Submitted:
21 October 2024
Posted:
24 October 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
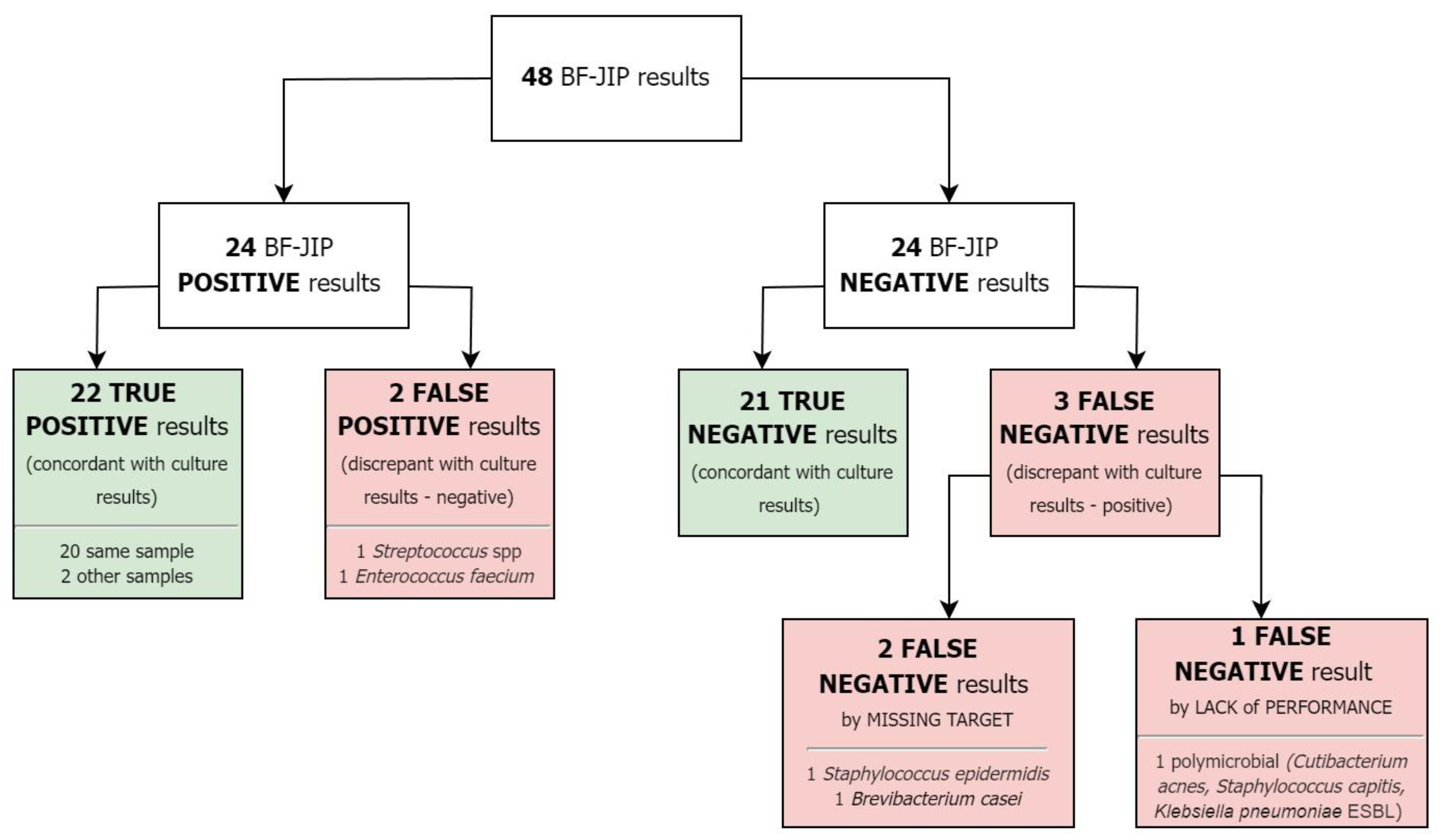
2. Results
2.1. Study Population
2.2. Samples
2.3. Microbiology
3. Discussion
4. Methods
4.1. Study Design
4.2. Data Collection
4.3. Microbiology
4.4. Data Analysis
4.5. Ethics Approval
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Muzzi M., Viaggi B., Fabbri S., Bechi L., Scirè-Calabrisotto C., Villa G., et al. The Impact of Fast Microbiology in Intensive Care Units in the Era of Antibiotic Resistance: An Observational Retrospective Study. Curr. Microbiol. [Internet] 2022 31;79:79. [CrossRef]
- Botan A., Campisciano G., Zerbato V., Di Bella S., Simonetti O., Busetti M., et al. Performance of 16S rRNA Gene Next-Generation Sequencing and the Culture Method in the Detection of Bacteria in Clinical Specimens. Diagnostics (Basel) [Internet] 2024 21;14. [CrossRef]
- Lagier J.-C., Edouard S., Pagnier I., Mediannikov O., Drancourt M., Raoult D. Current and Past Strategies for Bacterial Culture in Clinical Microbiology. Clin. Microbiol. Rev. [Internet] 2015 [cited 2024 21]. [CrossRef]
- Miller J.M., Binnicker M.J., Campbell S., Carroll K.C., Chapin K.C., Gonzalez, et al. Guide to Utilization of the Microbiology Laboratory for Diagnosis of Infectious Diseases: 2024 Update by the Infectious Diseases Society of America (IDSA) and the American Society for Microbiology (ASM). Clin. Infect. Dis. [Internet] 2024 5 [cited 2024 21]. [CrossRef]
- Hawkins S.F.C., Guest P.C. Multiplex Analyses Using Real-Time Quantitative PCR. Methods Mol. Biol. [Internet] 2017 [cited 2024 21];1546. [CrossRef]
- Trotter A.J., Aydin A., Strinden M.J., O’Grady J. Recent and emerging technologies for the rapid diagnosis of infection and antimicrobial resistance. Curr. Opin. Microbiol. [Internet] 2019;51:39–45. [CrossRef]
- FilmArray® Panels—Infectious Disease Diagnostics [Internet]. BioFire Diagnostics 2016 2 [Accessed 21 July 2024]; https://www.biofiredx.com/products/the-filmarray-panels/.
- BIOFIRE® Joint Infection (JI) Panel [Internet]. bioMérieux Website [Accessed 21 July 2024]; https://www.biomerieux.com/us/en/our-offer/clinical-products/biofire-joint-infection-panel.html.
- Esteban J., Salar-Vidal L., Schmitt B.H., Waggoner A., Laurent F., Abad L., et al. Multicenter evaluation of the BIOFIRE Joint Infection Panel for the detection of bacteria, yeast, and AMR genes in synovial fluid samples. J. Clin. Microbiol. [Internet] 2023 21;61:e0035723. [CrossRef]
- Hoffman T., Kriger O., Cohen S., Gefen-Halevi S., Yahav D., Amit S. Real-Life Experience and Diagnostic Utility of the BioFire Joint Infection PCR Panel in Bone and Joint Infections: Analysis of a Prospective Validation Study. Infect Dis Ther [Internet] 2023;12:1437–43. [CrossRef]
- Gaillard T., Dupieux-Chabert C., Roux A.-L., Tessier E., Boutet-Dubois A., Courboulès C., et al. A prospective multicentre evaluation of BioFire® Joint Infection Panel for the rapid microbiological documentation of acute arthritis. Clin. Microbiol. Infect. [Internet] 2024;30:905–10. [CrossRef]
- Moran A., Arellano J., Bregman K., McElvania E. Evaluation of a BioFire multiplex PCR panel for detection of joint infections using retrospective and prospectively collected specimens. J. Clin. Microbiol. [Internet] 2024 17;e0018224. [CrossRef]
- Micó M., Navarro F., de Miniac D., González Y., Brell A., López C., et al. Efficacy of the FilmArray blood culture identification panel for direct molecular diagnosis of infectious diseases from samples other than blood. J. Med. Microbiol. [Internet] 2015;64:1481–8. [CrossRef]
- Hirai J., Mori N., Sakanashi D., Morishita Y., Kuge Y., Kishino T., et al. Usefulness of the FilmArray blood culture identification panel for identifying causative pathogens of bone and joint infections. J. Infect. Chemother. [Internet] 2023 [cited 2024 21];29. [CrossRef]
- Di Bella S., Antonello R.M., Sanson G., Maraolo A.E., Giacobbe D.R., Sepulcri C., et al. Anaerobic bloodstream infections in Italy (ITANAEROBY): A 5-year retrospective nationwide survey. Anaerobe [Internet] 2022;75:102583. [CrossRef]
- Soriano G., Esparcia O., Montemayor M., Guarner-Argente C., Pericas R., Torras X., et al. Bacterial DNA in the diagnosis of spontaneous bacterial peritonitis. Aliment. Pharmacol. Ther. [Internet] 2011 [cited 2024 22];33. [CrossRef]
| MICROORGANISMS DETECTED BY BF-JIP | ||
|---|---|---|
| Gram + | Gram - | Yeasts |
| Anaerococcus prevotii/vaginalis | Bacteroides fragilis | Candida |
| Clostridium perfringens | Citrobacter | Candida albicans |
| Cutibacterium avidum-granulosum | Enterobacter cloacae complex | |
| Enterococcus faecalis | Escherichia coli | |
| Enterococcus faecium | Haemophilus influenzae | |
| Finegoldia magna | Kingella kingae | |
| Parvimonas micra | Klebsiella aerogenes | |
| Peptoniphilus | Klebsiella pneumonia group | |
| Peptostreptococcus anaerobius | Morganella morganii | |
| Staphylococcus aureus | Neisseria gonorrhoeae | |
| Staphylococcus lugdunensis | Proteus spp. | |
| Streptococcus spp. | Pseudomonas aeruginosa | |
| Streptococcus agalactiae | Salmonella spp. | |
| Streptococcus pneumonia | Serratia marcescens | |
| Streptococcus pyogenes | ||
| Resistance genes: CTX-M, IMP, KPC, mecA/C and MREJ (MRSA), NDM, OXA-48 like, vanA/B, VIM | ||
| POPULATION CHARACTERISTICS | |
|---|---|
| N° samples | 48 |
| N° patients | 45 |
| Age (median) | 63 (IQR 45-74) |
| Sex | |
| Male | 24 (53%) |
| Female | 21 (47%) |
| Setting | |
| Surgical ward | 21 (44%) |
| Medical ward | 14 (29%) |
| ER | 6 (13%) |
| Outpatients | 6 (13%) |
| ICU | 1 (<1%) |
| Antimicrobial therapy | |
| Yes | 32 (67%) |
| No | 15 (33%) |
| Non known | 1 (<1%) |
| CCI (median) | 3 (IQR 1-5) |
| Final diagnosis | |
| SSTI | 9 (19%) |
| BJI | 7 (15%) |
| Abdominal abscess | 6 (13%) |
| Pleural empyema | 5 (10%) |
| Breast implant infection | 5 (10%) |
| Meningitis | 5 (10%) |
| No evidence of infection | 4 (8%) |
| Others | 7 (15%) |
| SAMPLE TYPES | |
| Abscess drainage fluid | 24 (50%) |
| Biopsy | 10 (21%) |
| Pleural fluid | 6 (13%) |
| CSF | 5 (10%) |
| Ascitic fluid | 2 (<1%) |
| Vitreous humor/aqueous humor | 1 (<1%) |
| BF-JIP AND CULTURES COMPARISON | |||||
|---|---|---|---|---|---|
| N° | SAMPLE | BF-JIP | CULTURE (same sample) | OTHER CULTURES | FINAL DIAGNOSIS |
| 1 | Biopsy | Parvimonas micra; Peptostreptococcus anaerobius; Streptococcus spp | Streptococcus anginosus | SSTI | |
| 2 | Abscess drainage liquid |
Staphylococcus aureus mecA/B and MREJ |
Staphylococcus aureus | Brain abscess | |
| 3 | Ascitic fluid | Peritonitis | |||
| 4 | Abscess drainage liquid | Streptococcus spp | No infection | ||
| 5 | Abscess drainage liquid | Escherichia coli | Escherichia coli | BJI | |
| 6 | Abscess drainage liquid | Anaerococcus prevotii; Finegoldia magna; Peptoniphilus; Proteus spp | Proteus mirabilis | SSTI | |
| 7 | Abscess drainage liquid | Staphylococcus aureus | Staphylococcus aureus | BJI | |
| 8 | Pleural fluid | Pulmonary aspergillosis | |||
| 9 | Pleural fluid | Streptococcus pyogenes | Streptococcus pyogenes | Pleural empyema | |
| 10 | Ascitic fluid |
Enterococcus faecium; Candida albicans VanA/B |
Candida albicans | Peritonitis | |
| 11 | Pleural fluid | Staphylococcus aureus | Staphylococcus aureus (blood culture and BAL) | Pleural empyema | |
| 12 | Abscess drainage liquid | Escherichia coli; Peptoniphilus | Escherichia coli | Abdominal abscess | |
| 13 | Pleural fluid | Pleural empyema | |||
| 14 | Biopsy | Staphylococcus epidermidis | SSTI | ||
| 15 | Abscess drainage liquid | Abdominal abscess | |||
| 16 | CSF | Meningitis | |||
| 17 | Abscess drainage liquid |
Anaerococcus prevotii; Enterococcus faecium; Finegoldia magna; Parvimonas micra; Streptococcus spp; Bacteroides fragilis; Citrobacter; Enterobacter cloacae; Escherichia coli; Haemophilus influenzae; Klebsiella pneumoniae; Candida albicans CTX-M, vanA/B, VIM |
Pseudomonas aeruginosa; Klebsiella pneumoniae; Candida albicans | Abdominal abscess | |
| 18 | Biopsy | Pseudomonas aeruginosa | Pseudomonas aeruginosa | SSTI | |
| 19 | Pleural fluid | Enterococcus faecium | Pleural empyema | ||
| 20 | Biopsy | Bacteroides fragilis | Bacteroides fragilis | SSTI | |
| 21 | Abscess drainage liquid | Escherichia coli | Escherichia coli | Abdominal abscess | |
| 22 | Abscess drainage liquid | Streptococcus pyogenes | Streptococcus pyogenes | Necrotizing fasciitis | |
| 23 | Abscess drainage liquid | Staphylococcus aureus | Staphylococcus aureus | Muscle hematoma | |
| 24 | Abscess drainage liquid | SSTI | |||
| 25 | Abscess drainage liquid | No infection | |||
| 26 | Biopsy | BJI | |||
| 27 | Abscess drainage liquid | Escherichia coli | Candida glabrata (abscess drainage fluid and blood culture); Escherichia coli (blood culture); Pseudomonas aeruginosa (abscess drainage fluid); Klebsiella pneumoniae (abscess-drainage fluid) | Abdominal abscess | |
| 28 | Abscess drainage liquid | BJI | |||
| 29 | Abscess drainage liquid | No infection | |||
| 30 | Abscess drainage liquid | Abdominal abscess | |||
| 31 | CSF | Meningitis | |||
| 32 | vitreous umor/aqueus umor | Endophthalmitis | |||
| 33 | CSF | Streptococcus spp | Streptococcus salivarius | Meningitis | |
| 34 | Biopsy | Cutibacterium acnes; Staphylococcus capitis; Klebsiella pneumoniae | BJI | ||
| 35 | Biopsy | Enterococcus faecalis | Enterococcus faecalis | SSTI | |
| 36 | Pleural fluid | Pleural empyema | |||
| 37 | Abscess drainage liquid | Breast implant infection | |||
| 38 | Abscess drainage liquid | Breast implant infection | |||
| 39 | CSF | Meningitis | |||
| 40 | Abscess drainage liquid | Parvimonas micra; Streptococcus spp | Streptococcus anginosus | SSTI | |
| 41 | Abscess drainage liquid | No infection | |||
| 42 | Abscess drainage liquid | Staphylococcus aureus | Staphylococcus aureus | Breast implant infection | |
| 43 | Abscess drainage liquid | Staphylococcus aureus | Staphylococcus aureus | SSTI | |
| 44 | Biopsy | BJI | |||
| 45 | CSF | Haemophilus influenzae | Haemophilus influenzae | Meningitis | |
| 46 | Biopsy | BJI | |||
| 47 | Biopsy | Enterococcus faecium | Brevibacterium casei | Breast implant infection | |
| 48 | Abscess drainage liquid | Parvimonas micra; Peptostreptococcus anaerobius; Streptococcus spp | Breast implant infection | ||
| BF-JIPPERFORMANCES | ||||||
|---|---|---|---|---|---|---|
| PPA | NPA | PPV | NPV | C | A | |
| All samples (n=48) | 88.0% | 91.3% | 91.7% | 87.5% | 85.4% | 89.6% |
| Abscess drainage fluid (n=24) | 100% | 90.9% | 92.9% | 100% | 91.7% | 95.8% |
| Biopsy (n=10) | 57.1% | 100% | 100% | 50% | 70.0% | 70.0% |
| Pleural fluid (n=6) | 100% | 75% | 66.7% | 100% | 66.7% | 83.3% |
| Cerebrospinal fluid(n=5) | 100% | 100% | 100% | 100% | 100% | 100% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





