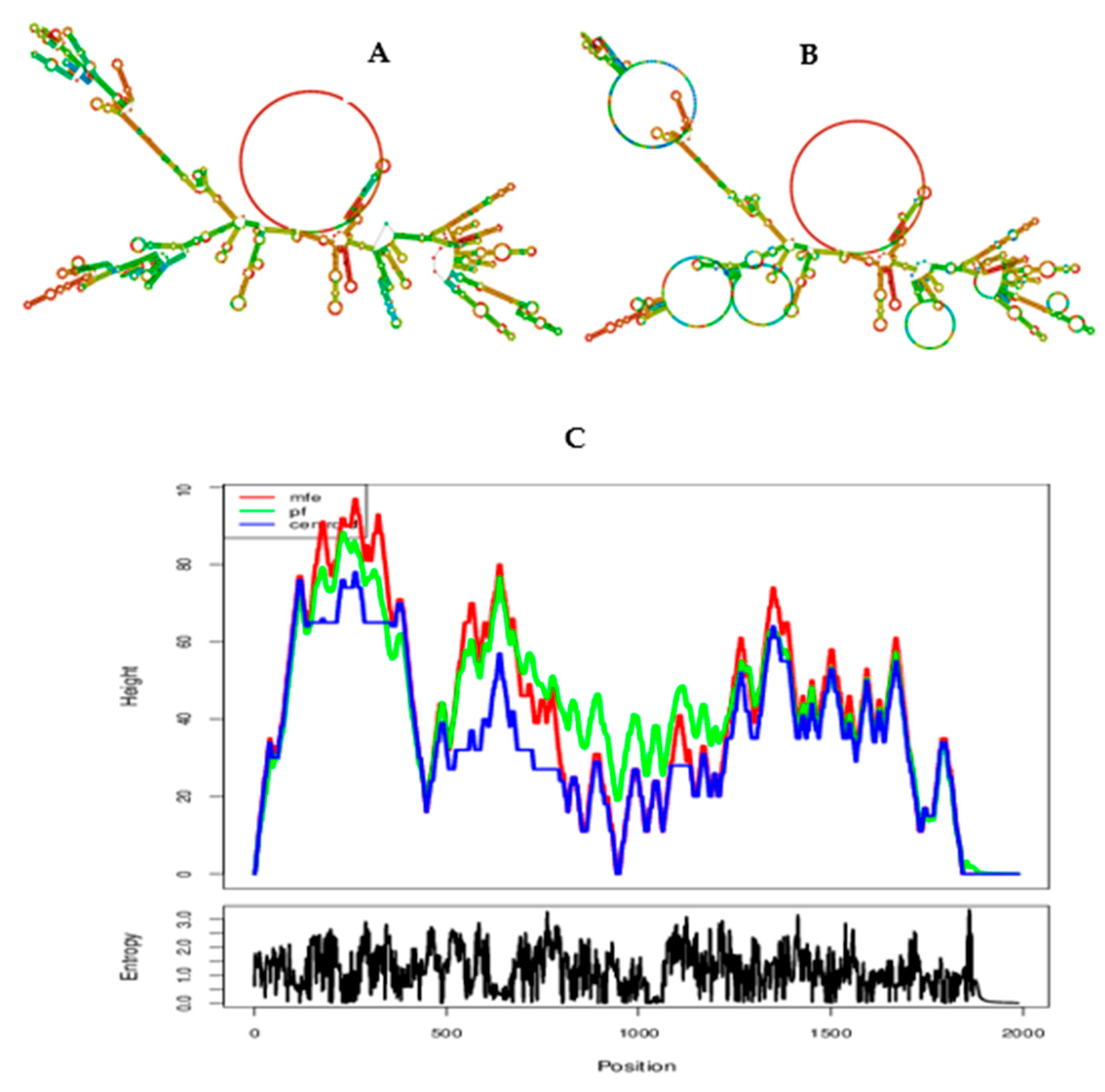
3.5. Secondary (2D) structure prediction of the chimeric L1/L2 mRNA–based vaccine
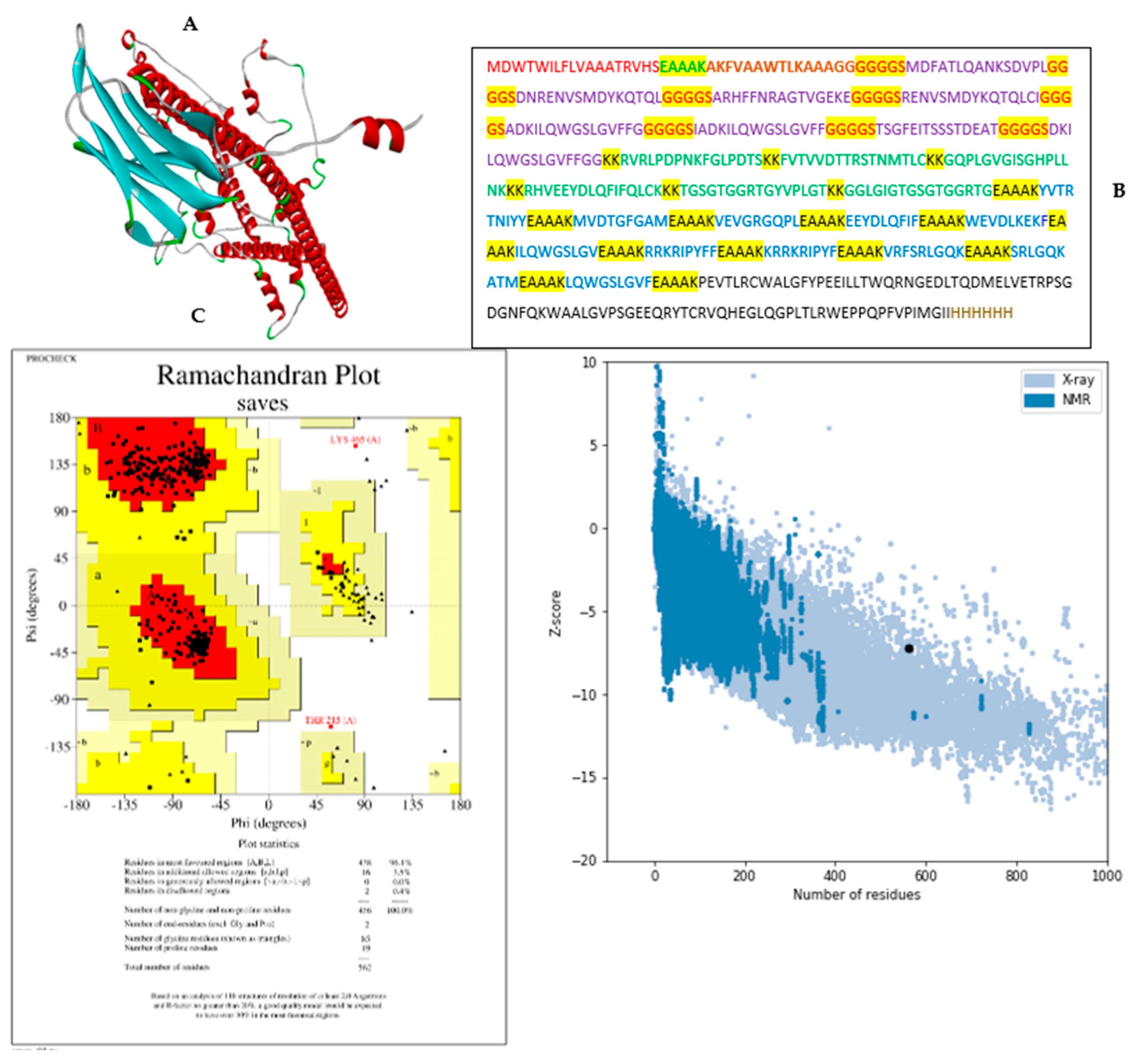
3.5. Modelling and validation of tertiary (3D) structure of the vaccine construct
3.10. Identification of discontinuous B-cell epitopes
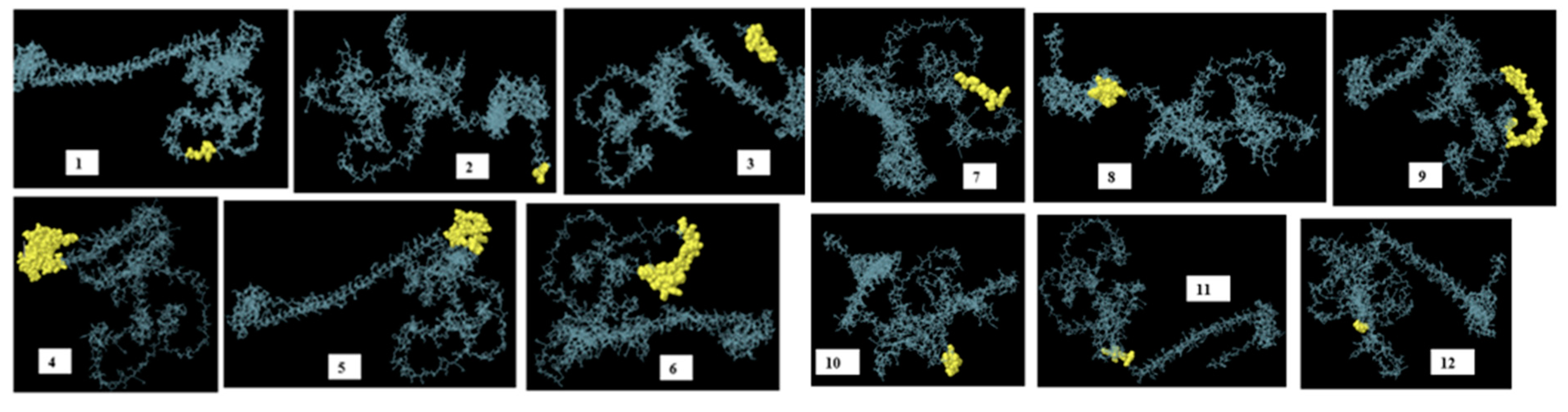
| No. | Residues | Number of residues | score |
| 1 | A:N106, A:V107, A:S108, A:M109, A:D110, A:Y111 | 6 | 0.979 |
| 2 | A:H560, A:H561, A:H562 | 3 | 0.969 |
| 3 | A:F549, A:V550, A:P551, A:I552, A:M553, A:G554, A:I555, A:I556, A:H557, A:H558, A:H559 | 11 | 0.895 |
| 4 | A:S438, A:R439, A:L440, A:G441, A:Q442, A:K443, A:A444, A:T445, A:M446, A:E447, A:A448, A:A449, A:A450, A:K451, A:L452, A:Q453, A:W454, A:G455, A:S456, A:L457, A:G458, A:V459, A:F460, A:E461, A:A462, A:A463, A:A464, A:K465, A:P466, A:E467, A:V468, A:T469, A:L470, A:R471, A:C472, A:W473, A:A474, A:L475, A:G476, A:F477, A:Y478, A:P479, A:E480, A:E481, A:I482, A:L483, A:L484, A:T485, A:W486, A:Q487, A:R488, A:N489, A:G490, A:E491, A:D492, A:L493, A:T494, A:Q495, A:D496, A:M497, A:E498, A:L499, A:V500, A:E501, A:T502, A:R503, A:P504, A:S505, A:G506, A:D507, A:G508, A:N509, A:F510, A:Q511, A:K512, A:W513, A:A514, A:A515, A:L516, A:G517, A:V518, A:P519, A:S520, A:G521, A:E522, A:E523, A:Q524, A:R525, A:Y526, A:T527, A:C528, A:R529, A:V530, A:Q531, A:H532, A:E533, A:G534, A:L535, A:Q536, A:G537, A:P538, A:L539, A:T540, A:L541, A:R542, A:W543, A:E544, A:P545, A:P546, A:Q547, A:P548 | 111 | 0.776 |
| 5 | A:T232, A:L233, A:C234, A:K235, A:K236, A:G237, A:Q238, A:P239, A:L240, A:G241, A:V242, A:G243, A:I244, A:S245, A:G246, A:H247, A:P248, A:L249, A:L250, A:N251, A:K252, A:K253, A:K254, A:R255 | 24 | 0.741 |
| 6 | A:D64, A:N65, A:E67, A:N68, A:V69, A:S70, A:M71, A:D72, A:Y73, A:K74, A:Q75, A:T76, A:Q77, A:L78, A:G79, A:G82, A:S83, A:A84, A:H86, A:F87, A:F88, A:N89, A:R90, A:A91, A:G92, A:T93, A:V94, A:G95, A:E96, A:K97, A:G99, A:G100, A:G101, A:G102, A:S103, A:R104, A:E105 | 37 | 0.74 |
| 7 | A:K112, A:Q113, A:T114, A:Q115, A:L116, A:C117, A:I118, A:G119 | 8 | 0.724 |
| 8 | A:G345, A:Q346, A:P347, A:L348, A:E349, A:A350, A:A351, A:A352, A:K353, A:E354, A:E355, A:Y356, A:D357, A:L358, A:Q359, A:F360, A:I361, A:E363, A:A364, A:K367 | 20 | 0.701 |
| 9 | A:W151, A:G152, A:S153, A:L154, A:G155, A:V156, A:F157, A:F158, A:G159, A:G160, A:G161, A:G162, A:S163, A:T164, A:S165, A:G166, A:F167, A:E168, A:I169, A:T170, A:S171, A:S172, A:S173, A:T174, A:D175, A:E176, A:A177, A:T178, A:G179, A:G180, A:G181 | 31 | 0.682 |
| 10 | A:G282, A:Y283, A:V284, A:P285, A:L286, A:G287, A:T288, A:K289, A:G291, A:G292, A:L293, A:G294, A:I295, A:G296 | 14 | 0.594 |
| 11 | A:A338, A:K339, A:V340, A:E341, A:V342, A:G343, A:R344 | 7 | 0.528 |
| 12 | A:A36, A:G37, A:G38, A:G39, A:G40 | 5 | 0.511 |
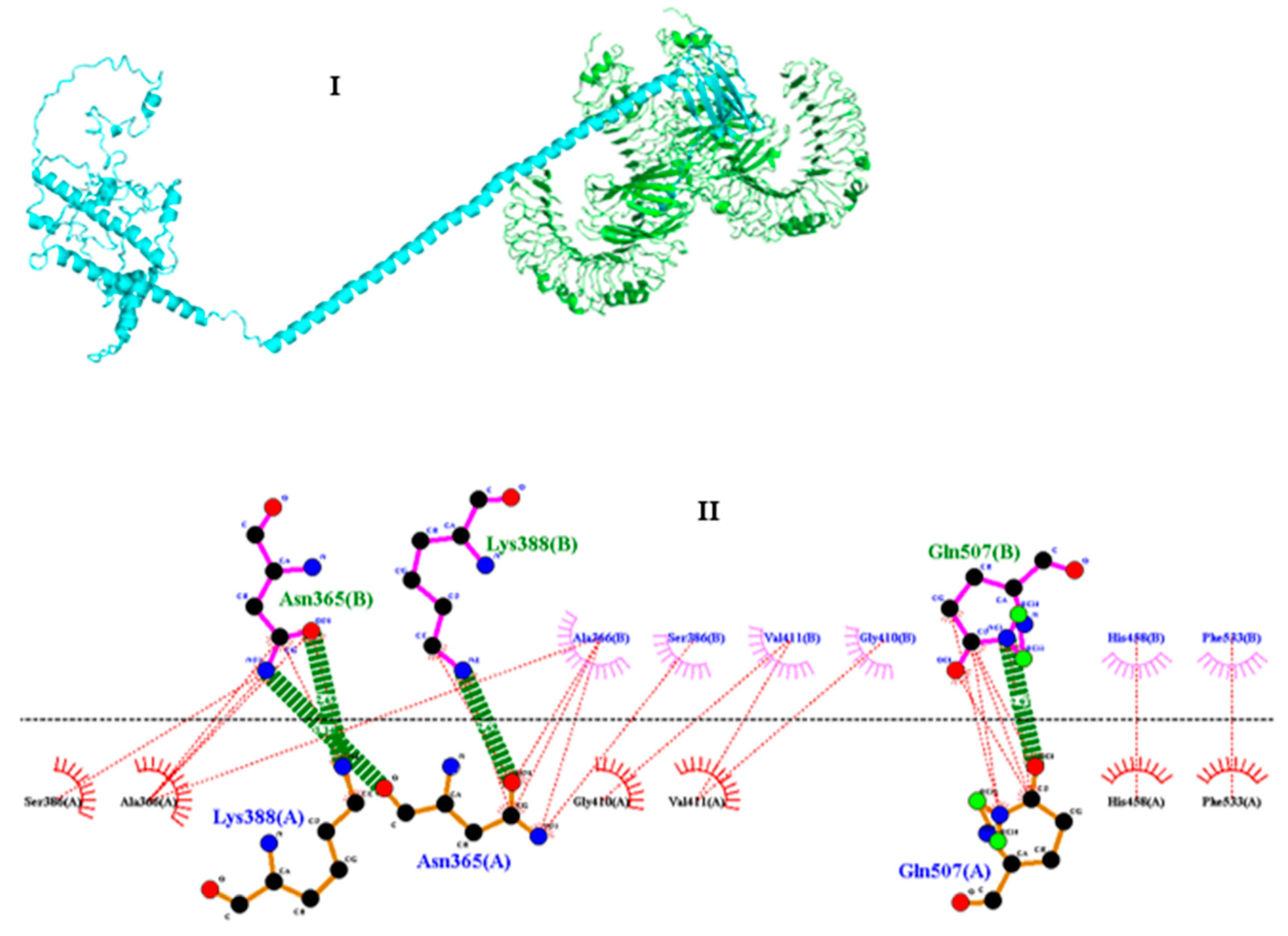
3.11. Molecular docking of vaccine construct with TLR4
3.12. Molecular dynamics simulation (MD)
3.13. Immune simulation of the chimeric vaccine
3.14. Codon-optimization and in Silico cloning
3.14. Prediction of secondary structure and minimum free enery of the chimeric vaccine
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mahmoudvand, S.; Shokri, S.; Makvandi, M.; Taherkhani, R.; Rashno, M.; Jalilian, F. A.; Angali, K. A. In Silico Prediction of T-Cell and B-Cell Epitopes of Human Papillomavirus Type 16 L1 Protein. Biotechnol. Appl. Biochem. 2022, 69, 514–525. [Google Scholar] [CrossRef]
- Singh, D.; Vignat, J.; Lorenzoni, V.; Eslahi, M.; Ginsburg, O.; Lauby-Secretan, B.; Arbyn, M.; Basu, P.; Bray, F.; Vaccarella, S. Global Estimates of Incidence and Mortality of Cervical Cancer in 2020: A Baseline Analysis of the WHO Global Cervical Cancer Elimination Initiative. Lancet Glob. Heal. 2023, 11, e197–e206. [Google Scholar] [CrossRef] [PubMed]
- Jedy-Agba, E.; Joko, W. Y.; Liu, B.; Buziba, N. G.; Borok, M.; Korir, A.; Masamba, L.; Manraj, S. S.; Finesse, A.; Wabinga, H.; Somdyala, N.; Parkin, D. M. Trends in Cervical Cancer Incidence in Sub-Saharan Africa. Br. J. Cancer 2020, 123, 148–154. [Google Scholar] [CrossRef]
- Drokow, E. K.; Fangninou, F. F.; Effah, C. Y.; Agboyibor, C.; Zhang, Y.; Arboh, F.; Deku, M. A.; Xinyin, W.; Wang, Y.; Sun, K. Cervical Cancer Survival Times in Africa. Front. Public Heal. 2022, 10. [Google Scholar] [CrossRef]
- Bruni, L.; Diaz, M.; Castellsagué, X.; Ferrer, E.; Bosch, F. X.; De Sanjosé, S. Cervical Human Papillomavirus Prevalence in 5 Continents: Meta-Analysis of 1 Million Women with Normal Cytological Findings. J. Infect. Dis. 2010, 202, 1789–1799. [Google Scholar] [CrossRef] [PubMed]
- Haręża, D. A.; Wilczyński, J. R.; Paradowska, E. Human Papillomaviruses as Infectious Agents in Gynecological Cancers―Oncogenic Properties of Viral Proteins. Int. J. Mol. Sci. 2022, 23. [Google Scholar] [CrossRef] [PubMed]
- Olczak, P.; Roden, R. B. S. Progress in L2-Based Prophylactic Vaccine Development for Protection against Diverse Human Papillomavirus Genotypes and Associated Diseases. Vaccines 2020, 8, 1–22. [Google Scholar] [CrossRef]
- Namvar, A.; Bolhassani, A.; Javadi, G.; Noormohammadi, Z. In Silico/In Vivo Analysis of High-Risk Papillomavirus L1 and L2 Conserved Sequences for Development of Cross-Subtype Prophylactic Vaccine. Sci. Rep. 2019, 9, 1–22. [Google Scholar] [CrossRef]
- Boxus, M.; Fochesato, M.; Miseur, A.; Mertens, E.; Dendouga, N.; Brendle, S.; Balogh, K. K.; Christensen, N. D.; Giannini, S. L. Broad Cross-Protection Is Induced in Preclinical Models by a Human Papillomavirus Vaccine Composed of L1/L2 Chimeric Virus-Like Particles. J. Virol. 2016, 90, 6314–6325. [Google Scholar] [CrossRef]
- Huber, B.; Wang, J. W.; Roden, R. B. S.; Kirnbauer, R. Rg1-Vlp and Other L2-Based, Broad-Spectrum Hpv Vaccine Candidates. J. Clin. Med. 2021, 10, 1–21. [Google Scholar] [CrossRef]
- Begum Akuzum1,†, Sinae Kim2, 3,†, Tam Thanh Nguyen2, 3,†, J. H.; Lee3, S.;, Eunhye Kim2, 3, J. K.; Choi1, Y.;, Hyunjhung Jhun3, 4; Lee5, Y.; Kim6, H.; Sohn7, D. H.;, Soohyun Kim1, 2. L1 Recombinant Proteins of HPV Tested for Antibody Forming Using Sera of HPV Quadrivalent Vaccine. Immune Netw. 2018, 18, 1–13. [Google Scholar]
- Huber, B.; Schellenbacher, C.; Shafti-Keramat, S.; Jindra, C.; Christensen, N.; Kirnbauer, R. Chimeric L2-Based Virus-like Particle (VLP) Vaccines Targeting Cutaneous Human Papillomaviruses (HPV). PLoS One 2017, 12, 1–27. [Google Scholar] [CrossRef]
- Schellenbacher, C.; Roden, R. B. S.; Kirnbauer, R. Developments in L2-Based Human Papillomavirus (HPV) Vaccines. Virus Res. 2017, 231, 166–175. [Google Scholar] [CrossRef] [PubMed]
- Tumban, E.; Peabody, J.; Peabody, D. S.; Chackerian, B. A Pan-HPV Vaccine Based on Bacteriophage PP7 VLPs Displaying Broadly Cross-Neutralizing Epitopes from the HPV Minor Capsid Protein, L2. PLoS One 2011, 6, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Varsani, A.; Williamson, A.-L.; de Villiers, D.; Becker, I.; Christensen, N. D.; Rybicki, E. P. Chimeric Human Papillomavirus Type 16 (HPV-16) L1 Particles Presenting the Common Neutralizing Epitope for the L2 Minor Capsid Protein of HPV-6 and HPV-16. J. Virol. 2003, 77, 8386–8393. [Google Scholar] [CrossRef] [PubMed]
- McGrath, M.; de Villiers, G. K.; Shephard, E.; Hitzeroth, I. I.; Rybicki, E. P. Development of Human Papillomavirus Chimaeric L1/L2 Candidate Vaccines. Arch. Virol. 2013, 158, 2079–2088. [Google Scholar] [CrossRef]
- Wang, J. W.; Roden, R. B. S. L2, the Minor Capsid Protein of Papillomavirus. Virology 2013, 445, 175–186. [Google Scholar] [CrossRef]
- Sanami, S.; Nazarian, S.; Ahmad, S.; Raeisi, E.; ul Qamar, M. T.; Tahmasebian, S.; Pazoki-Toroudi, H.; Fazeli, M.; Ghatreh Samani, M. In Silico Design and Immunoinformatics Analysis of a Universal Multi-Epitope Vaccine against Monkeypox Virus. PLoS One 2023, 18. [Google Scholar] [CrossRef]
- Oladipo, E. K.; Adeniyi, M. O.; Ogunlowo, M. T.; Irewolede, B. A.; Adekanola, V. O.; Oluseyi, G. S.; Omilola, J. A.; Udoh, A. F.; Olufemi, S. E.; Adediran, D. A.; Olonade, A.; Idowu, U. A.; Kolawole, O. M.; Oloke, J. K.; Onyeaka, H. Bioinformatics Designing and Molecular Modelling of a Universal MRNA Vaccine for SARS-CoV-2 Infection. Vaccines 2022, 10. [Google Scholar] [CrossRef]
- Maruggi, G.; Zhang, C.; Li, J.; Ulmer, J. B.; Yu, D. MRNA as a Transformative Technology for Vaccine Development to Control Infectious Diseases. Mol. Ther. 2019, 27. [Google Scholar] [CrossRef]
- Rcheulishvili, N.; Mao, J.; Papukashvili, D.; Feng, S.; Liu, C.; Yang, X. Candidate for Monkeypox, Smallpox, and Vaccinia Viruses : 2023.
- Hall, T. Bio Edit. Pdf. 2004. [Google Scholar]
- Waterhouse, A. M.; Procter, J. B.; Martin, D. M. A.; Clamp, M.; Barton, G. J. Jalview Version 2-A Multiple Sequence Alignment Editor and Analysis Workbench. Bioinformatics 2009, 25, 1189–1191. [Google Scholar] [CrossRef] [PubMed]
- Stranzl, T.; Larsen, M. V.; Lundegaard, C.; Nielsen, M. NetCTLpan: Pan-Specific MHC Class I Pathway Epitope Predictions. Immunogenetics 2010, 62, 357–368. [Google Scholar] [CrossRef]
- Jensen, K. K.; Andreatta, M.; Marcatili, P.; Buus, S.; Greenbaum, J. A.; Yan, Z.; Sette, A.; Peters, B.; Nielsen, M. Improved Methods for Predicting Peptide Binding Affinity to MHC Class II Molecules. Immunology 2018, 154, 394–406. [Google Scholar] [CrossRef]
- Dehghani, A.; Mamizadeh, M.; Karimi, A.; Hosseini, S. A.; Siamian, D.; Shams, M.; Ghiabi, S.; Basati, G.; Abaszadeh, A. Multi-Epitope Vaccine Design against Leishmaniasis Using IFN-γ Inducing Epitopes from Immunodominant Gp46 and G63 Proteins. J. Genet. Eng. Biotechnol. 2024, 22, 100355. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Sahu, U.; Kumari, P.; Dixit, A.; Khare, P. Designing of Multi-Epitope Chimeric Vaccine Using Immunoinformatic Platform by Targeting Oncogenic Strain HPV 16 and 18 against Cervical Cancer. Sci. Rep. 2022, 12, 1–16. [Google Scholar] [CrossRef]
- Sanchez, G. Las Instituciones de Ciencia y Tecnología En Los Procesos de Aprendizaje de La Producción Agroalimentaria En Argentina. El Sist. argentino innovación Inst. Empres. y redes. El desafío la creación y apropiación Conoc. 2013, 14, 659–664. [Google Scholar] [CrossRef]
- Naveed, M.; Hassan, J. U.; Ahmad, M.; Naeem, N.; Mughal, M. S.; Rabaan, A. A.; Aljeldah, M.; Shammari, B. R. A.; Alissa, M.; Sabour, A. A.; Alaeq, R. A.; Alshiekheid, M. A.; Turkistani, S. A.; Elmi, A. H.; Ahmed, N. Designing MRNA- and Peptide-Based Vaccine Construct against Emerging Multidrug-Resistant Citrobacter Freundii: A Computational-Based Subtractive Proteomics Approach. Med. 2022, 58. [Google Scholar] [CrossRef]
- De Groot, A. S.; Moise, L.; Terry, F.; Gutierrez, A. H.; Hindocha, P.; Richard, G.; Hoft, D. F.; Ross, T. M.; Noe, A. R.; Takahashi, Y.; Kotraiah, V.; Silk, S. E.; Nielsen, C. M.; Minassian, A. M.; Ashfield, R.; Ardito, M.; Draper, S. J.; Martin, W. D. Better Epitope Discovery, Precision Immune Engineering, and Accelerated Vaccine Design Using Immunoinformatics Tools. Front. Immunol. 2020, 11, 1–13. [Google Scholar] [CrossRef]
- Chen, C.; Li, Z.; Huang, H.; Suzek, B. E.; Wu, C. H. A Fast Peptide Match Service for UniProt Knowledgebase. Bioinformatics 2013, 29, 2808–2809. [Google Scholar] [CrossRef]
- Hasan, M.; Ahmed, S.; Imranuzzaman, M.; Bari, R.; Roy, S.; Hasan, M. M.; Mia, M. M. Designing and Development of Efficient Multi-Epitope-Based Peptide Vaccine Candidate against Emerging Avian Rotavirus Strains: A Vaccinomic Approach. J. Genet. Eng. Biotechnol. 2024, 22, 100398. [Google Scholar] [CrossRef]
- Shawan, M. M. A. K.; Sharma, A. R.; Halder, S. K.; Arian, T. Al; Shuvo, M. N.; Sarker, S. R.; Hasan, M. A. Advances in Computational and Bioinformatics Tools and Databases for Designing and Developing a Multi-Epitope-Based Peptide Vaccine. Int. J. Pept. Res. Ther. 2023, 29, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Kakakhel, S.; Ahmad, A.; Mahdi, W. A.; Alshehri, S.; Aiman, S.; Begum, S.; Shams, S.; Kamal, M.; Imran, M.; Shakeel, F.; Khan, A. Annotation of Potential Vaccine Targets and Designing of MRNA-Based Multi-Epitope Vaccine against Lumpy Skin Disease Virus via Reverse Vaccinology and Agent-Based Modeling. Bioengineering 2023, 10, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Mohammadi, Y.; Nezafat, N.; Negahdaripour, M.; Eskandari, S.; Zamani, M. In Silico Design and Evaluation of a Novel MRNA Vaccine against BK Virus: A Reverse Vaccinology Approach. Immunol. Res. 2023, 71, 422–441. [Google Scholar] [CrossRef] [PubMed]
- Ghaffari-Nazari, H.; Tavakkol-Afshari, J.; Jaafari, M. R.; Tahaghoghi-Hajghorbani, S.; Masoumi, E.; Jalali, S. A. Improving Multi-Epitope Long Peptide Vaccine Potency by Using a Strategy That Enhances CD4+ T Help in BALB/c Mice. PLoS One 2015, 10, 1–12. [Google Scholar] [CrossRef]
- Pichon, C.; Perche, F. Design and Delivery of Messenger RNA-Based Vaccines. Biochem. (Lond). 2021, 43, 4–7. [Google Scholar] [CrossRef]
- Du, Z.; Su, H.; Wang, W.; Ye, L.; Wei, H.; Peng, Z.; Anishchenko, I.; Baker, D.; Yang, J. The TrRosetta Server for Fast and Accurate Protein Structure Prediction. Nat. Protoc. 2021, 16, 5634–5651. [Google Scholar] [CrossRef]
- Ponomarenko, J.; Bui, H. H.; Li, W.; Fusseder, N.; Bourne, P. E.; Sette, A.; Peters, B. ElliPro: A New Structure-Based Tool for the Prediction of Antibody Epitopes. BMC Bioinformatics 2008, 9, 1–8. [Google Scholar] [CrossRef]
- Akbay, B.; Abidi, S. H.; Ibrahim, M. A. A.; Mukhatayev, Z.; Ali, S. Multi-Subunit Sars-Cov-2 Vaccine Design Using Evolutionarily Conserved t-and b-Cell Epitopes. Vaccines 2021, 9, 1–27. [Google Scholar] [CrossRef]
- Hollingsworth, S. A.; Dror, R. O. Molecular Dynamics Simulation for All. Neuron 2018, 99, 1129–1143. [Google Scholar] [CrossRef]
- López-Blanco, J. R.; Aliaga, J. I.; Quintana-Ortí, E. S.; Chacón, P. IMODS: Internal Coordinates Normal Mode Analysis Server. Nucleic Acids Res. 2014, 42, 271–276. [Google Scholar] [CrossRef] [PubMed]
- Rapin, N.; Lund, O.; Castiglione, F. Immune System Simulation Online. Bioinformatics 2011, 27, 2013–2014. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Yang, Q.; Zhao, F. Synonymous but Not Silent: The Codon Usage Code for Gene Expression and Protein Folding. Annu. Rev. Biochem. 2021, 90, 375–401. [Google Scholar] [CrossRef]
- Gruber, A. R.; Lorenz, R.; Bernhart, S. H.; Neuböck, R.; Hofacker, I. L. The Vienna RNA Websuite. Nucleic Acids Res. 2008, 36, 70–74. [Google Scholar] [CrossRef]
- Morris, A. L.; MacArthur, M. W.; Hutchinson, E. G.; Thornton, J. M. Stereochemical Quality of Protein Structure Coordinates. Proteins Struct. Funct. Bioinforma. 1992, 12, 345–364. [Google Scholar] [CrossRef]
- Adam, K. M. Immunoinformatics Approach for Multi-Epitope Vaccine Design against Structural Proteins and ORF1a Polyprotein of Severe Acute Respiratory Syndrome Coronavirus-2 (SARS-CoV-2). Trop. Dis. Travel Med. Vaccines 2021, 7, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Manuscript, A. Capsomeres to Broaden Protection against HPV Infection. Vaccination 2011, 28, 4478–4486. [Google Scholar]
- Gupta, S. K.; Singh, A.; Srivastava, M.; Gupta, S. K.; Akhoon, B. A. In Silico DNA Vaccine Designing against Human Papillomavirus (HPV) Causing Cervical Cancer. Vaccine 2009, 28, 120–131. [Google Scholar] [CrossRef]
- Chen, X.; Zaro, J. L.; Shen, W. C. Fusion Protein Linkers: Property, Design and Functionality. Adv. Drug Deliv. Rev. 2013, 65, 1357–1369. [Google Scholar] [CrossRef]
- Tossberg, J. T.; Esmond, T. M.; Aune, T. M. A Simplified Method to Produce MRNAs and Functional Proteins from Synthetic Double-Stranded DNA Templates. Biotechniques 2020, 69, 281–288. [Google Scholar] [CrossRef]
- Niwa, T.; Ying, B. W.; Saito, K.; Jin, W.; Takada, S.; Ueda, T.; Taguchi, H. Bimodal Protein Solubility Distribution Revealed by an Aggregation Analysis of the Entire Ensemble of Escherichia Coli Proteins. Proc. Natl. Acad. Sci. U. S. A. 2009, 106, 4201–4206. [Google Scholar] [CrossRef] [PubMed]
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Duvaud, S.; Wilkins, M. R.; Appel, R. D.; Bairoch, A. Protein Identification and Analysis Tools on the ExPASy Server; The Proteomics Protocols Handbook. Humana Press 2005, 112, 531–52. [Google Scholar]
- Wiederstein, M.; Sippl, M. J. ProSA-Web: Interactive Web Service for the Recognition of Errors in Three-Dimensional Structures of Proteins. Nucleic Acids Res. 2007, 35, 407–410. [Google Scholar] [CrossRef] [PubMed]
- Sarvmeili, J.; Baghban Kohnehrouz, B.; Gholizadeh, A.; Shanehbandi, D.; Ofoghi, H. Immunoinformatics Design of a Structural Proteins Driven Multi-Epitope Candidate Vaccine against Different SARS-CoV-2 Variants Based on Fynomer. Sci. Rep. 2024, 14, 1–26. [Google Scholar] [CrossRef]
- Lu, Q.; Wu, H.; Meng, J.; Wang, J.; Wu, J.; Liu, S.; Tong, J.; Nie, J.; Huang, W. Multi-Epitope Vaccine Design for Hepatitis E Virus Based on Protein ORF2 and ORF3. Front. Microbiol. 2024, 15, 1–10. [Google Scholar] [CrossRef]

| Type of epitopes | Allele | Epitope sequence | Antigenicity score | Toxicity | Conservancy analysis at the max. identity ≤ 100% |
| CTL (5) | HLA-A*01:01,HLA-A*26:01 | YVTRTNIYY | 0.6843 | Non-toxin | 100.00%: HPV31,HPV35,HPV29 |
| HLA-A*01:01,HLA-A*26:01 | MVDTGFGAM | 1.6337 | Non-toxin | 100.00%: HPV31, HPV35, HPV66, HPV6, HPV11, HPV40, HPV43, HPV44 | |
| HLA-B*39:01,HLA-B*40:01 | VEVGRGQPL | 1.2418 | Non-toxin | 100.00%: HPV16, HPV35, HPV39, HPV51, HPV40, HPV27, HPV29, HPV57 | |
| HLA-B*40:01 | EEYDLQFIF | 1.7384 | Non-toxin | 100.00%:HPV16,HPV18,HPV35,HPV39,HPV45,HPV45,HPV68, HPV6, HPV43,HPV27, HPV29, HPV57 | |
| HLA-B*40:01 | WEVDLKEKF | 1.2621 | Non-toxin | 100.00%: HPV35, HPV52, HPV33 | |
| HTL (4) | HLA-DRB1*01:01 | MDFATLQANKSDVPL | 0.7698 | Non-toxin | 93.33% |
| HLA-DRB1*03:01 | DNRENVSMDYKQTQL | 1.4352 | Non-toxin | 100%: HPV42 | |
| HLA-DRB1*04:05 | ARHFFNRAGTVGEKE | 0.7295 | Non-toxin | 86.67% | |
| HLA-DRB1*03:01 | RENVSMDYKQTQLCI | 1.6266 | Non-toxin | 93.33% | |
| FVTVVDTTRSTNMTLC | 1.0558 | Non-toxin | 100.00%: HPV52, HPV58, HPV6, HPV11, HPV33 | ||
| GQPLGVGISGHPLLNK | 0.5668 | Non-toxin | 100.00%: HPV16, HPV31, HPV35, HPV52, HPV42, HPV44, HPV33 | ||
| LBL (4) | RHVEEYDLQFIFQLCK | 0.4410 | Non-toxin | 93.75% | |
| RVRLPDPNKFGLPDTS | 0.4553 | Non-toxin | 100.00%: HPV16, HPV31, HPV35, HPV52, HPV42, HPV44, HPV33 |
| Type of epitopes | Allele | Epitope sequence | Antigenicity score at the threshold (≥ 0.4) | Toxicity | Conservancy analysis at the max. identity ≥ ≤ 100% |
| CTL (6) | HLA-A*02:01 | ILQWGSLGV | 1.8703 | Non-toxin | 100.00%: HPV42, HPV29 |
| HLA-B* 15:01 | LQWGSLGVF | 2.2096 | Non-toxin | 100%: HPV42 | |
| HLA-B*27:05 | RRKRIPYFF | 1.5931 | Non-toxin | 100.00%:HPV35,HPV56,HPV66, HPV51 | |
| HLA-B*27:05 | KRRKRIPYF | 1.7636 | Non-toxin | 100.00%: HPV51 | |
| HLA-B*27:05 | VRFSRLGQK | 2.1048 | Non-toxin | 100.00%:HPV51 | |
| HLA-B*27:05 | SRLGQKATM | 1.2853 | Non-toxin | 100.00%: HPV51 | |
| HTL (4) | HLA-DRB1*15:01 | ADKILQWGSLGVFFG | 0.8722 | Non-toxin | 100.00%: HPV42 |
| HLA-DRB1*15:01 | IADKILQWGSLGVFF | 0.9813 | Non-toxin | 93.33% | |
| HLA-DRB1*07:01 | TSGFEITSSSTDEAT | 0.6108 | Non-toxin | 73.33% | |
| HLA-DRB1*15:01 | DKILQWGSLGVFFGG | 0.6126 | Non-toxin | 100.00%: HPV42 | |
| LBL (2) | TGSGTGGRTGYVPLGT | 1.3789 | Non-toxin | 93.75% | |
| GGLGIGTGSGTGGRTG | 1.4744 | Non-toxin | 100.00%: HPV16, HPV18, HPV58, HPV59, HPV6, HPV43, HPV44, HPV27, HPV29,HPV 57 |
| Features | Value |
| Number of amino acids | 562aa |
| Molecular weigh | 60161.29~ 60kDa |
| Theoretical pI Solubility (≥ 0.45) Antigenicity score (≥ 0.4) Allergenicity Instability index Aliphatic index Grand average of hydropath city GRAVY) Formula Total number of atoms Estimated half-life |
9.44 0.451 0.9178 Non-allergen 36.89 68.49 -0.366 C2683H4187N761O786S15 8432 30 hours (mammalian reticulocytes, in vitro) > 20 hours (yeast, in vivo) > 10 hours (Escherichia coli, in vivo) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).





