Submitted:
21 October 2024
Posted:
22 October 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Antiretroviral Drugs
2.2. Vectors
2.3. Laboratory-Adapted Strains
2.4. HEK293T Cell Culture
2.5. Selection of NRTI Drug Resistance Mutations
2.6. Preparation of Mutant Pseudoviruses in Reference Isolates
2.7. Preparation of Mutant Pseudoviruses in Laboratory-Adapted Strains
2.8. In Vitro Phenotypic ISL Susceptibility Testing
2.9. Statistics
3. Results
3.1. Sequence database
3.1.1. Regimens
3.1.2. NRTI resistance mutations
3.2. In Vitro Phenotypic ISL Susceptibility Testing
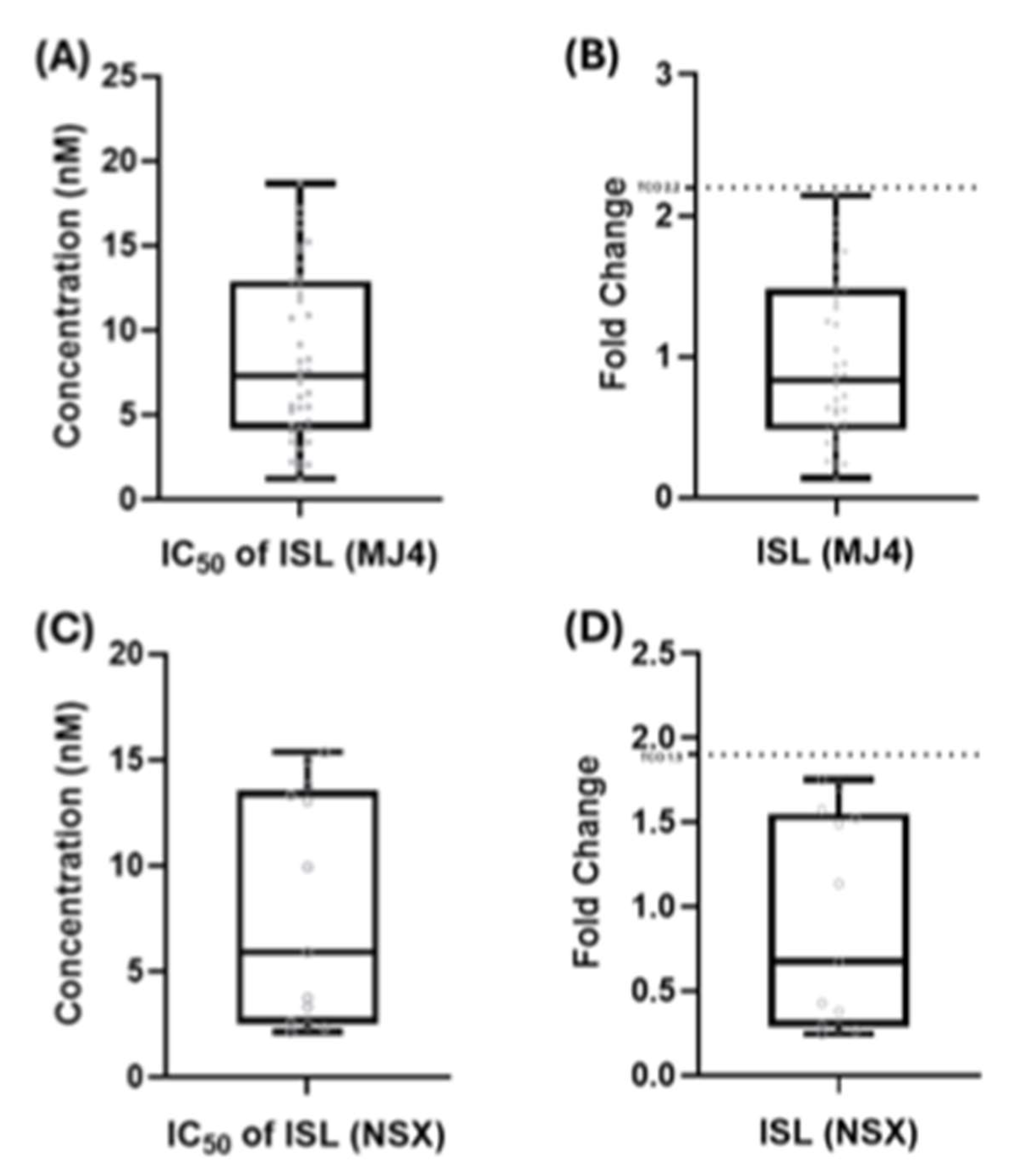
3.2.1. TCO for ISL and Assay-Defined Classifications
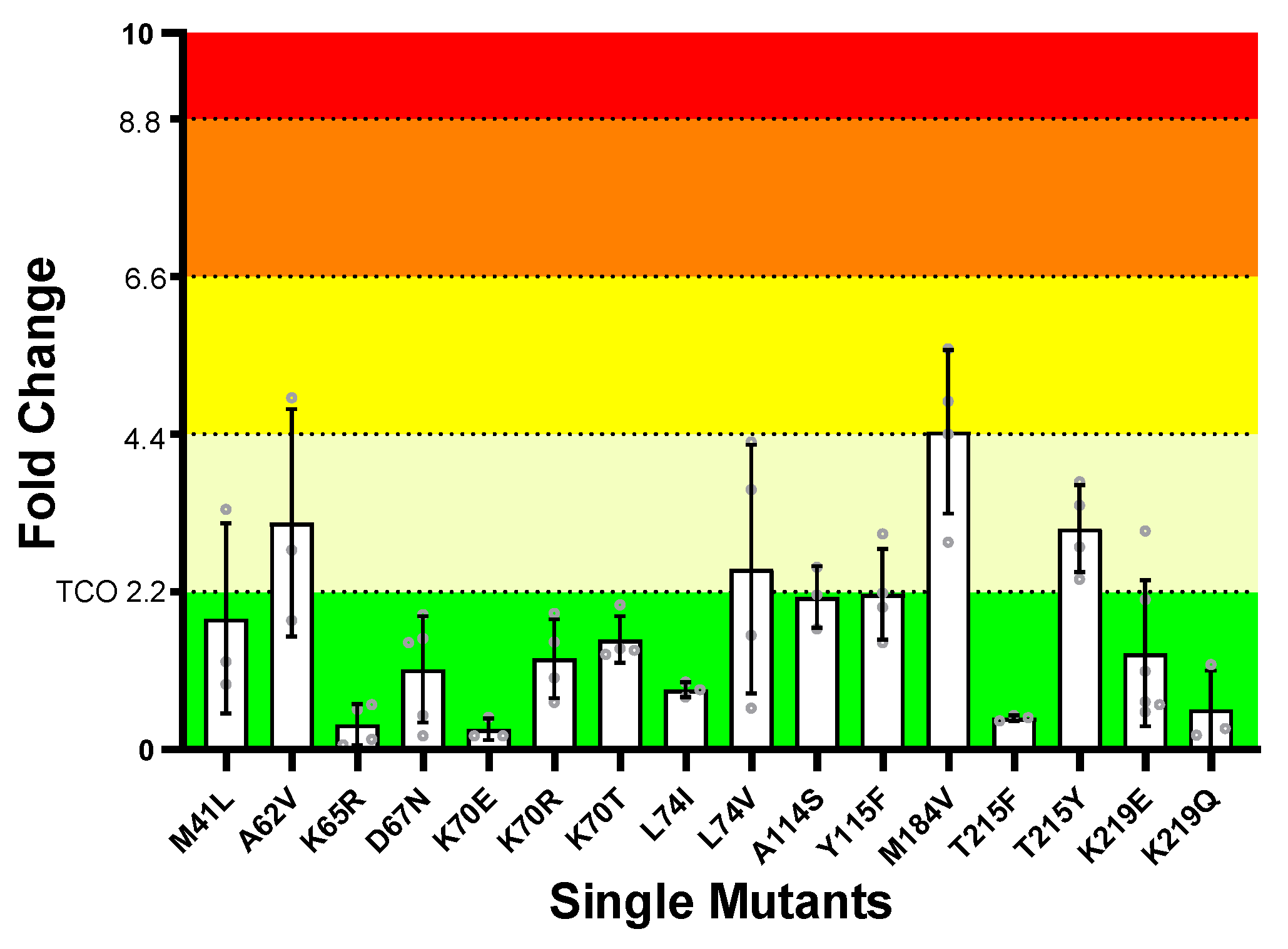
3.2.2. ISL Susceptibility of Single NRTI Drug Resistance Mutations
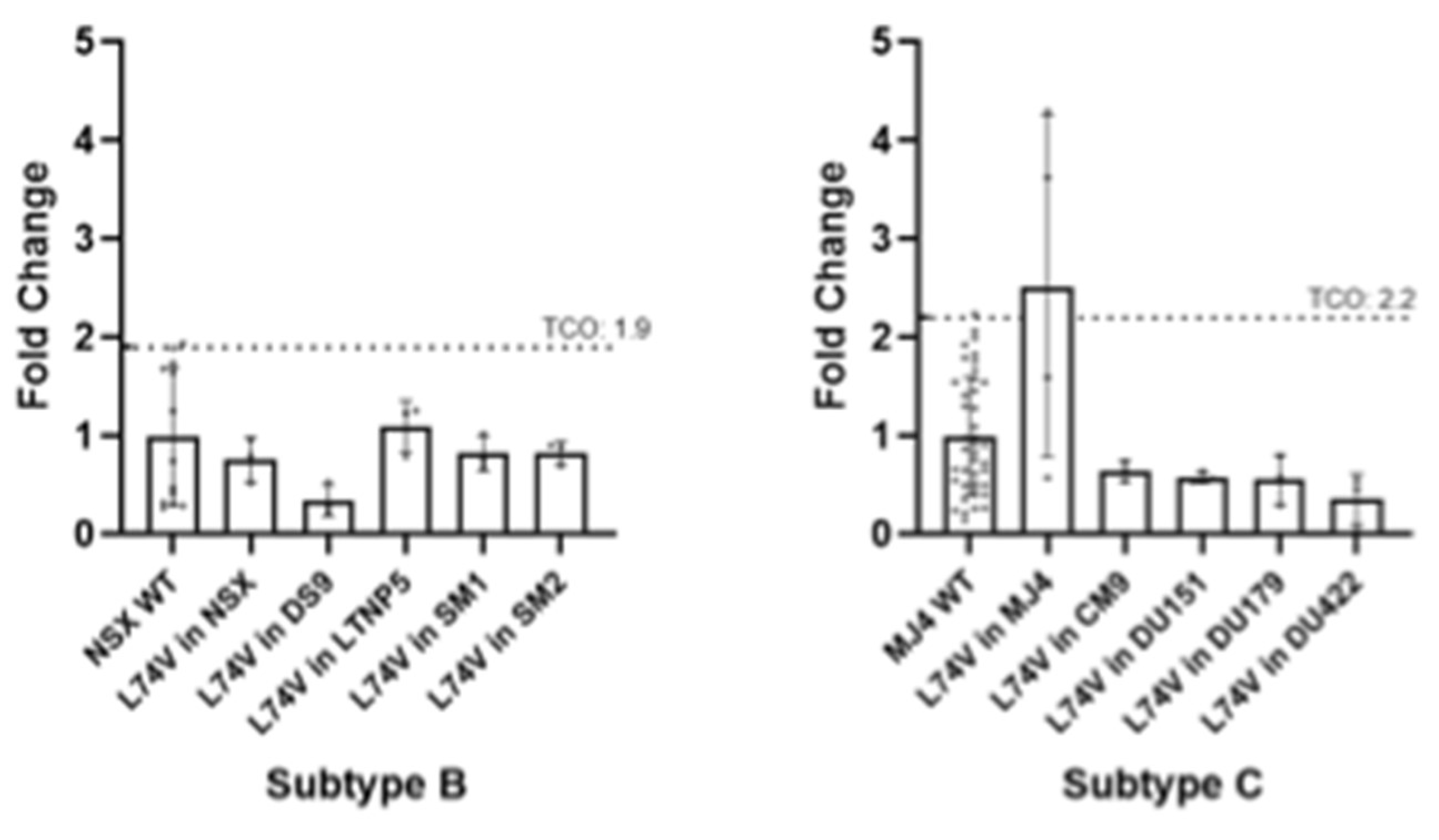
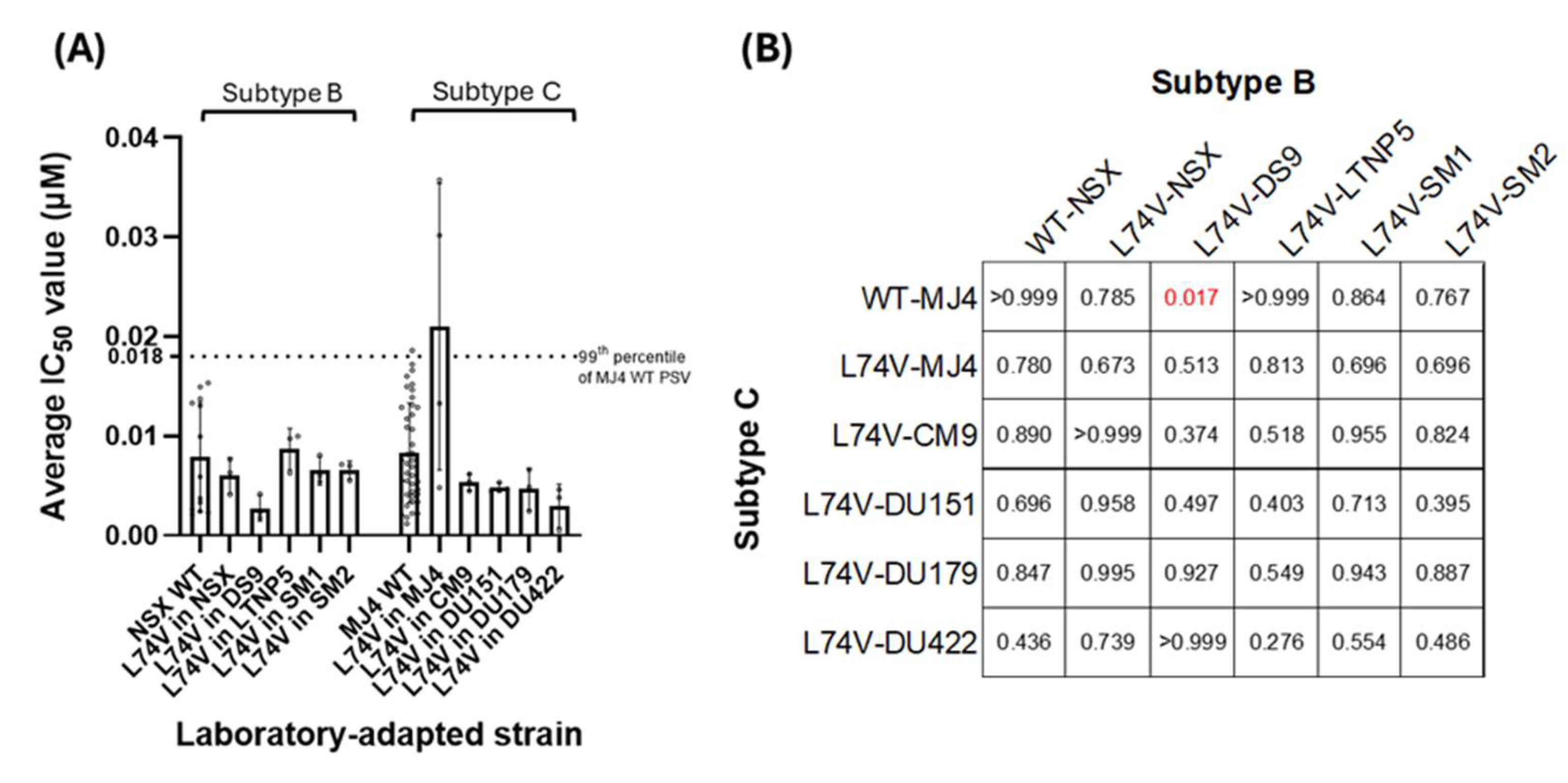
3.2.3. ISL Susceptibility of Combined NRTI Drug Resistance Mutations
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- WHO Fact Sheets: HIV and AIDS Available online:. Available online: https://www.who.int/news-room/fact-sheets/detail/hiv-aids (accessed on 18 September 2024).
- Lee, L.M.; Karon, J.M.; Selik, R.; Neal, J.J.; Fleming, P.L. Survival after AIDS Diagnosis in Adolescents and Adults during the Treatment Era, United States, 1984-1997. JAMA 2001, 285, 1308–1315. [Google Scholar] [CrossRef] [PubMed]
- Nieuwkerk, P.T.; Gisolf, E.H.; Reijers, M.H.; Lange, J.M.; Danner, S.A.; Sprangers, M.A. ; NATIVE Study Group; PROMETHEUS Study Group; ADAM Study Group Long-Term Quality of Life Outcomes in Three Antiretroviral Treatment Strategies for HIV-1 Infection. AIDS 2001, 15, 1985–1991. [Google Scholar] [CrossRef]
- Mate, K.K.V.; Engler, K.; Lessard, D.; Lebouché, B. Barriers to Adherence to Antiretroviral Therapy: Identifying Priority Areas for People with HIV and Healthcare Professionals. Int J STD AIDS 2023, 34, 677–686. [Google Scholar] [CrossRef]
- Kheswa, J.G. Exploring the Factors and Effects of Non-Adherence to Antiretroviral Treatment by People Living with HIV/AIDS. Indo-Pacific Journal of Phenomenology 2017, 17, 1–11. [Google Scholar] [CrossRef]
- Michailidis E; Marchand B; Kodama EN; Singh K; Matsuoka M; Kirby KA; Ryan EM; Sawani AM; Nagy E; Ashida N; et al. Mechanism of Inhibition of HIV-1 Reverse Transcriptase by 4’-Ethynyl-2-Fluoro-2’-Deoxyadenosine Triphosphate, a Translocation-Defective Reverse Transcriptase Inhibitor. The Journal of biological chemistry 2009, 284. [Google Scholar] [CrossRef]
- Matthews, R.P.; Rudd, D.J.; Fillgrove, K.L.; Zhang, S.; Tomek, C.; Stoch, S.A.; Iwamoto, M. A Phase 1 Study to Evaluate the Drug Interaction Between Islatravir (MK-8591) and Doravirine in Adults Without HIV. Clinical Drug Investigation 2021, 41, 629. [Google Scholar] [CrossRef]
- Amy Colson; Gordon Crofoot; Peter J. Ruane; Moti Ramgopal; Alexandra W. Dretler; Ronald G. Nahass; Gary Sinclair; Mezgebe Berhe; Chris Deaton; Angela S. Liu; et al. Efficacy and Safety of Weekly Islatravir Plus Lenacapavir in PWH at 24 Weeks: A Phase II Study - CROI Conference.; Denver, Colorado, USA, 2024; Vol. Abstract 208. 7 March.
- Correll T; Molina JM; Klopfer S; Grandhi A; Lahoulou R; Zhou Y-P; Eves K; Squires K O46 Total Lymphocyte and CD4+ T-Cell Count Changes in Participants Receiving Islatravir (0. 25, 0.75 and 2.25 Mg QD) and Doravirine +/- Lamivudine: Post-Hoc Analysis from a Phase 2b Dose- Ranging Study (P011). Journal of the International AIDS Society 2022, 25, e26009. [Google Scholar] [CrossRef]
- Mills AM; Rizzardini G; Ramgopal MN; Osiyemi OO; Bogner JR; Hagins DP; Paredes R; Reynes J; Rockstroh JK; Carr A; et al. Switch to Fixed-Dose Doravirine (100 Mg) with Islatravir (0·75 Mg) Once Daily in Virologically Suppressed Adults with HIV-1 on Bictegravir, Emtricitabine, and Tenofovir Alafenamide: 48-Week Results of a Phase 3, Randomised, Controlled, Double-Blind, Non-Inferiority Trial. The lancet. HIV 2024, 11. [Google Scholar] [CrossRef]
- Diamond, T.L.; Goh, S.L.; Ngo, W.; Rodriguez, S.; Xu, M.; Klein, D.J.; Grobler, J.A.; Asante-Appiah, E. No Antagonism or Cross-Resistance and a High Barrier to the Emergence of Resistance in Vitro for the Combination of Islatravir and Lenacapavir. Antimicrobial Agents and Chemotherapy 2024, 68, e00334–24. [Google Scholar] [CrossRef]
- Diamond, T.L.; Ngo, W.; Xu, M.; Goh, S.L.; Rodriguez, S.; Lai, M.-T.; Asante-Appiah, E.; Grobler, J.A. Islatravir Has a High Barrier to Resistance and Exhibits a Differentiated Resistance Profile from Approved Nucleoside Reverse Transcriptase Inhibitors (NRTIs). Antimicrob Agents Chemother 2022, 66, e0013322. [Google Scholar] [CrossRef]
- Singh, K.; Flores, J.A.; Kirby, K.A.; Neogi, U.; Sonnerborg, A.; Hachiya, A.; Das, K.; Arnold, E.; McArthur, C.; Parniak, M.; et al. Drug Resistance in Non-B Subtype HIV-1: Impact of HIV-1 Reverse Transcriptase Inhibitors. Viruses 2014, 6, 3535. [Google Scholar] [CrossRef]
- Hemelaar, J.; Elangovan, R.; Yun, J.; Dickson-Tetteh, L.; Fleminger, I.; Kirtley, S.; Williams, B.; Gouws-Williams, E.; Ghys, P.D.; Abimiku, A.G.; et al. Global and Regional Molecular Epidemiology of HIV-1, 1990–2015: A Systematic Review, Global Survey, and Trend Analysis. The Lancet Infectious Diseases 2019, 19, 143–155. [Google Scholar] [CrossRef]
- UNAIDS Country Facts Sheet: South Africa (2023). 2024.
- South Africa National Department of Health 2023 ART Clinical Guidelines for the Management of HIV in Adults, Pregnancy and Breastfeeding, Adolescents, Children, Infants and Neonates. 2023.
- Liu TF; Shafer RW Web Resources for HIV Type 1 Genotypic-Resistance Test Interpretation. Clinical infectious diseases : an official publication of the Infectious Diseases Society of America 2006, 42. [CrossRef]
- Yang, Z.; Chen, Z.; Zhang, Y. A Simple and Economical Site-Directed Mutagenesis Method for Large Plasmids by Direct Transformation of Two Overlapping PCR Fragments. Biotechniques 2022, 73, 239–245. [Google Scholar] [CrossRef]
- Gupta, R.K.; Kohli, A.; McCormick, A.L.; Towers, G.J.; Pillay, D.; Parry, C.M. Full-Length HIV-1 Gag Determines Protease Inhibitor Susceptibility within in Vitro Assays. AIDS 2010, 24, 1651–1655. [Google Scholar] [CrossRef]
- Naldini, L.; Blömer, U.; Gallay, P.; Ory, D.; Mulligan, R.; Gage, F.H.; Verma, I.M.; Trono, D. In Vivo Gene Delivery and Stable Transduction of Nondividing Cells by a Lentiviral Vector. Science 1996, 272, 263–267. [Google Scholar] [CrossRef]
- Parry, C.M.; Kohli, A.; Boinett, C.J.; Towers, G.J.; McCormick, A.L.; Pillay, D. Gag Determinants of Fitness and Drug Susceptibility in Protease Inhibitor-Resistant Human Immunodeficiency Virus Type 1. J Virol 2009, 83, 9094–9101. [Google Scholar] [CrossRef]
- Diamond, T.L.; Ngo, W.; Xu, M.; Goh, S.L.; Rodriguez, S.; Lai, M.-T.; Asante-Appiah, E.; Grobler, J.A. Islatravir Has a High Barrier to Resistance and Exhibits a Differentiated Resistance Profile from Approved Nucleoside Reverse Transcriptase Inhibitors (NRTIs). Antimicrob Agents Chemother 66. [CrossRef]
- Grobler, J.A.; Huang, Q.; Hazuda, D.J.; Lai, M. Efficacy of MK-8591 against Diverse HIV-1 Subtypes and NRTI-Resistant Clinical Isolates. In Proceedings of the Presented at HIV Drug Therapy, Glasgow, UK; October 28 2018. [Google Scholar]
- SANDOH National Department of Health, SA. 2015 National Consolidated for the Prevention of Mother-to-Child Transmission of HIV (PMTCT) and the Management of HIV in Children, Adolescents and Adulta 2015.
- SANDOH National Department of Health, SA. 2019 ART Clinical Guidelines for the Management of HIV in Adults, Pregnancy, Adolescents, Children, Infants and Neonates. 2019.
- Marcelin AG; Delaugerre C; Wirden M; Viegas P; Simon A; Katlama C; Calvez V Thymidine Analogue Reverse Transcriptase Inhibitors Resistance Mutations Profiles and Association to Other Nucleoside Reverse Transcriptase Inhibitors Resistance Mutations Observed in the Context of Virological Failure. Journal of medical virology 2004, 72. [CrossRef]
- Tisdale, M.; Alnadaf, T.; Cousens, D. Combination of Mutations in Human Immunodeficiency Virus Type 1 Reverse Transcriptase Required for Resistance to the Carbocyclic Nucleoside 1592U89. Antimicrob Agents Chemother 1997, 41, 1094–1098. [Google Scholar] [CrossRef]
- Boucher, C.A.; Cammack, N.; Schipper, P.; Schuurman, R.; Rouse, P.; Wainberg, M.A.; Cameron, J.M. High-Level Resistance to (-) Enantiomeric 2’-Deoxy-3’-Thiacytidine in Vitro Is Due to One Amino Acid Substitution in the Catalytic Site of Human Immunodeficiency Virus Type 1 Reverse Transcriptase. Antimicrob Agents Chemother 1993, 37, 2231–2234. [Google Scholar] [CrossRef]
- Tisdale, M.; Kemp, S.D.; Parry, N.R.; Larder, B.A. Rapid in Vitro Selection of Human Immunodeficiency Virus Type 1 Resistant to 3’-Thiacytidine Inhibitors Due to a Mutation in the YMDD Region of Reverse Transcriptase. Proceedings of the National Academy of Sciences of the United States of America 1993, 90, 5653. [Google Scholar] [CrossRef]
- Brenner, B.G.; Oliveira, M.; Doualla-Bell, F.; Moisi, D.D.; Ntemgwa, M.; Frankel, F.; Essex, M.; Wainberg, M.A. HIV-1 Subtype C Viruses Rapidly Develop K65R Resistance to Tenofovir in Cell Culture. AIDS 2006, 20, F9–13. [Google Scholar] [CrossRef]
- Brenner, B.G.; Coutsinos, D. The K65R Mutation in HIV-1 Reverse Transcriptase: Genetic Barriers, Resistance Profile and Clinical Implications. HIV therapy 2009, 3, 583. [Google Scholar] [CrossRef]
- Margot, N.A.; Isaacson, E.; McGowan, I.; Cheng, A.K.; Schooley, R.T.; Miller, M.D. Genotypic and Phenotypic Analyses of HIV-1 in Antiretroviral-Experienced Patients Treated with Tenofovir DF. AIDS 2002, 16, 1227–1235. [Google Scholar] [CrossRef]
- Maldonado, J.O.; Mansky, L.M. The HIV-1 Reverse Transcriptase A62V Mutation Influences Replication Fidelity and Viral Fitness in the Context of Multi-Drug-Resistant Mutations. Viruses 2018, 10, 376. [Google Scholar] [CrossRef]
- Steegen, K.; Bronze, M.; Papathanasopoulos, M.A.; van Zyl, G.; Goedhals, D.; Variava, E.; MacLeod, W.; Sanne, I.; Stevens, W.S.; Carmona, S. HIV-1 Antiretroviral Drug Resistance Patterns in Patients Failing NNRTI-Based Treatment: Results from a National Survey in South Africa. Journal of Antimicrobial Chemotherapy 2017, 72, 210–219. [Google Scholar] [CrossRef]
- Melikian Gl; Rhee SY; Taylor J; Fessel WJ; Kaufman D; W, T. ; Pv, T.-C.; A, Z.; Gk, R.; R, K.; et al. Standardized Comparison of the Relative Impacts of HIV-1 Reverse Transcriptase (RT) Mutations on Nucleoside RT Inhibitor Susceptibility. Antimicrobial agents and chemotherapy 2012, 56. [Google Scholar] [CrossRef]
- Weber, J.; Chakraborty, B.; Weberova, J.; Miller, M.D.; Quiñones-Mateu, M.E. Diminished Replicative Fitness of Primary Human Immunodeficiency Virus Type 1 Isolates Harboring the K65R Mutation. Journal of Clinical Microbiology 2005, 43, 1395. [Google Scholar] [CrossRef]
- J. A. Grobler; Q. Huang; D.J. Hazuda; M.-T. Lai Efficacy of MK-8591 against Diverse HIV-1 Subtypes and NRTI-Resistant Clinical Isolates. Journal of the International AIDS Society 2022, 25, e26009. [Google Scholar] [CrossRef]
- Murphey-Corb M; Rajakumar P; Michael H; Nyaundi J; Didier PJ; Ab, R. ; H, M.; Sg, S.; Ma, P. Response of Simian Immunodeficiency Virus to the Novel Nucleoside Reverse Transcriptase Inhibitor 4’-Ethynyl-2-Fluoro-2’-Deoxyadenosine in Vitro and in Vivo. Antimicrobial agents and chemotherapy 2012, 56. [Google Scholar] [CrossRef]
- Borroto-Esoda, K.; Parkin, N.; Miller, M.D. A Comparison of the Phenotypic Susceptibility Profiles of Emtricitabine and Lamivudine. Antivir Chem Chemother 2007, 18, 297–300. [Google Scholar] [CrossRef]
- Whitcomb JM; Parkin NT; Chappey C; Hellmann NS; Petropoulos CJ Broad Nucleoside Reverse-Transcriptase Inhibitor Cross-Resistance in Human Immunodeficiency Virus Type 1 Clinical Isolates. The Journal of infectious diseases 2003, 188. [CrossRef]
- Cilento, M.E.; Reeve, A.B.; Michailidis, E.; Ilina, T.V.; Nagy, E.; Mitsuya, H.; Parniak, M.A.; Tedbury, P.R.; Sarafianos, S.G. Development of Human Immunodeficiency Virus Type 1 Resistance to 4′-Ethynyl-2-Fluoro-2′-Deoxyadenosine Starting with Wild-Type or Nucleoside Reverse Transcriptase Inhibitor-Resistant Strains. Antimicrobial Agents and Chemotherapy 2021, 65. [Google Scholar] [CrossRef]
- Invernizzi, C.F.; Coutsinos, D.; Oliveira, M.; Moisi, D.; Brenner, B.G.; Wainberg, M.A. Signature Nucleotide Polymorphisms at Positions 64 and 65 in Reverse Transcriptase Favor the Selection of the K65R Resistance Mutation in HIV-1 Subtype C. J Infect Dis 2009, 200, 1202–1206. [Google Scholar] [CrossRef]
- TenoRes Study Group Global Epidemiology of Drug Resistance after Failure of WHO Recommended First-Line Regimens for Adult HIV-1 Infection: A Multicentre Retrospective Cohort Study. The Lancet. Infectious Diseases 2016, 16, 565. [CrossRef]
- White KL; Margot NA; Wrin T; Petropoulos CJ; Miller MD; Naeger LK Molecular Mechanisms of Resistance to Human Immunodeficiency Virus Type 1 with Reverse Transcriptase Mutations K65R and K65R+M184V and Their Effects on Enzyme Function and Viral Replication Capacity. Antimicrobial agents and chemotherapy 2002, 46. [CrossRef]
- Cilento ME; Wen X; Reeve AB; Ukah OB; Snyder AA; Carrillo CM; Smith CP; Edwards K; Wahoski CC; Kitzler DR; et al. HIV-1 Resistance to Islatravir/Tenofovir Combination Therapy in Wild-Type or NRTI-Resistant Strains of Diverse HIV-1 Subtypes. Viruses 2023, 15. [Google Scholar] [CrossRef]
- Harrigan, P.R.; Kinghorn, I.; Bloor, S.; Kemp, S.D.; Nájera, I.; Kohli, A.; Larder, B.A. Significance of Amino Acid Variation at Human Immunodeficiency Virus Type 1 Reverse Transcriptase Residue 210 for Zidovudine Susceptibility. J Virol 1996, 70, 5930–5934. [Google Scholar] [CrossRef]
- Kellam, P.; Boucher, C.A.; Larder, B.A. Fifth Mutation in Human Immunodeficiency Virus Type 1 Reverse Transcriptase Contributes to the Development of High-Level Resistance to Zidovudine. Proceedings of the National Academy of Sciences of the United States of America 1992, 89, 1934. [Google Scholar] [CrossRef]
- Larder, B.A.; Kemp, S.D. Multiple Mutations in HIV-1 Reverse Transcriptase Confer High-Level Resistance to Zidovudine (AZT). Science 1989, 246, 1155–1158. [Google Scholar] [CrossRef] [PubMed]
- Cozzi-Lepri, A.; Ruiz, L.; Loveday, C.; Phillips, A.N.; Clotet, B.; Reiss, P.; Ledergerber, B.; Holkmann, C.; Staszewski, S.; Lundgren, J.D.; et al. Thymidine Analogue Mutation Profiles: Factors Associated with Acquiring Specific Profiles and Their Impact on the Virological Response to Therapy. Antiviral Therapy 2005, 10, 791–802. [Google Scholar] [CrossRef] [PubMed]
- Flandre, P.; Descamps, D.; Joly, V.; Meiffrédy, V.; Tamalet, C.; Izopet, J.; Aboulker, J.-P.; Brun-Vézinet, F. Predictive Factors and Selection of Thymidine Analogue Mutations by Nucleoside Reverse Transcriptase Inhibitors According to Initial Regimen Received. Antiviral Therapy 2003, 8, 65–72. [Google Scholar] [CrossRef]
- Brenner BG; Oliveira M; Ibanescu RI; Routy JP; Thomas R Doravirine Responses to HIV-1 Viruses Bearing Mutations to NRTIs and NNRTIs under in Vitro Selective Drug Pressure. The Journal of antimicrobial chemotherapy 2023, 78. [CrossRef]
- Svarovskaia, E.S.; Feng, J.Y.; Margot, N.A.; Myrick, F.; Goodman, D.; Ly, J.K.; White, K.L.; Kutty, N.; Wang, R.; Borroto-Esoda, K.; et al. The A62V and S68G Mutations in HIV-1 Reverse Transcriptase Partially Restore the Replication Defect Associated with the K65R Mutation. J Acquir Immune Defic Syndr 2008, 48, 428–436. [Google Scholar] [CrossRef]
- Rhee, S.; Gonzales, M.J.; Kantor, R.; Betts, B.J.; Ravela, J.; Shafer, R.W. Human Immunodeficiency Virus Reverse Transcriptase and Protease Sequence Database. Nucleic Acids Research 2003, 31, 298–303. [Google Scholar] [CrossRef]
- Shafer, R.W. Rationale and Uses of a Public HIV Drug-Resistance Database. J INFECT DIS 2006, 194, S51–S58. [Google Scholar] [CrossRef]
- Iacobucci, G. HIV: Dolutegravir Should Be Preferred Treatment Option in All Populations, Says WHO. BMJ 2019, 366, l4831. [Google Scholar] [CrossRef]
- Paton NI; Musaazi J; Kityo C; Walimbwa S; Hoppe A; Balyegisawa A; Asienzo J; Kaimal A; Mirembe G; Lugemwa A; et al. Efficacy and Safety of Dolutegravir or Darunavir in Combination with Lamivudine plus Either Zidovudine or Tenofovir for Second-Line Treatment of HIV Infection (NADIA): Week 96 Results from a Prospective, Multicentre, Open-Label, Factorial, Randomised, Non-Inferiority Trial. The Lancet. HIV 2022, 9. [Google Scholar] [CrossRef]
- Obasa, A.E.; Engelbrecht, S.; Jacobs, G.B. Near Full-Length HIV-1 Subtype B Sequences from the Early South African Epidemic, Detecting a BD Unique Recombinant Form (URF) from a Sample in 1985. Sci Rep 2019, 9, 6227. [Google Scholar] [CrossRef]
- Mikhail, M.; Wang, B.; Lemey, P.; Beckholdt, B.; Vandamme, A.-M.; Gill, M.J.; Saksena, N.K. Full-Length HIV Type 1 Genome Analysis Showing Evidence for HIV Type 1 Transmission from a Nonprogressor to Two Recipients Who Progressed to AIDS. AIDS Res Hum Retroviruses 2005, 21, 575–579. [Google Scholar] [CrossRef]
- Ledwaba, J.; Sayed, Y.; Pillay, V.; Morris, L.; Hunt, G. Low Frequency of Protease Inhibitor Resistance Mutations and Insertions in HIV-1 Subtype C Protease Inhibitor-Naïve Sequences. AIDS Res Hum Retroviruses 2019, 35, 673–678. [Google Scholar] [CrossRef]
- Maldarelli, F.; Kearney, M.; Palmer, S.; Stephens, R.; Mican, J.; Polis, M.A.; Davey, R.T.; Kovacs, J.; Shao, W.; Rock-Kress, D.; et al. HIV Populations Are Large and Accumulate High Genetic Diversity in a Nonlinear Fashion. J Virol 2013, 87, 10313–10323. [Google Scholar] [CrossRef]
- Papathanasopoulos, M.A.; Cilliers, T.; Morris, L.; Mokili, J.L.; Dowling, W.; Birx, D.L.; McCutchan, F.E. Full-Length Genome Analysis of HIV-1 Subtype C Utilizing CXCR4 and Intersubtype Recombinants Isolated in South Africa. AIDS Res Hum Retroviruses 2002, 18, 879–886. [Google Scholar] [CrossRef]
- Williamson, C.; Morris, L.; Maughan, M.F.; Ping, L.-H.; Dryga, S.A.; Thomas, R.; Reap, E.A.; Cilliers, T.; van Harmelen, J.; Pascual, A.; et al. Characterization and Selection of HIV-1 Subtype C Isolates for Use in Vaccine Development. AIDS Research and Human Retroviruses 2003, 19, 133–144. [Google Scholar] [CrossRef]
- Van Harmelen, J.; Williamson, C.; Kim, B.; Morris, L.; Carr, J.; Abdool Karim, S.S.; McCutchan, F. Characterization of Full-Length HIV Type 1 Subtype C Sequences from South Africa. AIDS Research and Human Retroviruses 2001, 17, 1527–1531. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




