I. Introduction
Studying biological networks provides invaluable insights into the complex interactions and regulatory mechanisms underpinning cellular functions. Recent advancements in bioinformatics and computational biology have allowed for the detailed mapping and analysis of these networks, particularly in the context of gene expression and regulation [
1,
2]. One emerging area of interest is the exploration of network topology through geometric and topological measures, such as network curvature. Network curvature, a concept borrowed from differential geometry, has shown potential in highlighting key features and differences in the structural organization of networks, which may be linked to biological function or disease states. Gene expression varies significantly between genders, a crucial factor influencing susceptibility, onset, and the progression of numerous diseases. Understanding these differences at the network level can provide deeper insights into gender-specific medical treatments and interventions [
3,
4,
5,
6].
However, traditional network analysis methods often overlook the subtleties that may distinguish male-specific networks from female-specific ones in gene expression data. Here, we propose the use of network curvature—specifically [
7], Forman-Ricci and Ollivier-Ricci [
8] curvatures—as discriminant features to explore and compare the correlation networks of gene expressions between genders, underscoring the importance of gender-specific research in our understanding of biological networks [
9].
The curvature of a network provides a quantitative measure of how the network deviates from being "flat" or "tree-like" in its local or global connectivity patterns. Forman-Ricci curvature, which considers edge weights and the degree of nodes, offers insights into edge-centric properties [
10]. For instance, it can help identify hubs or bottleneck nodes in a network. On the other hand, Ollivier-Ricci curvature focuses on the distribution of shortest paths and can be interpreted in terms of network transport efficiency and robustness. It can reveal how easily information or signals can flow through a network. Both these curvature measures have been applied in various domains, such as analyzing brain networks and social networks, and have shown their capability to reveal critical structural information that is not apparent through traditional network metrics .
This paper introduces a novel concept of applying network curvature to gender-specific gene correlation networks. This approach aims to identify key differences in network topology that correlate with gender. By applying these curvature measures to publicly available gene expression datasets, we aim to demonstrate how they can serve as powerful tools for uncovering gender-specific characteristics in gene regulatory mechanisms. Our innovative approach not only enhances the understanding of genetic differences influenced by gender but also has the potential to revolutionize the field of network science by integrating geometric analysis into the study of biological networks. The remainder of this paper is organized as follows.
II. Network Curvature Measures
The Forman-Ricci curvature for an edge
e in a weighted graph connecting vertices
u and
v with weights
,
, and
is given by: “`latex
where
is an edge adjacent to
e and
and
are the vertices connected by
.
The Ollivier-Ricci curvature for an edge
is defined as:
where
and
are probability measures concentrated at vertices
u and
v, respectively, and
is the 1-Wasserstein distance between these measures.
These formulas encapsulate the essence of the curvature concepts applied to graph edges in the context of network analysis, useful for understanding connectivity and topology within graph structures.
III. A Case Study
In this case study we explore the possibility to use network curvature measures for discriminating correlation netwroks based on their health status and gender of the related patients. We constructed correlation networks for various tissues, categorizing them into distinct groups.
Each network models the correlation of gene expression values extracted from the GTEx database and stratified by gender to create separate networks for males and females. We employed standard correlation techniques available on the PyWGCMA package to identify connections between genes.
To analyze the network structure, we calculated the Ollivier-Ricci curvature a metric derived from Riemannian geometry. This measure assesses the "shape" of the network at each node and edge, providing insights into the overall topological structure.
Areas of high curvature indicate tightly interconnected gene clusters, while low curvature regions may suggest gene expression divergence potentially related to disease processes.
Our primary hypothesis was that network curvature measures could effectively discriminate between the four defined patient groups.
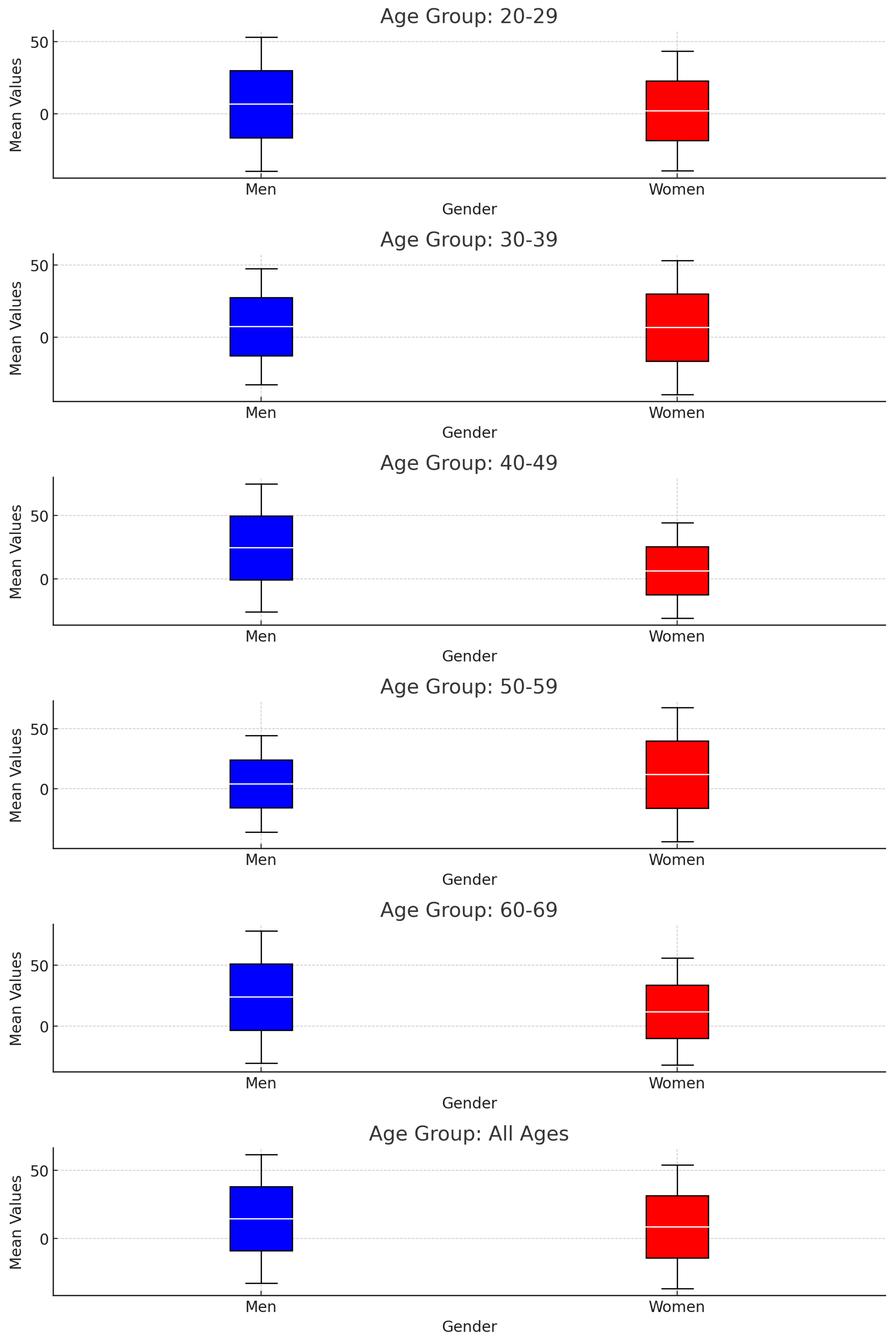
We here show preliminary results for a single tissue, as summarised in
Table 1.
Figure 1.
Boxplots showing the Comparison Between Men and Women Across Age Groups
Figure 1.
Boxplots showing the Comparison Between Men and Women Across Age Groups
IV. Conclusion
This study demonstrates the potential of network curvature measures as discriminative tools for categorizing gene expression profiles across gender and health conditions. The distinct curvature patterns observed between male and female networks, as well as healthy and diseased states, highlight the intricate biological differences that can be quantitatively captured through topological analysis.
Acknowledgments
The Authors are partially funded by the Next Generation EU - Italian NRRP, Mission 4, Component 2, Investment 1.5, call for the creation and strengthening of “Innovation Ecosystems”, building “Territorial R&D Leaders” (Directorial Decree n. 2021/3277) - project Tech4You - Technologies for climate change adaptation and quality of life improvement, n. ECS0000009. This work reflects only the authors’ views and opinions, neither the Ministry for University and Research nor the European Commission can be considered responsible for them. This work was funded by the Next Generation EU - Italian NRRP, Mission 4, Component 2, Investment 1.5, call for the creation and strengthening of ’Innovation Ecosystems’, building ’Territorial R&D Leaders’ (Directorial Decree n. 2021/3277) - project Tech4You - Technologies for climate change adaptation and quality of life improvement, n. ECS0000009. This work reflects only the authors’ views and opinions, neither the Ministry for University and Research nor the European Commission can be considered responsible for them. We acknowledge the support of the PNRR project FAIR - Future AI Research (PE00000013), Spoke 9 - Green-aware AI, under the NRRP MUR program funded by the NextGenerationEU.
References
- P. H. Guzzi and S. Roy, “Biological network analysis: Trends, approaches, graph theory, and algorithms,” 2020.
- M. Zitnik, M. M. Li, A. Wells, K. Glass, D. M. Gysi, A. Krishnan, T. Murali, P. Radivojac, S. Roy, A. Baudot, et al. arXiv preprint arXiv:2309.08478, arXiv:2309.08478, 2023.
- F. Mauvais-Jarvis, “Sex differences in metabolic homeostasis, diabetes, and obesity,” Biology of sex differences, vol. 6, no. 1, pp. 1–9, 2015.
- C. M. Lopes-Ramos, C.-Y. Chen, M. L. Kuijjer, J. N. Paulson, A. R. Sonawane, M. Fagny, J. Platig, K. Glass, J. Quackenbush, and D. L. DeMeo, “Sex differences in gene expression and regulatory networks across 29 human tissues,” Cell reports, vol. 31, no. 12, 2020.
- A. R. Sonawane, J. Platig, M. Fagny, C.-Y. Chen, J. N. Paulson, C. M. Lopes-Ramos, D. L. DeMeo, J. Quackenbush, K. Glass, and M. L. Kuijjer, “Understanding tissue-specific gene regulation,” Cell reports, vol. 21, no. 4, pp. 1077–1088, 2017.
- D. Mercatelli, E. Pedace, F. M. Giorgi, and P. H. Guzzi, “Exploiting molecular basis of age and gender differences in outcomes of sars-cov-2 infections.,” medRxiv, 2021.
- H. Li, J. Cao, J. Zhu, Y. Liu, Q. Zhu, and G. Wu, “Curvature graph neural network,” Information Sciences, vol. 592, pp. 50–66, 2022.
- Y. Lin, L. Lu, and S.-T. Yau, “Ricci curvature of graphs,” Tohoku Mathematical Journal, Second Series, vol. 63, no. 4, pp. 605–627, 2011.
- P. Hiram Guzzi, F. Petrizzelli, and T. Mazza, “Disease spreading modeling and analysis: A survey,” Briefings in Bioinformatics, vol. 23, no. 4, p. bbac230, 2022.
- T. Chatterjee, B. DasGupta, and R. Albert, “A review of two network curvature measures,” Nonlinear Analysis and Global Optimization, pp. 51–69, 2021.
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).