Submitted:
21 October 2024
Posted:
21 October 2024
You are already at the latest version
Abstract
Keywords:
Pathogen Shedding and Wastewater Surveillance
Potential Role of a Non-Fecal Source in Near-Source Wastewater Surveillance
SARS-CoV-2 RNA in Urine: Literature Review and Data Extraction
Previous Reviews of SARS-CoV-2 RNA Shedding in Urine
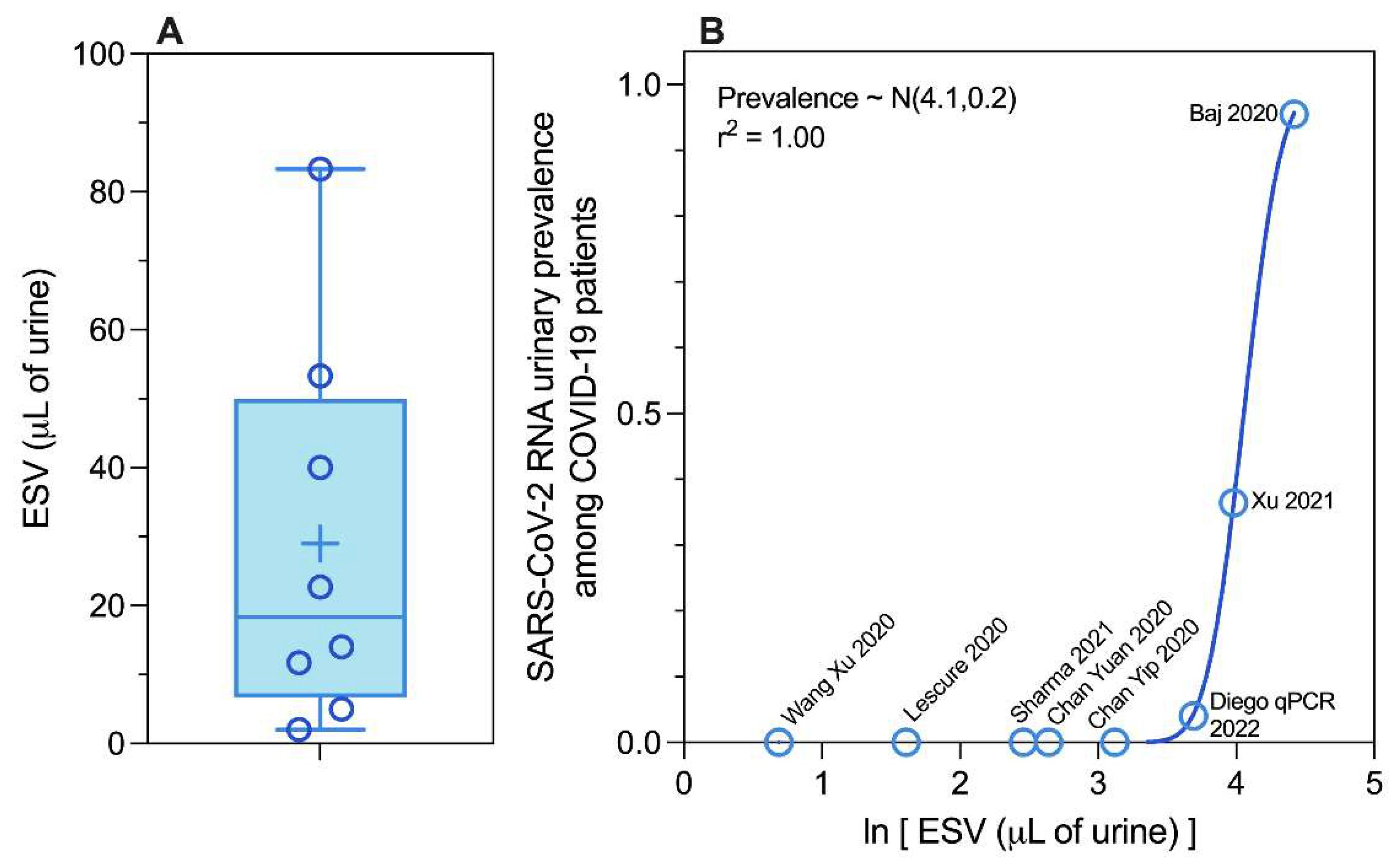
SARS-CoV-2 RNA Urinary Shedding Prevalence Revisited
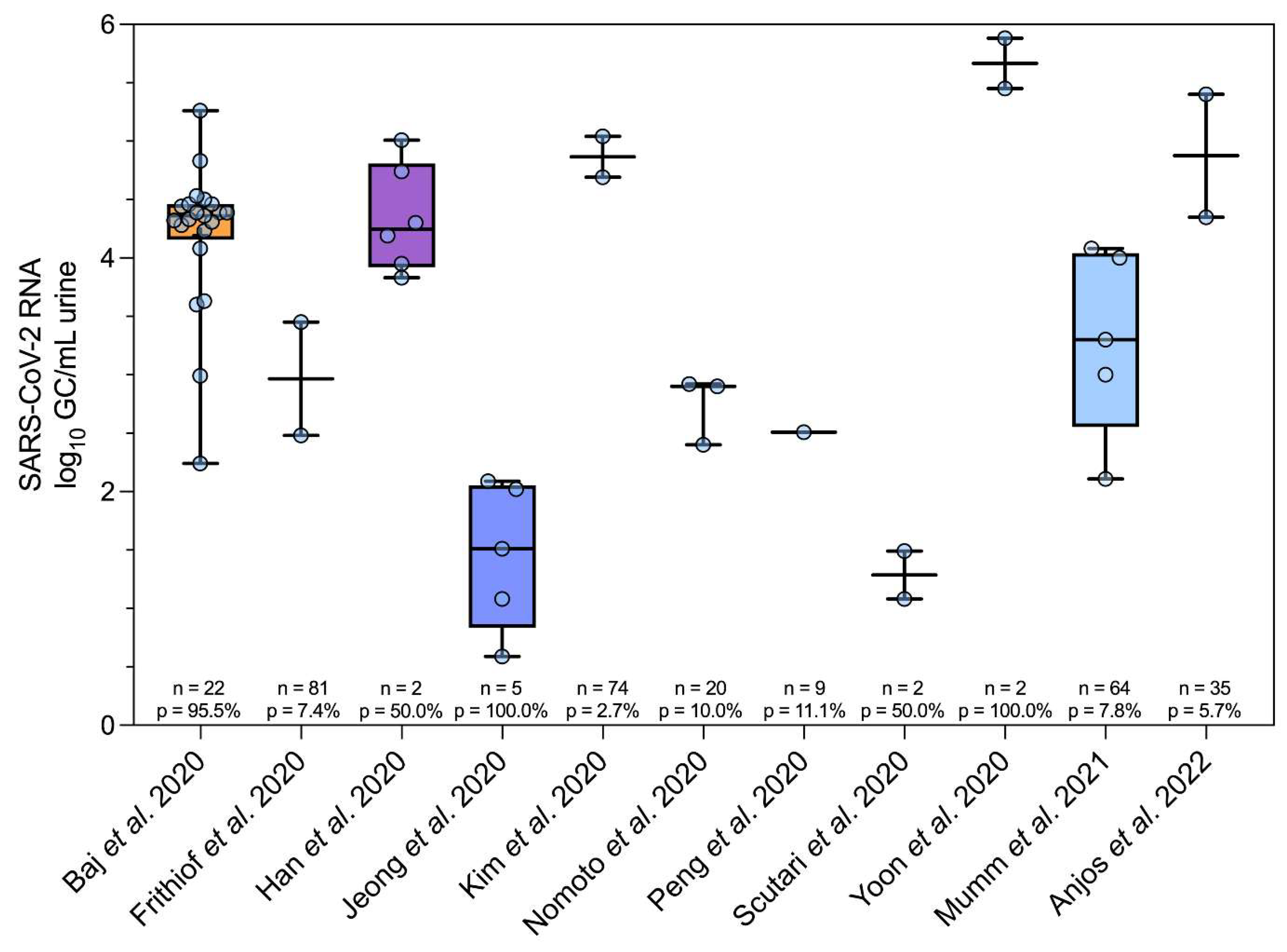
Clinical Diversity: Study Population
Methodological Diversity: Sample Type Equivalent Sample Volume
Methodological Diversity: Analytical System
Methodological Diversity: Equivalent Sample Volume
SARS-CoV-2 RNA Urinary Abundance Revisited
Additional Evidence for SARS-CoV-2 Constituents in Urine
Implications for Wastewater Surveillance of Infectious Disease (631 words)
Supplementary Materials
References
- Wilson, W.J. Isolation of Enteric Bacilli from Sewage and Water and Its Bearing on Epidemiology. Br Med J 1933, 2, 560–562. [Google Scholar] [CrossRef]
- Paul, J.R.; Trask, J.D.; Gard, S. II. POLIOMYELITIC VIRUS IN URBAN SEWAGE. J. Exp. Med. 1940, 71, 765–777. [Google Scholar] [CrossRef] [PubMed]
- Heijnen, L.; Medema, G. Surveillance of Influenza A and the Pandemic Influenza A (H1N1) 2009 in Sewage and Surface Water in the Netherlands. J. Water Health 2011, 9, 434–442. [Google Scholar] [CrossRef] [PubMed]
- Wastewater-Based Disease Surveillance for Public Health Action; National Academies Press: Washington, DC, USA, 2023. [CrossRef]
- Boehm, A.B.; Hughes, B.; Duong, D.; Chan-Herur, V.; Buchman, A.; Wolfe, M.K.; White, B.J. Wastewater Concentrations of Human Influenza, Metapneumovirus, Parainfluenza, Respiratory Syncytial Virus, Rhinovirus, and Seasonal Coronavirus Nucleic-Acids during the COVID-19 Pandemic: A Surveillance Study. Lancet Microbe 2023, 4, e340–e348. [Google Scholar] [CrossRef] [PubMed]
- Moore, B.; Perry, C.E.L.; Chard, S.T. A Survey by the Sewage Swab Method of Latent Enteric Infection in an Urban Area. Epidemiol. Infect. 1952, 50, 137–156. [Google Scholar] [CrossRef]
- Wölfel, R.; Corman, V.M.; Guggemos, W.; Seilmaier, M.; Zange, S.; Müller, M.A.; Niemeyer, D.; Jones, T.C.; Vollmar, P.; Rothe, C.; Hoelscher, M.; Bleicker, T.; Brünink, S.; Schneider, J.; Ehmann, R.; Zwirglmaier, K.; Drosten, C.; Wendtner, C. Virological Assessment of Hospitalized Patients with COVID-2019. Nature 2020, 581, 465–469. [Google Scholar] [CrossRef]
- He, X.; Lau, E.H.Y.; Wu, P.; Deng, X.; Wang, J.; Hao, X.; Lau, Y.C.; Wong, J.Y.; Guan, Y.; Tan, X.; Mo, X.; Chen, Y.; Liao, B.; Chen, W.; Hu, F.; Zhang, Q.; Zhong, M.; Wu, Y.; Zhao, L.; Zhang, F.; Cowling, B.J.; Li, F.; Leung, G.M. Temporal Dynamics in Viral Shedding and Transmissibility of COVID-19. Nat. Med. 2020, 26, 672–675. [Google Scholar] [CrossRef]
- Hart, O.E.; Halden, R.U. Computational Analysis of SARS-CoV-2/COVID-19 Surveillance by Wastewater-Based Epidemiology Locally and Globally: Feasibility, Economy, Opportunities and Challenges. Sci. Total Environ. 2020, 730, 138875. [Google Scholar] [CrossRef]
- Ahmed, W.; Angel, N.; Edson, J.; Bibby, K.; Bivins, A.; O’Brien, J.W.; Choi, P.M.; Kitajima, M.; Simpson, S.L.; Li, J.; Tscharke, B.; Verhagen, R.; Smith, W.J.M.; Zaugg, J.; Dierens, L.; Hugenholtz, P.; Thomas, K.V.; Mueller, J.F. First Confirmed Detection of SARS-CoV-2 in Untreated Wastewater in Australia: A Proof of Concept for the Wastewater Surveillance of COVID-19 in the Community. Sci. Total Environ. 2020, 728, 138764. [Google Scholar] [CrossRef]
- Crank, K.; Chen, W.; Bivins, A.; Lowry, S.; Bibby, K. Contribution of SARS-CoV-2 RNA Shedding Routes to RNA Loads in Wastewater. Sci. Total Environ. 2022, 806, 150376. [Google Scholar] [CrossRef]
- Arts, P.J.; Kelly, J.D.; Midgley, C.M.; Anglin, K.; Lu, S.; Abedi, G.R.; Andino, R.; Bakker, K.M.; Banman, B.; Boehm, A.B.; Briggs-Hagen, M.; Brouwer, A.F.; Davidson, M.C.; Eisenberg, M.C.; Garcia-Knight, M.; Knight, S.; Peluso, M.J.; Pineda-Ramirez, J.; Diaz Sanchez, R.; Saydah, S.; Tassetto, M.; Martin, J.N.; Wigginton, K.R. Longitudinal and Quantitative Fecal Shedding Dynamics of SARS-CoV-2, Pepper Mild Mottle Virus, and crAssphage. mSphere 2023, 8, e00132–23. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, W.; Bivins, A.; Simpson, S.L.; Bertsch, P.M.; Ehret, J.; Hosegood, I.; Metcalfe, S.S.; Smith, W.J.; Thomas, K.V.; Tynan, J. Wastewater Surveillance Demonstrates High Predictive Value for COVID-19 Infection on Board Repatriation Flights to Australia. Environ. Int. 2022, 158, 106938. [Google Scholar] [CrossRef] [PubMed]
- Jones, D.L.; Rhymes, J.M.; Wade, M.J.; Kevill, J.L.; Malham, S.K.; Grimsley, J.M.S.; Rimmer, C.; Weightman, A.J.; Farkas, K. Suitability of Aircraft Wastewater for Pathogen Detection and Public Health Surveillance. Sci. Total Environ. 2023, 856, 159162. [Google Scholar] [CrossRef] [PubMed]
- Farkas, K.; Williams, R.; Alex-Sanders, N.; Grimsley, J.M.S.; Pântea, I.; Wade, M.J.; Woodhall, N.; Jones, D.L. Wastewater-Based Monitoring of SARS-CoV-2 at UK Airports and Its Potential Role in International Public Health Surveillance. PLOS Glob. Public Health 2023, 3, e0001346. [Google Scholar] [CrossRef]
- Morfino, R.C. Notes from the Field: Aircraft Wastewater Surveillance for Early Detection of SARS-CoV-2 Variants — John F. Kennedy International Airport, New York City, August–September 2022. MMWR Morb. Mortal. Wkly. Rep. 2023, 72. [Google Scholar] [CrossRef]
- Bivins, A.; Morfino, R.; Franklin, A.; Simpson, S.; Ahmed, W. The Lavatory Lens: Tracking the Global Movement of Pathogens via Aircraft Wastewater. Preprints May 9, 2023. [CrossRef]
- Hassard, F.; Lundy, L.; Singer, A.C.; Grimsley, J.; Cesare, M.D. Innovation in Wastewater Near-Source Tracking for Rapid Identification of COVID-19 in Schools. Lancet Microbe 2021, 2, e4–e5. [Google Scholar] [CrossRef]
- Hassard, F.; Singh, S.; Coulon, F.; Yang, Z. Can Wastewater Monitoring Protect Public Health in Schools? Lancet Reg. Health Am. 2023, 20. [Google Scholar] [CrossRef]
- Wolken, M.; Sun, T.; McCall, C.; Schneider, R.; Caton, K.; Hundley, C.; Hopkins, L.; Ensor, K.; Domakonda, K.; Kalvapalle, P.; Persse, D.; Williams, S.; Stadler, L.B. Wastewater Surveillance of SARS-CoV-2 and Influenza in preK-12 Schools Shows School, Community, and Citywide Infections. Water Res. 2023, 231, 119648. [Google Scholar] [CrossRef]
- Crowe, J.; Schnaubelt, A.T.; SchmidtBonne, S.; Angell, K.; Bai, J.; Eske, T.; Nicklin, M.; Pratt, C.; White, B.; Crotts-Hannibal, B.; Staffend, N.; Herrera, V.; Cobb, J.; Conner, J.; Carstens, J.; Tempero, J.; Bouda, L.; Ray, M.; Lawler, J.V.; Campbell, W.S.; Lowe, J.-M.; Santarpia, J.; Bartelt-Hunt, S.; Wiley, M.; Brett-Major, D.; Logan, C.; Broadhurst, M.J. Assessment of a Program for SARS-CoV-2 Screening and Environmental Monitoring in an Urban Public School District. JAMA Netw. Open 2021, 4, e2126447. [Google Scholar] [CrossRef]
- Hassard, F.; Vu, M.; Rahimzadeh, S.; Castro-Gutierrez, V.; Stanton, I.; Burczynska, B.; Wildeboer, D.; Baio, G.; Brown, M.R.; Garelick, H.; Hofman, J.; Kasprzyk-Hordern, B.; Majeed, A.; Priest, S.; Denise, H.; Khalifa, M.; Bassano, I.; Wade, M.J.; Grimsley, J.; Lundy, L.; Singer, A.C.; Cesare, M.D. Wastewater Monitoring for Detection of Public Health Markers during the COVID-19 Pandemic: Near-Source Monitoring of Schools in England over an Academic Year. PLoS ONE 2023, 18, e0286259. [Google Scholar] [CrossRef]
- Kim, S.; Boehm, A.B. Wastewater Monitoring of SARS-CoV-2 RNA at K-12 Schools: Comparison to Pooled Clinical Testing Data. PeerJ 2023, 11, e15079. [Google Scholar] [CrossRef] [PubMed]
- Lopez Marin, M.A.; Zdenkova, K.; Bartackova, J.; Cermakova, E.; Dostalkova, A.; Demnerova, K.; Vavruskova, L.; Novakova, Z.; Sykora, P.; Rumlova, M.; Bartacek, J. Monitoring COVID-19 Spread in Selected Prague’s Schools Based on the Presence of SARS-CoV-2 RNA in Wastewater. Sci. Total Environ. 2023, 871, 161935. [Google Scholar] [CrossRef] [PubMed]
- Fielding-Miller, R.; Karthikeyan, S.; Gaines, T.; Garfein, R.S.; Salido, R.A.; Cantu, V.J.; Kohn, L.; Martin, N.K.; Wynn, A.; Wijaya, C.; Flores, M.; Omaleki, V.; Majnoonian, A.; Gonzalez-Zuniga, P.; Nguyen, M.; Vo, A.V.; Le, T.; Duong, D.; Hassani, A.; Tweeten, S.; Jepsen, K.; Henson, B.; Hakim, A.; Birmingham, A.; Hoff, P.D.; Mark, A.M.; Nasamran, C.A.; Rosenthal, S.B.; Moshiri, N.; Fisch, K.M.; Humphrey, G.; Farmer, S.; Tubb, H.M.; Valles, T.; Morris, J.; Kang, J.; Khaleghi, B.; Young, C.; Akel, A.D.; Eilert, S.; Eno, J.; Curewitz, K.; Laurent, L.C.; Rosing, T.; Knight, R.; Baer, N.A.; Barber, T.; Castro-Martinez, A.; Chacón, M.; Cheung, W.; Crescini, E.S.; Eisner, E.R.; Vargas, L.F.; Hakim, A.; Hobbs, C.; Lastrella, A.L.; Lawrence, E.S.; Matteson, N.L.; Gangavarapu, K.; Ngo, T.T.; Seaver, P.; Smoot, E.W.; Tsai, R.; Xia, B.; Aigner, S.; Anderson, C.; Belda-Ferre, P.; Sathe, S.; Zeller, M.; Andersen, K.G.; Yeo, G.W.; Kurzban, E. Safer at School Early Alert: An Observational Study of Wastewater and Surface Monitoring to Detect COVID-19 in Elementary Schools. Lancet Reg. Health Am. 2023, 19. [Google Scholar] [CrossRef] [PubMed]
- Castro-Gutierrez, V.; Hassard, F.; Vu, M.; Leitao, R.; Burczynska, B.; Wildeboer, D.; Stanton, I.; Rahimzadeh, S.; Baio, G.; Garelick, H.; Hofman, J.; Kasprzyk-Hordern, B.; Kwiatkowska, R.; Majeed, A.; Priest, S.; Grimsley, J.; Lundy, L.; Singer, A.C.; Cesare, M.D. Monitoring Occurrence of SARS-CoV-2 in School Populations: A Wastewater-Based Approach. PLoS ONE 2022, 17, e0270168. [Google Scholar] [CrossRef]
- Jones, D.L.; Baluja, M.Q.; Graham, D.W.; Corbishley, A.; McDonald, J.E.; Malham, S.K.; Hillary, L.S.; Connor, T.R.; Gaze, W.H.; Moura, I.B.; Wilcox, M.H.; Farkas, K. Shedding of SARS-CoV-2 in Feces and Urine and Its Potential Role in Person-to-Person Transmission and the Environment-Based Spread of COVID-19. Sci. Total Environ. 2020, 749, 141364. [Google Scholar] [CrossRef]
- Johnson, H.; Garg, M.; Shantikumar, S.; Thachil, J.; Rai, B.; Aboumarzouk, O.M.; Hashim, H.; Philip, J. COVID-19 (SARS-CoV-2) in Non-Airborne Body Fluids: A Systematic Review & Meta-Analysis. Turk. J. Urol. 2021, 47, 87–97. [Google Scholar] [CrossRef]
- Böger, B.; Fachi, M.M.; Vilhena, R.O.; Cobre, A.F.; Tonin, F.S.; Pontarolo, R. Systematic Review with Meta-Analysis of the Accuracy of Diagnostic Tests for COVID-19. Am. J. Infect. Control 2021, 49, 21–29. [Google Scholar] [CrossRef]
- Bwire, G.M.; Majigo, M.V.; Njiro, B.J.; Mawazo, A. Detection Profile of SARS-CoV-2 Using RT-PCR in Different Types of Clinical Specimens: A Systematic Review and Meta-Analysis. J. Med. Virol. 2021, 93, 719–725. [Google Scholar] [CrossRef]
- Morone, G.; Palomba, A.; Iosa, M.; Caporaso, T.; De Angelis, D.; Venturiero, V.; Savo, A.; Coiro, P.; Carbone, D.; Gimigliano, F.; Iolascon, G.; Paolucci, S. Incidence and Persistence of Viral Shedding in COVID-19 Post-Acute Patients With Negativized Pharyngeal Swab: A Systematic Review. Front. Med. 2020, 7, 562. [Google Scholar] [CrossRef]
- Roshandel, M.R.; Nateqi, M.; Lak, R.; Aavani, P.; Sari Motlagh, R.; F Shariat, S.; Aghaei Badr, T.; Sfakianos, J.; Kaplan, S.A.; Tewari, A.K. Diagnostic and Methodological Evaluation of Studies on the Urinary Shedding of SARS-CoV-2, Compared to Stool and Serum: A Systematic Review and Meta-Analysis. Cell. Mol. Biol. Noisy--Gd. Fr. 2020, 66, 148–156. [Google Scholar] [CrossRef]
- Brönimann, S.; Rebhan, K.; Lemberger, U.; Misrai, V.; Shariat, S.F.; Pradere, B. Secretion of Severe Acute Respiratory Syndrome Coronavirus 2 in Urine. Curr. Opin. Urol. 2020, 30, 735–739. [Google Scholar] [CrossRef] [PubMed]
- Chan, V.W.-S.; Chiu, P.K.-F.; Yee, C.-H.; Yuan, Y.; Ng, C.-F.; Teoh, J.Y.-C. A Systematic Review on COVID-19: Urological Manifestations, Viral RNA Detection and Special Considerations in Urological Conditions. World J. Urol. 2021, 39, 3127–3138. [Google Scholar] [CrossRef] [PubMed]
- Mishra, C.; Meena, S.; Meena, J.K.; Tiwari, S.; Mathur, P. Detection of Three Pandemic Causing Coronaviruses from Non-Respiratory Samples: Systematic Review and Meta-Analysis. Sci. Rep. 2021, 11, 16131. [Google Scholar] [CrossRef] [PubMed]
- Kashi, A.H.; De la Rosette, J.; Amini, E.; Abdi, H.; Fallah-Karkan, M.; Vaezjalali, M. Urinary Viral Shedding of COVID-19 and Its Clinical Associations: A Systematic Review and Meta-Analysis of Observational Studies. Urol. J. 2020, 17, 433–441. [Google Scholar] [CrossRef]
- Trypsteen, W.; Van Cleemput, J.; Snippenberg, W.v.; Gerlo, S.; Vandekerckhove, L. On the Whereabouts of SARS-CoV-2 in the Human Body: A Systematic Review. PLoS Pathog. 2020, 16, e1009037. [Google Scholar] [CrossRef]
- DerSimonian, R.; Laird, N. Meta-Analysis in Clinical Trials. Control. Clin. Trials 1986, 7, 177–188. [Google Scholar] [CrossRef]
- Wallace, B.C.; Dahabreh, I.J.; Trikalinos, T.A.; Lau, J.; Trow, P.; Schmid, C.H. Closing the Gap between Methodologists and End-Users: R as a Computational Back-End. J. Stat. Softw. 2012, 49, 1–15. [Google Scholar] [CrossRef]
- Hasanoglu, I.; Korukluoglu, G.; Asilturk, D.; Cosgun, Y.; Kalem, A.K.; Altas, A.B.; Kayaaslan, B.; Eser, F.; Kuzucu, E.A.; Guner, R. Higher Viral Loads in Asymptomatic COVID-19 Patients Might Be the Invisible Part of the Iceberg. Infection 2021, 49, 117–126. [Google Scholar] [CrossRef]
- Han, M.S.; Seong, M.-W.; Heo, E.Y.; Park, J.H.; Kim, N.; Shin, S.; Cho, S.I.; Park, S.S.; Choi, E.H. Sequential Analysis of Viral Load in a Neonate and Her Mother Infected With Severe Acute Respiratory Syndrome Coronavirus 2. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 2020, 71, 2236–2239. [Google Scholar] [CrossRef]
- Jiehao, C.; Jin, X.; Daojiong, L.; Zhi, Y.; Lei, X.; Zhenghai, Q.; Yuehua, Z.; Hua, Z.; Ran, J.; Pengcheng, L.; Xiangshi, W.; Yanling, G.; Aimei, X.; He, T.; Hailing, C.; Chuning, W.; Jingjing, L.; Jianshe, W.; Mei, Z. A Case Series of Children With 2019 Novel Coronavirus Infection: Clinical and Epidemiological Features. Clin. Infect. Dis. 2020, 71, 1547–1551. [Google Scholar] [CrossRef]
- Liu, P.; Cai, J.; Jia, R.; Xia, S.; Wang, X.; Cao, L.; Zeng, M.; Xu, J. Dynamic Surveillance of SARS-CoV-2 Shedding and Neutralizing Antibody in Children with COVID-19. Emerg. Microbes Infect. 2020, 9, 1254–1258. [Google Scholar] [CrossRef] [PubMed]
- Baj, A.; Azzi, L.; Dalla Gasperina, D.; Genoni, A.; Tamborini, A.; Gambarini, C.; Carcano, G.; Grossi, P.; Sessa, F. Pilot Study: Long-Term Shedding of SARS-CoV-2 in Urine: A Threat for Dispersal in Wastewater. Front. Public Health 2020, 8, 569209. [Google Scholar] [CrossRef] [PubMed]
- Couturier, A.; Ferlicot, S.; Chevalier, K.; Guillet, M.; Essig, M.; Jauréguiberry, S.; Collarino, R.; Dargelos, M.; Michaut, A.; Geri, G.; Roque-Afonso, A.-M.; Zaidan, M.; Massy, Z.A. Indirect Effects of Severe Acute Respiratory Syndrome Coronavirus 2 on the Kidney in Coronavirus Disease Patients. Clin. Kidney J. 2020, 13, 347–353. [Google Scholar] [CrossRef] [PubMed]
- Frithiof, R.; Bergqvist, A.; Järhult, J.D.; Lipcsey, M.; Hultström, M. Presence of SARS-CoV-2 in Urine Is Rare and Not Associated with Acute Kidney Injury in Critically Ill COVID-19 Patients. Crit. Care Lond. Engl. 2020, 24, 587. [Google Scholar] [CrossRef] [PubMed]
- Tong, Y.; Bao, A.; Chen, H.; Huang, J.; Lv, Z.; Feng, L.; Cheng, Y.; Wang, Y.; Bai, L.; Rao, W.; Zheng, H.; Wu, Z.; Qiao, B.; Zhao, Z.; Wang, H.; Li, Y. Necessity for Detection of SARS-CoV-2 RNA in Multiple Types of Specimens for the Discharge of the Patients with COVID-19. J. Transl. Med. 2020, 18, 411. [Google Scholar] [CrossRef]
- Tejerina, F.; Catalan, P.; Rodriguez-Grande, C.; Adan, J.; Rodriguez-Gonzalez, C.; Muñoz, P.; Aldamiz, T.; Diez, C.; Perez, L.; Fanciulli, C.; Garcia de Viedma, D. Post-COVID-19 Syndrome. SARS-CoV-2 RNA Detection in Plasma, Stool, and Urine in Patients with Persistent Symptoms after COVID-19. BMC Infect. Dis. 2022, 22, 211. [Google Scholar] [CrossRef]
- Fang, Z.; Zhang, Y.; Hang, C.; Ai, J.; Li, S.; Zhang, W. Comparisons of Viral Shedding Time of SARS-CoV-2 of Different Samples in ICU and Non-ICU Patients. J. Infect. 2020, 81, 147–178. [Google Scholar] [CrossRef]
- Zheng, S.; Fan, J.; Yu, F.; Feng, B.; Lou, B.; Zou, Q.; Xie, G.; Lin, S.; Wang, R.; Yang, X.; Chen, W.; Wang, Q.; Zhang, D.; Liu, Y.; Gong, R.; Ma, Z.; Lu, S.; Xiao, Y.; Gu, Y.; Zhang, J.; Yao, H.; Xu, K.; Lu, X.; Wei, G.; Zhou, J.; Fang, Q.; Cai, H.; Qiu, Y.; Sheng, J.; Chen, Y.; Liang, T. Viral Load Dynamics and Disease Severity in Patients Infected with SARS-CoV-2 in Zhejiang Province, China, January-March 2020: Retrospective Cohort Study. BMJ 2020, 369, m1443. [Google Scholar] [CrossRef]
- Perrella, A.; Brita, M.; Coletta, F.; Cotena, S.; De Marco, G.; Longobardi, A.; Sala, C.; Sannino, D.; Tomasello, A.; Perrella, M.; Russo, G.; Tarsitano, M.; Chetta, M.; Della Monica, M.; Orlando, V.; Coscioni, E.; Villani, R. SARS-CoV-2 in Urine May Predict a Severe Evolution of COVID-19. J. Clin. Med. 2021, 10, 4061. [Google Scholar] [CrossRef]
- Mendes-Correa, M.C.; Salomão, M.C.; Ghilardi, F.; Tozetto-Mendoza, T.R.; Santos Villas-Boas, L.; de Paula, A.V.; Paiao, H.G.O.; da Costa, A.C.; Leal, F.E.; Ferraz, A. de B. C.; Sales, F.C.S.; Claro, I.M.; Ferreira, N.E.; Pereira, G.M.; da Silva, A.R.J.; Freire, W.; Espinoza, E.P.S.; Manuli, E.R.; Romano, C.M.; de Jesus, J.G.; Sabino, E.C.; Witkin, S.S. SARS-CoV-2 Detection and Culture in Different Biological Specimens from Immunocompetent and Immunosuppressed COVID-19 Patients Infected with Two Different Viral Strains. Viruses 2023, 15. [Google Scholar] [CrossRef]
- Anjos, D.; Fiaccadori, F.S.; Servian, C.d.P.; da Fonseca, S.G.; Guilarde, A.O.; Borges, M.A.S.B.; Franco, F.C.; Ribeiro, B.M.; Souza, M. SARS-CoV-2 Loads in Urine, Sera and Stool Specimens in Association with Clinical Features of COVID-19 Patients. J. Clin. Virol. Plus 2022, 2, 100059. [Google Scholar] [CrossRef] [PubMed]
- Maltezou, H.C.; Raftopoulos, V.; Vorou, R.; Papadima, K.; Mellou, K.; Spanakis, N.; Kossyvakis, A.; Gioula, G.; Exindari, M.; Froukala, E.; Martinez-Gonzalez, B.; Panayiotakopoulos, G.; Papa, A.; Mentis, A.; Tsakris, A. Association Between Upper Respiratory Tract Viral Load, Comorbidities, Disease Severity, and Outcome of Patients With SARS-CoV-2 Infection. J. Infect. Dis. 2021, 223, 1132–1138. [Google Scholar] [CrossRef]
- Fajnzylber, J.; Regan, J.; Coxen, K.; Corry, H.; Wong, C.; Rosenthal, A.; Worrall, D.; Giguel, F.; Piechocka-Trocha, A.; Atyeo, C.; Fischinger, S.; Chan, A.; Flaherty, K.T.; Hall, K.; Dougan, M.; Ryan, E.T.; Gillespie, E.; Chishti, R.; Li, Y.; Jilg, N.; Hanidziar, D.; Baron, R.M.; Baden, L.; Tsibris, A.M.; Armstrong, K.A.; Kuritzkes, D.R.; Alter, G.; Walker, B.D.; Yu, X.; Li, J.Z. SARS-CoV-2 Viral Load Is Associated with Increased Disease Severity and Mortality. Nat. Commun. 2020, 11, 5493. [Google Scholar] [CrossRef]
- Dadras, O.; Afsahi, A.M.; Pashaei, Z.; Mojdeganlou, H.; Karimi, A.; Habibi, P.; Barzegary, A.; Fakhfouri, A.; Mirzapour, P.; Janfaza, N.; Dehghani, S.; Afroughi, F.; Dashti, M.; Khodaei, S.; Mehraeen, E.; Voltarelli, F.; Sabatier, J.-M.; SeyedAlinaghi, S. The Relationship between COVID-19 Viral Load and Disease Severity: A Systematic Review. Immun. Inflamm. Dis. 2022, 10, e580. [Google Scholar] [CrossRef]
- Zuin, M.; Gentili, V.; Cervellati, C.; Rizzo, R.; Zuliani, G. Viral Load Difference between Symptomatic and Asymptomatic COVID-19 Patients: Systematic Review and Meta-Analysis. Infect. Dis. Rep. 2021, 13, 645–653. [Google Scholar] [CrossRef]
- Zhang, L.; Tian, M.; Song, Y.; Liang, W.; Li, X.; Tong, Y.; Wang, H. Urinary SARS-CoV-2 RNA Is an Indicator for the Progression and Prognosis of COVID-19. Diagn. Basel Switz. 2021, 11. [Google Scholar] [CrossRef]
- Wang, W.; Xu, Y.; Gao, R.; Lu, R.; Han, K.; Wu, G.; Tan, W. Detection of SARS-CoV-2 in Different Types of Clinical Specimens. JAMA 2020, 323, 1843–1844. [Google Scholar] [CrossRef]
- Kim, J.-M.; Kim, H.M.; Lee, E.J.; Jo, H.J.; Yoon, Y.; Lee, N.-J.; Son, J.; Lee, Y.-J.; Kim, M.S.; Lee, Y.-P.; Chae, S.-J.; Park, K.R.; Cho, S.-R.; Park, S.; Kim, S.J.; Wang, E.; Woo, S.; Lim, A.; Park, S.-J.; Jang, J.; Chung, Y.-S.; Chin, B.S.; Lee, J.-S.; Lim, D.; Han, M.-G.; Yoo, C.K. Detection and Isolation of SARS-CoV-2 in Serum, Urine, and Stool Specimens of COVID-19 Patients from the Republic of Korea. Osong Public Health Res. Perspect. 2020, 11, 112–117. [Google Scholar] [CrossRef]
- Shinde, M.; Lavania, M.; Rawal, J.; Chavan, N.; Shinde, P. Evaluation of Droplet Digital qRT-PCR (Dd qRT-PCR) for Quantification of SARS CoV-2 RNA in Stool and Urine Specimens of COVID-19 Patients. Front. Med. 2023, 10, 1148688. [Google Scholar] [CrossRef]
- Jeong, H.W.; Kim, S.-M.; Kim, H.-S.; Kim, Y.-I.; Kim, J.H.; Cho, J.Y.; Kim, S.-H.; Kang, H.; Kim, S.-G.; Park, S.-J.; Kim, E.-H.; Choi, Y.K. Viable SARS-CoV-2 in Various Specimens from COVID-19 Patients. Clin. Microbiol. Infect. Off. Publ. Eur. Soc. Clin. Microbiol. Infect. Dis. 2020, 26, 1520–1524. [Google Scholar] [CrossRef]
- Chan, J.F.-W.; Yuan, S.; Kok, K.-H.; To, K.K.-W.; Chu, H.; Yang, J.; Xing, F.; Liu, J.; Yip, C.C.-Y.; Poon, R.W.-S.; Tsoi, H.-W.; Lo, S.K.-F.; Chan, K.-H.; Poon, V.K.-M.; Chan, W.-M.; Ip, J.D.; Cai, J.-P.; Cheng, V.C.-C.; Chen, H.; Hui, C.K.-M.; Yuen, K.-Y. A Familial Cluster of Pneumonia Associated with the 2019 Novel Coronavirus Indicating Person-to-Person Transmission: A Study of a Family Cluster. The Lancet 2020, 395, 514–523. [Google Scholar] [CrossRef] [PubMed]
- Xu, T.; Wang, J.; Hu, B.; Zhang, G.; Zhou, W.; Zheng, M.; Shen, B.; Sun, B.; Zhang, Y.; Chen, Y.; Yu, J.; Liang, M.; Pan, J.; Chen, C.; Chen, H.; Jiang, M.; Xu, L.; Qu, J.; Chen, J.-F. Identification of the RNase-Binding Site of SARS-CoV-2 RNA for Anchor Primer-PCR Detection of Viral Loading in 306 COVID-19 Patients. Brief. Bioinform. 2021, 22, 1215–1224. [Google Scholar] [CrossRef] [PubMed]
- García-Bernalt Diego, J.; Fernández-Soto, P.; Muñoz-Bellido, J.L.; Febrer-Sendra, B.; Crego-Vicente, B.; Carbonell, C.; López-Bernús, A.; Marcos, M.; Belhassen-García, M.; Muro, A. Detection of SARS-CoV-2 RNA in Urine by RT-LAMP: A Very Rare Finding. J. Clin. Med. 2022, 11, 158. [Google Scholar] [CrossRef] [PubMed]
- Scutari, R.; Piermatteo, L.; Ciancio Manuelli, M.; Iannetta, M.; Salpini, R.; Bertoli, A.; Alteri, C.; Saccomandi, P.; Bellocchi, M.C.; Malagnino, V.; Teti, E.; Sforza, D.; Siragusa, L.; Grande, M.; Sarmati, L.; Svicher, V.; Andreoni, M.; Ceccherini-Silberstein, F. Long-Term SARS-CoV-2 Infection Associated with Viral Dissemination in Different Body Fluids Including Bile in Two Patients with Acute Cholecystitis. Life Basel Switz. 2020, 10. [Google Scholar] [CrossRef] [PubMed]
- Yu, F.; Yan, L.; Wang, N.; Yang, S.; Wang, L.; Tang, Y.; Gao, G.; Wang, S.; Ma, C.; Xie, R.; Wang, F.; Tan, C.; Zhu, L.; Guo, Y.; Zhang, F. Quantitative Detection and Viral Load Analysis of SARS-CoV-2 in Infected Patients. Clin. Infect. Dis. 2020, 71, 793–798. [Google Scholar] [CrossRef]
- Whale, A.S.; von der Heide, E.K.; Kohlenberg, M.; Brinckmann, A.; Baedker, S.; Karalay, O.; Fernandez-Gonzalez, A.; Busby, E.J.; Bustin, S.A.; Hauser, H.; Missel, A.; O’Sullivan, D.M.; Huggett, J.F.; Pfaffl, M.W.; Nolan, T. Digital PCR Can Augment the Interpretation of RT-qPCR Cq Values for SARS-CoV-2 Diagnostics. Methods San Diego Calif 2022, 201, 5–14. [Google Scholar] [CrossRef]
- Amendola, A.; Canuti, M.; Bianchi, S.; Kumar, S.; Fappani, C.; Gori, M.; Colzani, D.; Kosakovsky Pond, S.L.; Miura, S.; Baggieri, M.; Marchi, A.; Borghi, E.; Zuccotti, G.; Raviglione, M.C.; Magurano, F.; Tanzi, E. Molecular Evidence for SARS-CoV-2 in Samples Collected from Patients with Morbilliform Eruptions since Late 2019 in Lombardy, Northern Italy. Environ. Res. 2022, 215 Pt 1, 113979. [Google Scholar] [CrossRef]
- Crank, K.; Papp, K.; Barber, C.; Wang, P.; Bivins, A.; Gerrity, D. Correspondence on “The Environmental Microbiology Minimum Information (EMMI) Guidelines: qPCR and dPCR Quality and Reporting for Environmental Microbiology. ” Environ. Sci. Technol. 2023, 57, 20448–20449. [Google Scholar] [CrossRef]
- Mumm, J.-N.; Ledderose, S.; Ostermann, A.; Rudelius, M.; Hellmuth, J.C.; Münchhoff, M.; Munker, D.; Scherer, C.; Volz, Y.; Ebner, B.; Giessen-Jung, C.; Lampert, C.; Vilsmaier, T.; Schneider, S.; Gapp, M.; Milger-Kneidinger, K.; Behr, J.; von Bergwelt-Baildon, M.; Keppler, O.T.; Stief, C.; Magistro, G.; Staehler, M.; Rodler, S. Dynamics of Urinary and Respiratory Shedding of Severe Acute Respiratory Syndrome Virus 2 (SARS-CoV-2) RNA Excludes Urine as a Relevant Source of Viral Transmission. Infection 2022, 50, 635–642. [Google Scholar] [CrossRef]
- Nomoto, H.; Ishikane, M.; Katagiri, D.; Kinoshita, N.; Nagashima, M.; Sadamasu, K.; Yoshimura, K.; Ohmagari, N. Cautious Handling of Urine from Moderate to Severe COVID-19 Patients. Am. J. Infect. Control 2020, 48, 969–971. [Google Scholar] [CrossRef]
- Peng, L.; Liu, J.; Xu, W.; Luo, Q.; Chen, D.; Lei, Z.; Huang, Z.; Li, X.; Deng, K.; Lin, B.; Gao, Z. SARS-CoV-2 Can Be Detected in Urine, Blood, Anal Swabs, and Oropharyngeal Swabs Specimens. J. Med. Virol. 2020, 92, 1676–1680. [Google Scholar] [CrossRef] [PubMed]
- Yoon, J.G.; Yoon, J.; Song, J.Y.; Yoon, S.-Y.; Lim, C.S.; Seong, H.; Noh, J.Y.; Cheong, H.J.; Kim, W.J. Clinical Significance of a High SARS-CoV-2 Viral Load in the Saliva. J. Korean Med. Sci. 2020, 35. [Google Scholar] [CrossRef] [PubMed]
- Bivins, A.; Kaya, D.; Bibby, K.; Simpson, S.L.; Bustin, S.A.; Shanks, O.C.; Ahmed, W. Variability in RT-qPCR Assay Parameters Indicates Unreliable SARS-CoV-2 RNA Quantification for Wastewater Surveillance. Water Res. 2021, 203, 117516. [Google Scholar] [CrossRef] [PubMed]
- Huggett, J.F.; Foy, C.A.; Benes, V.; Emslie, K.; Garson, J.A.; Haynes, R.; Hellemans, J.; Kubista, M.; Mueller, R.D.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; Vandesompele, J.; Wittwer, C.T.; Bustin, S.A. The Digital MIQE Guidelines: Minimum Information for Publication of Quantitative Digital PCR Experiments. Clin. Chem. 2013, 59, 892–902. [Google Scholar] [CrossRef] [PubMed]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; Vandesompele, J.; Wittwer, C.T. The MIQE Guidelines: Minimum Information for Publication of Quantitative Real-Time PCR Experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef]
- Liu, P.; Sablon, O.; Wang, Y.; Hilton, S.P.; Khalil, L.; Ingersoll, J.M.; Truell, J.; Edupuganti, S.; Alaaeddine, G.; Naji, A.; Monarrez, E.; Wolfe, M.; Rouphael, N.; Kraft, C.; Moe, C.L. Longitudinal Fecal Shedding of SARS-CoV-2, Pepper Mild Mottle Virus, and Human Mitochondrial DNA in COVID-19 Patients. Front. Med. 2024, 11. [Google Scholar] [CrossRef]
- Janovičová, Ľ.; Kmeťová, K.; Tóthová, Ľ.; Vlková, B.; Celec, P. DNA in Fresh Urine Supernatant Is Not Affected by Additional Centrifugation and Is Protected against Deoxyribonuclease. Mol. Cell. Probes 2023, 68, 101900. [Google Scholar] [CrossRef]
- Su, Y.-H.; Wang, M.; Brenner, D.E.; Ng, A.; Melkonyan, H.; Umansky, S.; Syngal, S.; Block, T.M. Human Urine Contains Small, 150 to 250 Nucleotide-Sized, Soluble DNA Derived from the Circulation and May Be Useful in the Detection of Colorectal Cancer. J. Mol. Diagn. 2004, 6, 101–107. [Google Scholar] [CrossRef]
- Wang, M.; Xiong, H.; Chen, H.; Li, Q.; Ruan, X.Z. Renal Injury by SARS-CoV-2 Infection: A Systematic Review. Kidney Dis. 2020, 7, 100–110. [Google Scholar] [CrossRef]
- Cheung, M.D.; Erman, E.N.; Liu, S.; Erdmann, N.B.; Ghajar-Rahimi, G.; Moore, K.H.; Edberg, J.C.; George, J.F.; Agarwal, A. Single-Cell RNA Sequencing of Urinary Cells Reveals Distinct Cellular Diversity in COVID-19-Associated AKI. Kidney360 2022, 3, 28–36. [Google Scholar] [CrossRef]
- Sun, J.; Zhu, A.; Li, H.; Zheng, K.; Zhuang, Z.; Chen, Z.; Shi, Y.; Zhang, Z.; Chen, S.-B.; Liu, X.; Dai, J.; Li, X.; Huang, S.; Huang, X.; Luo, L.; Wen, L.; Zhuo, J.; Li, Y.; Wang, Y.; Zhang, L.; Zhang, Y.; Li, F.; Feng, L.; Chen, X.; Zhong, N.; Yang, Z.; Huang, J.; Zhao, J.; Li, Y.-M. Isolation of Infectious SARS-CoV-2 from Urine of a COVID-19 Patient. Emerg. Microbes Infect. 2020, 9, 991–993. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Song, L.; Zheng, N.; Shi, J.; Wu, H.; Yang, X.; Xue, N.; Chen, X.; Li, Y.; Sun, C.; Chen, C.; Tang, L.; Ni, X.; Wang, Y.; Shi, Y.; Guo, J.; Wang, G.; Zhang, Z.; Qin, J. A Urinary Proteomic Landscape of COVID-19 Progression Identifies Signaling Pathways and Therapeutic Options. Sci. China Life Sci. 2022, 65, 1866–1880. [Google Scholar] [CrossRef] [PubMed]
- Wendt, R.; Thijs, L.; Kalbitz, S.; Mischak, H.; Siwy, J.; Raad, J.; Metzger, J.; Neuhaus, B.; Leyen, H.v.d.; Dudoignon, E.; Mebazaa, A.; Spasovski, G.; Milenkova, M.; Canevska-Talevska, A.; Czerwieńska, B.; Wiecek, A.; Peters, B.; Nilsson, Å.; Schwab, M.; Rothfuss, K.; Lübbert, C.; Staessen, J.A.; Beige, J. A Urinary Peptidomic Profile Predicts Outcome in SARS-CoV-2-Infected Patients. eClinicalMedicine 2021, 36. [Google Scholar] [CrossRef] [PubMed]
- Chavan, S.; Mangalaparthi, K.K.; Singh, S.; Renuse, S.; Vanderboom, P.M.; Madugundu, A.K.; Budhraja, R.; McAulay, K.; Grys, T.E.; Rule, A.D.; Alexander, M.P.; O’Horo, J.C.; Badley, A.D.; Pandey, A. Mass Spectrometric Analysis of Urine from COVID-19 Patients for Detection of SARS-CoV-2 Viral Antigen and to Study Host Response. J. Proteome Res. 2021, 20, 3404–3413. [Google Scholar] [CrossRef] [PubMed]
- Tampe, D.; Hakroush, S.; Bösherz, M.-S.; Franz, J.; Hofmann-Winkler, H.; Pöhlmann, S.; Kluge, S.; Moerer, O.; Stadelmann, C.; Ströbel, P.; Winkler, M.S.; Tampe, B. Urinary Levels of SARS-CoV-2 Nucleocapsid Protein Associate With Risk of AKI and COVID-19 Severity: A Single-Center Observational Study. Front. Med. 2021, 8. [Google Scholar] [CrossRef]
- George, S.; Pal, A.C.; Gagnon, J.; Timalsina, S.; Singh, P.; Vydyam, P.; Munshi, M.; Chiu, J.E.; Renard, I.; Harden, C.A.; Ott, I.M.; Watkins, A.E.; Vogels, C.B.F.; Lu, P.; Tokuyama, M.; Venkataraman, A.; Casanovas-Massana, A.; Wyllie, A.L.; Rao, V.; Campbell, M.; Farhadian, S.F.; Grubaugh, N.D.; Dela Cruz, C.S.; Ko, A.I.; Berna Perez, A.Z.; Akaho, E.H.; Moledina, D.G.; Testani, J.; John, A.R.; Ledizet, M.; Mamoun, C.B. Evidence for SARS-CoV-2 Spike Protein in the Urine of COVID-19 Patients. Kidney360 2021, 2, 924–936. [Google Scholar] [CrossRef]
- Markt, R.; Stillebacher, F.; Nägele, F.; Kammerer, A.; Peer, N.; Payr, M.; Scheffknecht, C.; Dria, S.; Draxl-Weiskopf, S.; Mayr, M.; Rauch, W.; Kreuzinger, N.; Rainer, L.; Bachner, F.; Zuba, M.; Ostermann, H.; Lackner, N.; Insam, H.; Wagner, A.O. Expanding the Pathogen Panel in Wastewater Epidemiology to Influenza and Norovirus. Viruses 2023, 15, 263. [Google Scholar] [CrossRef]
- Kilaru, P.; Hill, D.; Anderson, K.; Collins, M.B.; Green, H.; Kmush, B.L.; Larsen, D.A. Wastewater Surveillance for Infectious Disease: A Systematic Review. Am. J. Epidemiol. 2023, 192, 305–322. [Google Scholar] [CrossRef]
- Keshaviah, A.; Diamond, M.B.; Wade, M.J.; Scarpino, S.V.; Ahmed, W.; Amman, F.; Aruna, O.; Badilla-Aguilar, A.; Bar-Or, I.; Bergthaler, A.; et al. . Wastewater Monitoring Can Anchor Global Disease Surveillance Systems. Lancet Glob. Health 2023, 11, e976–e981. [Google Scholar] [CrossRef]
- Lee, W.L.; Gu, X.; Armas, F.; Leifels, M.; Wu, F.; Chandra, F.; Chua, F.J.D.; Syenina, A.; Chen, H.; Cheng, D.; Ooi, E.E.; Wuertz, S.; Alm, E.J.; Thompson, J. Monitoring Human Arboviral Diseases through Wastewater Surveillance: Challenges, Progress and Future Opportunities. Water Res. 2022, 223, 118904. [Google Scholar] [CrossRef]
- Chen, W.; Bibby, K. Model-Based Theoretical Evaluation of the Feasibility of Using Wastewater-Based Epidemiology to Monitor Monkeypox. Environ. Sci. Technol. Lett. 2022, 9, 772–778. [Google Scholar] [CrossRef]
- Chen, W.; Bibby, K. Making Waves: Establishing a Modeling Framework to Evaluate Novel Targets for Wastewater-Based Surveillance. Water Res. 2023, 245, 120573. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Bibby, K. A Model-Based Framework to Assess the Feasibility of Monitoring Zika Virus with Wastewater-Based Epidemiology. ACS EST Water 2023, 3, 1071–1081. [Google Scholar] [CrossRef]
- Lowry, S.A.; Wolfe, M.K.; Boehm, A.B. Respiratory Virus Concentrations in Human Excretions That Contribute to Wastewater: A Systematic Review and Meta-Analysis. J. Water Health 2023, 21, 831–848. [Google Scholar] [CrossRef]
- Rauch, W.; Brockmann, D.; Peters, I.; Larsen, T.A.; Gujer, W. Combining Urine Separation with Waste Design: An Analysis Using a Stochastic Model for Urine Production. Water Res. 2003, 37, 681–689. [Google Scholar] [CrossRef]
- Vo, V.; Harrington, A.; Chang, C.-L.; Baker, H.; Moshi, M.A.; Ghani, N.; Itorralba, J.Y.; Tillett, R.L.; Dahlmann, E.; Basazinew, N.; Gu, R.; Familara, T.D.; Boss, S.; Vanderford, F.; Ghani, M.; Tang, A.J.; Matthews, A.; Papp, K.; Khan, E.; Koutras, C.; Kan, H.-Y.; Lockett, C.; Gerrity, D.; Oh, E.C. Identification and Genome Sequencing of an Influenza H3N2 Variant in Wastewater from Elementary Schools during a Surge of Influenza A Cases in Las Vegas, Nevada. Sci. Total Environ. 2023, 872, 162058. [Google Scholar] [CrossRef]
- St-Onge, G.; Davis, J.T.; Hébert-Dufresne, L.; Allard, A.; Urbinati, A.; Scarpino, S.V.; Chinazzi, M.; Vespignani, A. Optimization and Performance Analytics of Global Aircraft-Based Wastewater Surveillance Networks. medRxiv August 4, 2024, p 2024.08.02.24311418. [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




