1. Introduction
Brain metastases represent a significant challenge in oncology, affecting a substantial proportion of cancer patients, particularly those with advanced-stage disease. According to recent statistics, approximately 20-40% of patients with systemic cancer will develop brain metastases, leading to increased morbidity and mortality [
1,
2]. Stereotactic radiosurgery (SRS) has emerged as a common treatment modality for patients with BM, offering a minimally invasive option that can effectively control tumor growth while preserving surrounding healthy brain tissue. [
3] However, the variability in patient response to SRS necessitates the development of predictive models that can identify responders and non-responders early in the treatment process [
4].The emergence of targeted treatment methods, particularly Gamma Knife radiosurgery (GKRS), is increasingly recognized as a preferred strategy for managing brain metastases (BMs) due to its enhanced ability to locally target tumors while minimizing side effects compared to traditional whole brain radiation therapy. Its effectiveness is comparable to that of surgical removal [
5,
6,
7,
8]. However, GKRS faces limitations when it comes to treating larger BMs (those exceeding 3 cm in diameter or 10 cc in volume) due to the risks of radiation-induced complications, such as radiation toxicity [
9,
10,
11,
12]. The Gamma Knife ICON, which utilizes mask fixation for patient positioning, offers an advanced solution for hypo-fractionated treatment of larger BMs. Despite these advancements, the median survival rate for patients with BMs remains around one year [
13,
14].In light of the complexity and variability of tumor dynamics following GKRS, there is a growing interest in employing advanced data analysis techniques to improve predictions of patient outcomes. Recent studies have explored various machine learning and deep learning approaches for predicting treatment responses in brain metastases, including convolutional neural networks (CNNs) and traditional statistical methods. However, these approaches often require extensive pre-processing and are limited by their inability to capture long-range dependencies within imaging data. Vision transformers (ViTs), leveraging self-attention mechanisms, present a promising alternative by directly modeling images as sequences of patches, thereby improving performance in medical imaging tasks.
Recent advancements in deep learning, particularly vision transformers (ViTs), have shown promise in medical imaging analysis due to their ability to model complex data distributions and capture intricate patterns within imaging data [
15]. ViTs leverage self-attention mechanisms, enabling them to focus on relevant image regions, which is essential for tasks such as tumor classification and response prediction [
16]. This study aims to evaluate the effectiveness of ViTs in predicting the early response of BM to SRS using a minimal pre-processing strategy on MRI images.Also, it evaluates the effectiveness of ViTs as a non-invasive predictive tool, aiming to bridge the gap in early response prediction and enhance clinical applicability.
This paper is structured as follows: In Chapter 2, we outline the materials and methods employed in our study, including patient characteristics, imaging protocols, and the deep learning framework utilized. Chapter 3 presents the results of our analyses, detailing patient demographics, model performance metrics, and qualitative evaluations of model predictions. In Chapter 4, we discuss the implications of our findings, highlighting the potential integration of ViTs into clinical practice and the importance of minimal pre-processing in enhancing the applicability of AI in oncology. Finally, Chapter 5 concludes the paper by summarizing our contributions and proposing directions for future research.
2. Materials and Methods
All experiments were conducted in compliance with applicable guidelines and regulations. The study utilized only pre-existing medical data, which eliminated the need for patient consent. Additionally, as this was a retrospective study, approval from the Ethics Committee of the Clinical Emergency Hospital “Prof. Dr. Nicolae Oblu” in Iasi was not necessary.
2.1. Study Population
This retrospective study analyzed MRI data from 19 patients diagnosed with brain metastases who underwent stereotactic radiosurgery (SRS) within three months of diagnosis. Patient demographic information, including age, sex, primary cancer type, and Karnofsky Performance Status (KPS), was collected from medical records. Among the 19 patients, there were 5 females and 14 males, aged between 43 and 80 years. All patients had a KPS of at least 70. The initial tumor volumes before the first treatment session ranged from 2 to 81 cm³, with an average of 16 cm³. The primary sites of the metastases included bronchopulmonary neoplasms in 14 cases, breast neoplasms in 3 cases, and one case each of laryngeal and prostate neoplasms.
2.2. Strategy for Gamma Knife Radiosurgery Implementation
The study was conducted from July 2022 to February 2023 at the Gamma Knife Stereotactic Radiosurgery Laboratory of the Prof. Dr. N. Oblu Emergency Clinical Hospital in Iasi. All patients underwent GKRS using the Leksell Gamma Knife ICON (Elekta AB, Stockholm, Sweden).All magnetic resonance images were registered with Leksell Gamma Plan (LGP, Version11.3.2, TMR algorithm) and any images with motion artifacts were excluded. The tumor volumes were calculated by LGP without margin. The treatment protocol involved administering a total dose of 30 Gy in three sessions (S1, S2, S3) of 10 Gy each, delivered at two-week intervals based on the linear quadratic model [
17,
18] and the work of Higuchi et al. from 2009 [
19]. The GKRS planning was determined through a consensus between the neurosurgeon, radiation oncologist, and expert medical physicist. Treatment response was assessed based on the Response Assessment in Neuro-Oncology Brain Metastases (RANO-BM) criteria, categorizing patients as responders (complete or partial response) or non-responders (stable or progressive disease) [
20]. Following treatment, only one patient exhibited a clear progression of the lesion, while three others demonstrated fluctuating patterns of progression and regression.
2.3. Medical Imaging Protocol
All MRI examinations were performed using a 1.5 Tesla whole-body scanner (GE SIGMA EXPLORER) equipped with a standard 16-channel head coil. The MRI study protocol included both conventional and advanced imaging techniques for the clinical routine diagnosis of brain tumors.
The conventional anatomical MRI (cMRI) protocol encompassed an axial fluid-attenuated inversion recovery (FLAIR) sequence and a high-resolution contrast-enhanced T1-weighted (CE T1w) sequence. The FLAIR sequence was utilized to visualize edema and non-enhancing tumor components, providing insights into the tumor microenvironment, while the CE T1w sequence was employed to delineate tumor boundaries and assess the degree of enhancement indicative of tumor activity. The selected sequences, FLAIR and CE T1w, are crucial for visualizing edema and tumor boundaries, respectively, making them particularly relevant for assessing treatment responses in brain metastases.
In addition to the conventional protocol, the advanced MRI (advMRI) protocol included axial diffusion-weighted imaging (DWI) with b values of 0 and 1000 s/mm², as well as a gradient echo dynamic susceptibility contrast (GE-DSC) perfusion MRI sequence. The GE-DSC sequence involved 60 dynamic measurements during the administration of 0.1 mmol/kg body weight gadoterate meglumine, enhancing the evaluation of tumor perfusion characteristics.
All imaging data were extracted from the tumor center, ensuring a standardized region of interest that minimized variability in tumor localization. This comprehensive imaging approach aimed to facilitate accurate assessment of tumor characteristics and treatment response in patients with brain metastases.
2.4. Image Dataset
The
brain_met_1 Image Dataset was stored in cloud on Google Drive, consisting of two subfolders: progression and regression. For training, the
train_ds is used and for validation, the
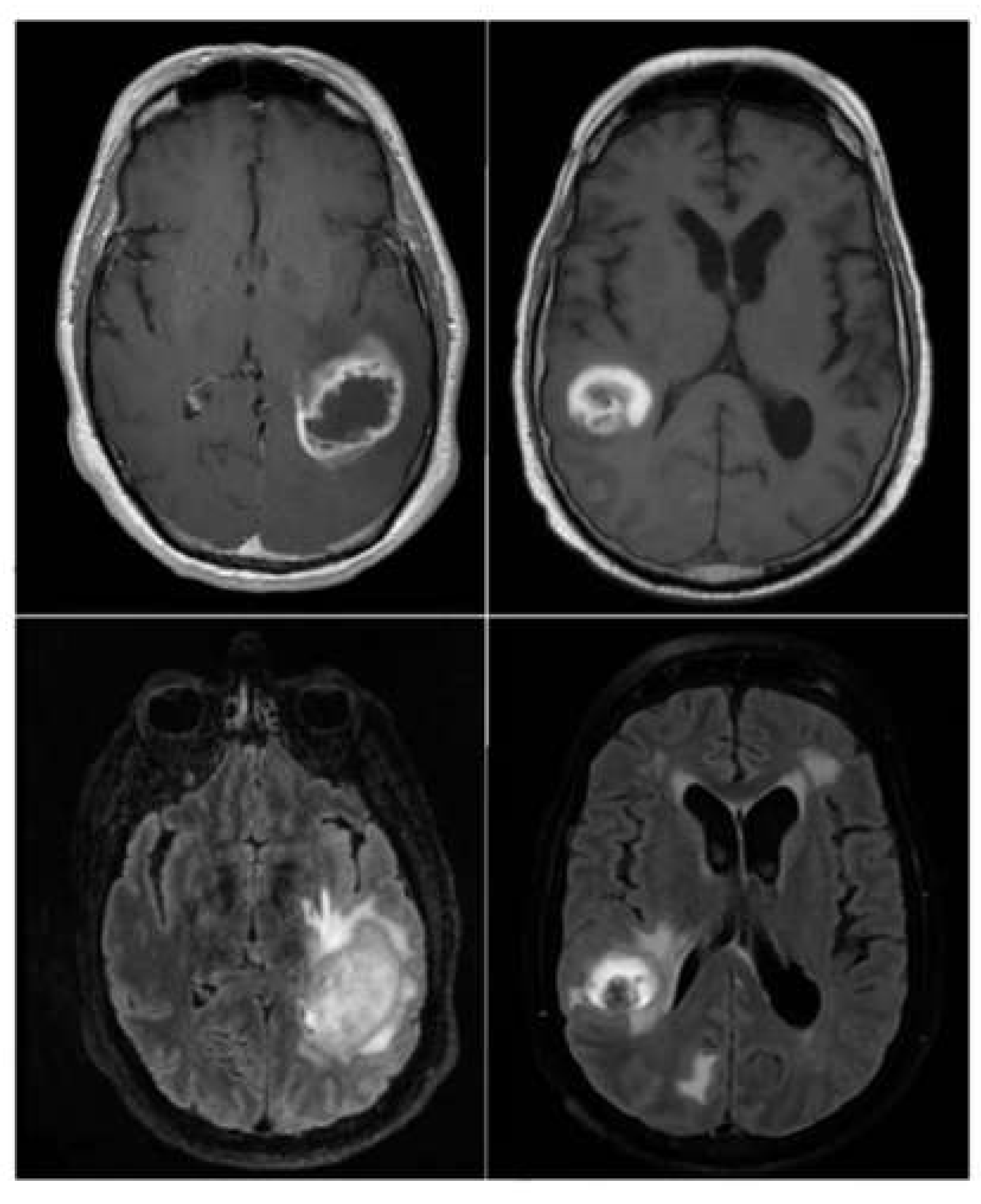
valid_ds is used. The Image Dataset contains 3194 MRI brain metastasis images, with 2320 images of regression class '1' and 874 images of progression class '0'. See example of images from the dataset in
Figure 1.
2.5. Project Workflow and Methodology Overview
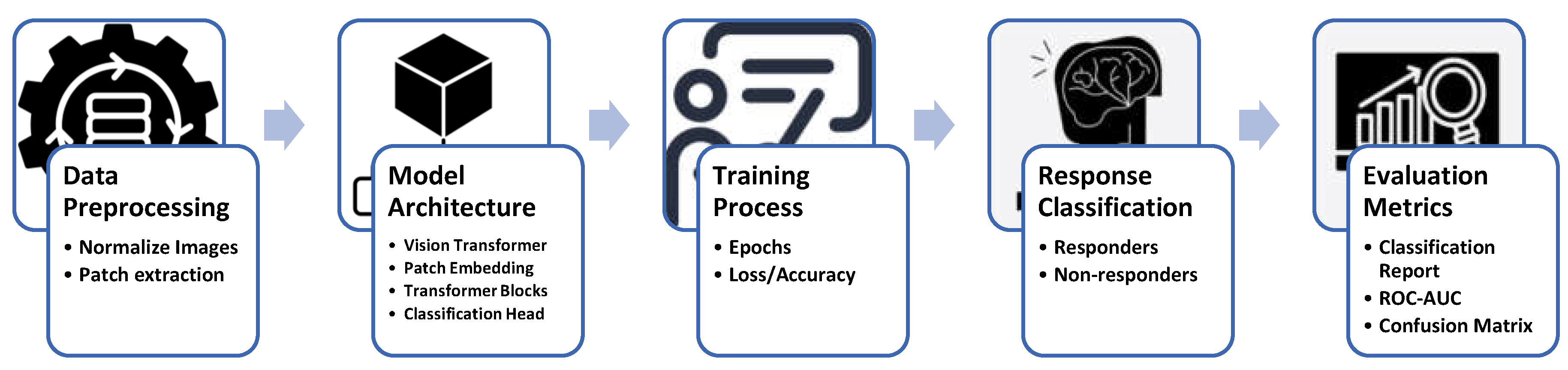
Figure 2 illustrates the comprehensive workflow employed in this study for predicting early responses of brain metastases to stereotactic radiosurgery (SRS) using Vision Transformers. The process begins with data preprocessing, which includes normalizing MRI images and extracting patches for model input. The architecture of the Vision Transformer is then defined, incorporating essential components such as patch embedding, transformer blocks, and a classification head. During the training process, the model undergoes multiple epochs to optimize its performance, tracking metrics like loss and accuracy. Following training, response classification distinguishes between responders and non-responders based on the model's predictions. Finally, evaluation metrics, including classification reports, ROC AUC, and confusion matrices, provide a thorough assessment of the model's effectiveness in clinical applications. This structured approach underscores the integration of advanced machine learning techniques in oncology research, aimed at enhancing patient outcomes.
2.6. Model Architecture
2.6.1. Overview of the Vision Transformer Model
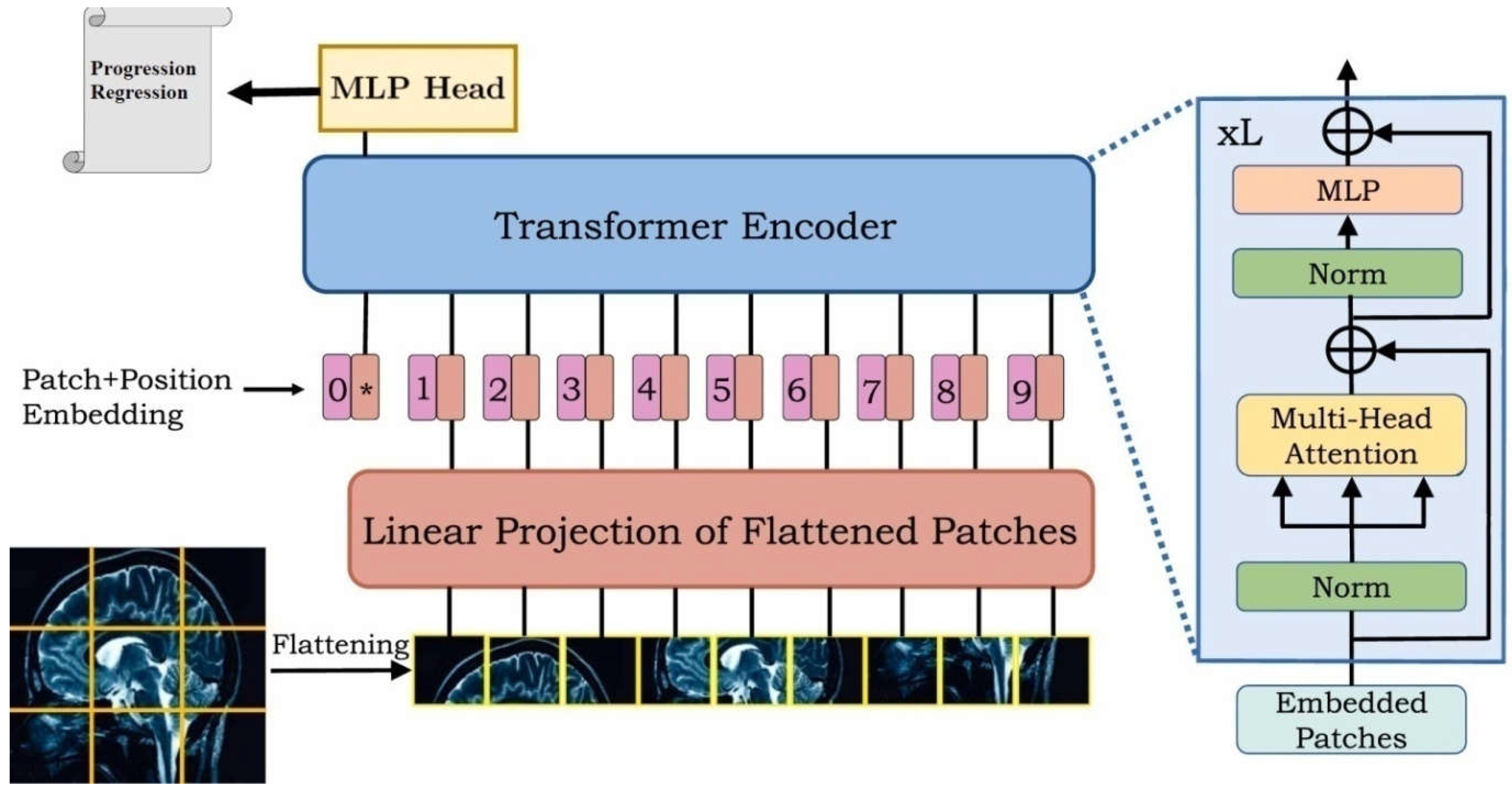
The Vision Transformer (ViT) model represents a significant advancement in computer vision, employing a transformer architecture originally designed for natural language processing. The key innovation of ViT is its ability to process images as sequences of patches, enabling the model to capture long-range dependencies effectively. This section outlines the ViT architecture tailored for the prediction of early responses in brain metastases (BM) following stereotactic radiosurgery (SRS) (
Figure 3).
2.6.2. Input Preprocessing
Before feeding images into the ViT model, the input data undergoes a series of preprocessing steps:
Image Acquisition: The MRI images utilized in this study were obtained from 19 patients, comprising axial fluid-attenuated inversion recovery (FLAIR) sequences and high-resolution contrast-enhanced T1-weighted (CE T1w) sequences.
Patch Extraction: Each image is divided into non-overlapping patches of size 25×25 pixels. This division converts the images into a sequence format, suitable for transformer processing. The total number of patches is determined by the formula:
where image_size=200 pixels.
- 3.
Normalization: The pixel values of the images are normalized to the range [0, 1] to facilitate effective training and convergence of the model.
2.6.3. Model Architecture
The Vision Transformer model architecture comprises several key components:
(num_patches, patch_size × patch_size × num_channels), where num_channels is 3 for RGB images.
- 2.
Patch Embedding: Each patch is linearly projected into a higher-dimensional space (hidden dimension), which allows the model to learn richer representations. This is achieved through a dense layer defined as:
- 3.
Positional Encoding: To retain the spatial information of the patches, positional embeddings are added to the patch embeddings. The positional embeddings are computed using an embedding layer that maps patch indices to dense vectors of the same dimensionality as the hidden representations.
- 4.
Class Token: A learnable class token is pre-pended to the sequence of patch embeddings. This token aggregates information from all patches and is used for the final classification task.
- 5.
-
Transformer Encoder Blocks: The core of the model consists of a stack of transformer encoder blocks, each comprising:
- o
Multi-Head Self-Attention Mechanism: This mechanism allows the model to attend to different parts of the input sequence simultaneously, capturing long-range dependencies.
- o
Multilayer Perceptron (MLP): After self-attention, the output is passed through a neural network with a non-linear activation function (GELU).
- o
Residual Connections and Layer Normalization: Each block includes residual connections that facilitate gradient flow during training and layer normalization that stabilizes the learning process.
The number of encoder layers in this architecture is set to 12, allowing for extensive learning capacity.
- 6.
Classification Head: The output corresponding to the class token is passed through a final dense layer with a softmax activation function to predict the class probabilities (responders or non-responders).
2.6.4. Summary of Hyperparameters
The following hyperparameters were employed in the model architecture:
Image Size: 200 pixels
Patch Size: 25 pixels
Hidden Dimension: 768
MLP Dimension: 3072
Number of Heads: 12
Number of Layers: 12
Dropout Rate: 0.1
The Vision Transformer architecture, with its innovative approach to image processing through patches and attention mechanisms, provides a robust framework for predicting early responses in brain metastases following radiosurgery. The subsequent chapters will discuss the training procedure, evaluation metrics, and results obtained from this model.
2.7. Training Evaluation
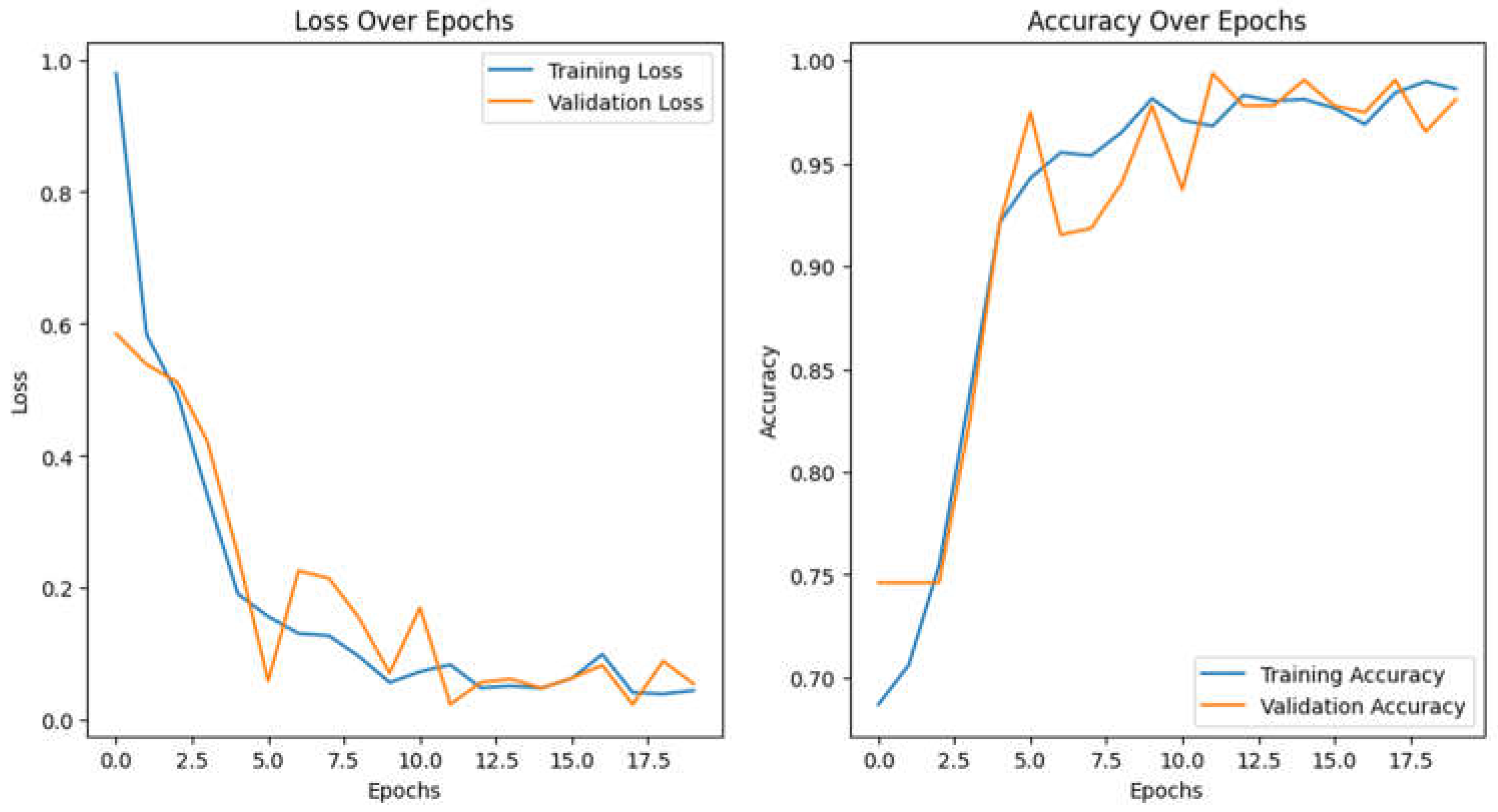
Training the Vision Transformer model requires substantial computational resources. In this study, we utilized a high-performance GPU (Google Colab) to facilitate efficient training over the 20 epochs. The batch size was set to 16, which balanced memory usage and training speed. The model was trained for 20 epochs with an Adam optimizer and used gradient clipping (clip value=1.0) to stabilize the training process. A learning rate of 1e-4 reduced the rate upon plateau, enhancing convergence. These configurations provided a stable and effective training setup, as shown in
Figure 4, which illustrates the model’s accuracy and loss curves.
Summary of Model Performance Curves
The performance curves for the Vision Transformer model indicate effective learning in predicting early responses of brain metastases to radiosurgery (
Figure 4).
Loss Over Epochs: The training loss declines significantly from about 1.0 to below 0.2, indicating effective weight adjustment, while the validation loss decreases more slowly, stabilizing around epoch 10 and remaining higher than training loss, suggesting mild overfitting without significant upward trends.
Accuracy Over Epochs: Training accuracy increases from approximately 70% to nearly 100%, reflecting strong performance on training data, whereas validation accuracy peaks near 98% by epoch 18, indicating good generalization to unseen data.
Overall, the model displays robust learning and generalization, making it a promising clinical tool.
3. Results
3.1. Patient Characteristics
The study population comprised 19 patients (5 females and 14 males), with ages from 43 to 80 years, with a median of 63. Tumor types included lung (n=14), breast (n=3), laryngeal (n=1) and prostate (n=1), reflecting the common primary cancers associated with BM. The distribution of treatment responses was characterized by 15 responders (78.9%) and 4 non-responders (21.1%).
3.2. Model Performance Evaluation
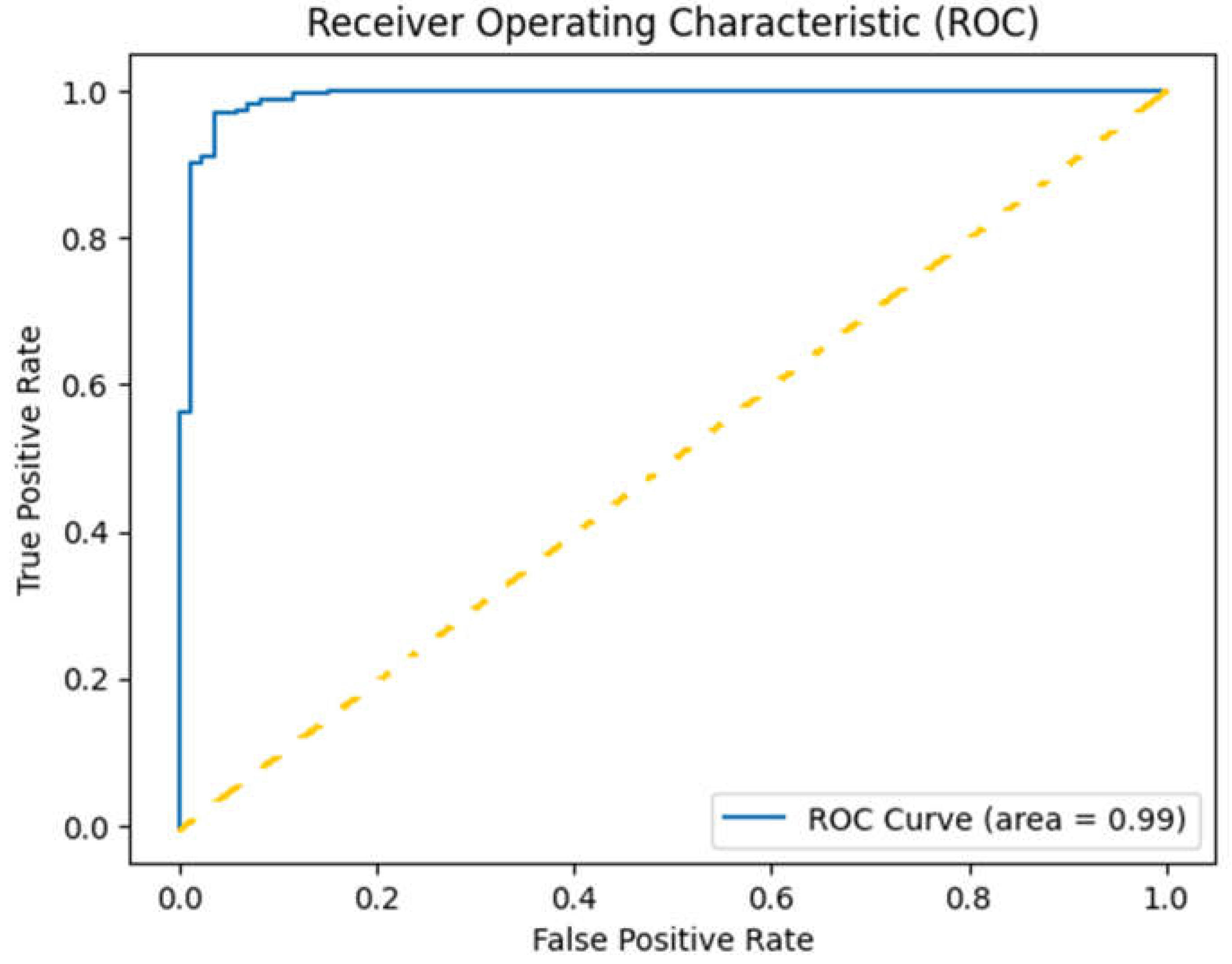
Evaluation metrics included accuracy, precision, recall, and F1-score (Table1), with a focus on the area under the receiver operating characteristic curve (AUC-ROC) for overall performance assessment (
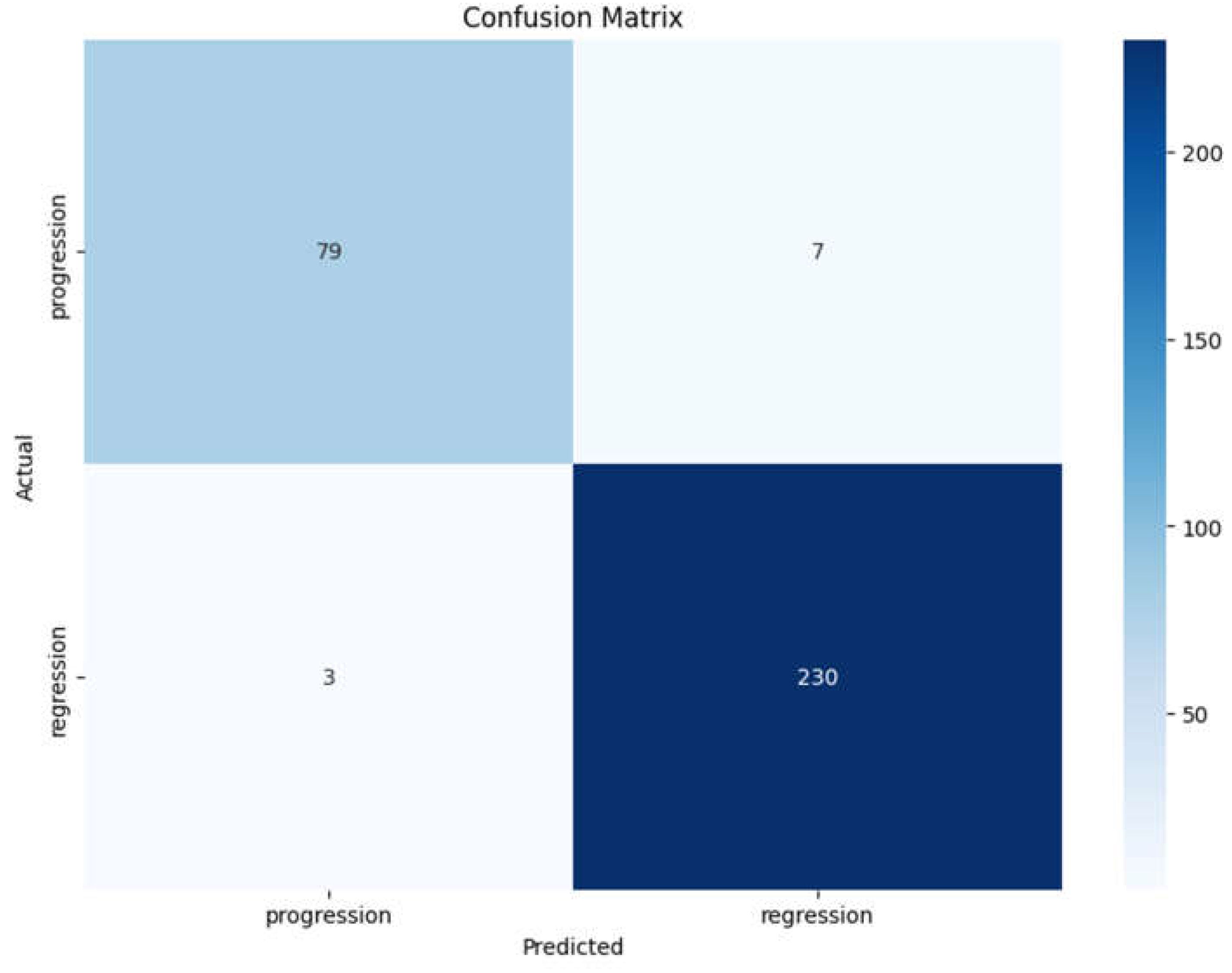
Figure 5). Additionally, confusion matrices were generated to visualize the true positive, false positive, true negative, and false negative rates (
Figure 6).
The classification report (Table1) summarizes the performance of the Vision Transformer model in predicting early responses of brain metastases, providing detailed metrics for both classes: progression and regression.
Precision
Progression: The precision for progression is 0.96, indicating that 96% of the instances predicted as progression were indeed correct. This high precision suggests the model is reliable in identifying true progression cases.
Regression: The precision for regression is 0.97, indicating that 97% of the instances predicted as regression were correct. This exceptionally high precision reflects the model's strong performance in classifying regression cases.
Recall
Progression: The recall for progression is 0.92, meaning the model correctly identifies 92% of actual progression cases. This high recall indicates the model's effectiveness in capturing most of the true positive cases for progression.
Regression: The recall for regression is 0.99, suggesting the model successfully identifies 99% of actual regression cases, demonstrating strong performance in recognizing positive instances.
F1-Score
Progression: The F1-score for progression is 0.94, reflecting a good balance between precision and recall, indicating the model's overall accuracy in identifying progression cases.
Regression: The F1-score for regression is 0.98, showing excellent performance, emphasizing both precision and recall for this class.
Overall Accuracy
The overall accuracy of the model is 0.97, demonstrating that the model correctly classified 97% of the total cases. This high accuracy, combined with the strong metrics for both classes, underscores the model's robustness and effectiveness in clinical predictions.
Averages
Weighted Average: The weighted average precision and recall are 0.97 and 0.97, showing that the model maintains high performance even when accounting for the unequal class sizes.
Here are the key points from the curve in
Figure 5:
True Positive Rate (Sensitivity): The y-axis represents the True Positive Rate (TPR), which indicates the proportion of actual positive cases correctly identified by the model. In this case, a TPR close to1.00 signifies that the model is highly effective in identifying responders.
False Positive Rate: The x-axis represents the False Positive Rate (FPR), indicating the proportion of negative cases that are incorrectly classified as positive. A lower FPR is desirable, as it means the model is making fewer mistakes in identifying non-responders.
Area Under the Curve (AUC): The area under the ROC curve (AUC) is reported as 0.99, indicating perfect classification performance. This suggests that the model correctly distinguishes between responders and non-responders without any overlap, effectively making accurate predictions across all thresholds.
Curve Shape: The ROC curve rises sharply and approaches the top left corner of the plot, reflecting that the model achieves high sensitivity with very few false positives. This shape indicates strong performance, particularly in clinical contexts where the cost of false negatives (missing a responder) can be high.
The confusion matrix displayed (
Figure 6) provides a clear overview of the classification performance of the Vision Transformer model in predicting the early response of brain metastases to radiosurgery. Here are the key observations:
True Positives (TP):The model correctly identified 79 cases of progression (top left cell), indicating a strong ability to classify patients who experienced disease progression.
True Negatives (TN):The model accurately classified 230 cases of regression (bottom right cell), demonstrating effective recognition of patients whose condition improved following treatment.
False Positives (FP):There are 7 cases where the model incorrectly predicted progression when the actual outcome was regression (top right cell). This low number of false positives indicates a high specificity of the model.
False Negatives (FN):The model misclassified 3 cases of progression as regression (bottom left cell). This suggests that the model has some limitations in identifying patients with progression, but the number is relatively low compared to the total number of cases.
To assess the robustness of the ViT model’s predictive performance, we used bootstrap resampling to compute 95% confidence intervals for both accuracy and AUC scores based on test set predictions. Using 1,000 bootstrap samples, we obtained confidence intervals for accuracy, with lower and upper bounds at 94.67% and 98.43%, respectively, and for AUC, at 97.87% and 99.85%. These intervals provide a statistically grounded measure of the model's reliability in classifying treatment responses.
3.3. Qualitative Analysis
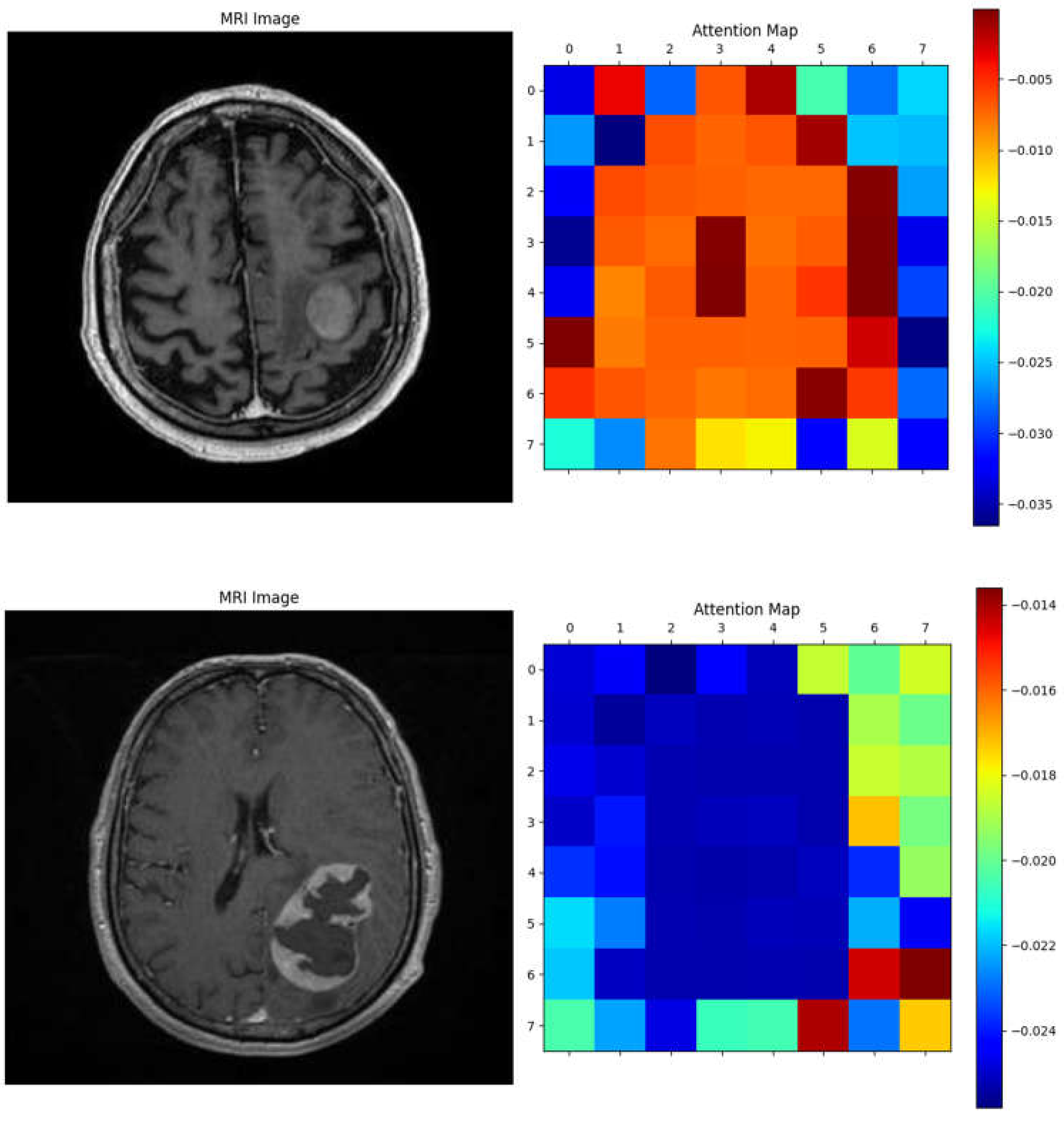
The Vision Transformer (ViT) model demonstrated the ability to focus on key tumor-related features, as shown by the attention maps in
Figure 7. In the MRI images, distinct tumor regions are visible, characterized by altered enhancement patterns and edema surrounding the lesion. The corresponding attention maps reveal that the model consistently highlights patches associated with these abnormal regions.
The attention maps indicate that the model assigns higher attention (marked by warmer colors like red and yellow) to areas likely containing tumor tissue, suggesting it recognizes significant pathological features. This interpretability reinforces the clinical relevance of the ViT model, as it not only distinguishes tumor areas effectively but also aligns its focus with known tumor characteristics, aiding in understanding the basis of its predictions. These examples illustrate the model's capacity to differentiate response patterns in brain metastasis, potentially contributing to improved diagnostic confidence in clinical settings.
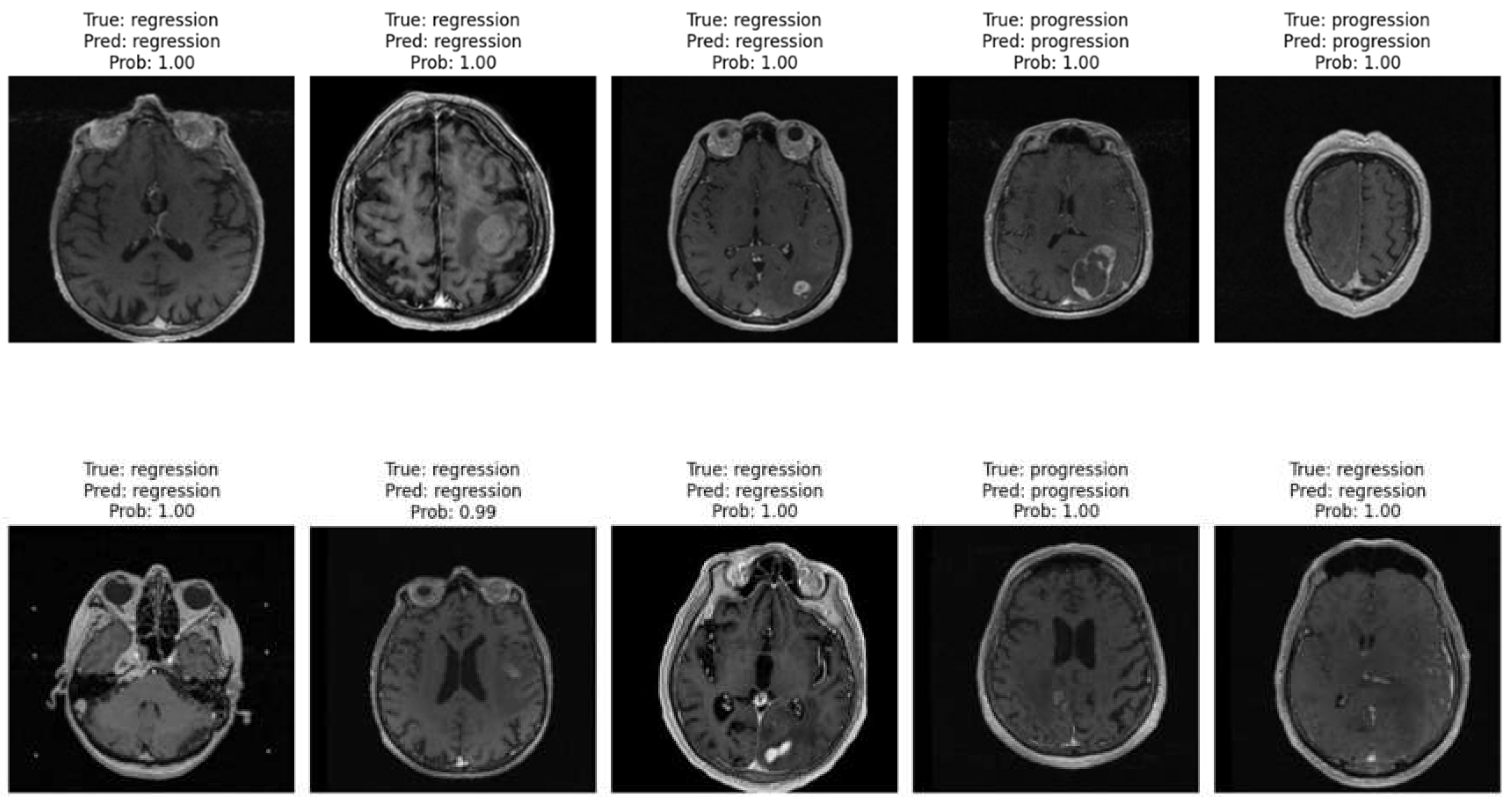
Figure 8 displays ten randomly selected MRI cases from the patient image database, showcasing the model’s performance in distinguishing between progression and regression patterns. Each image includes the ground truth label, the model’s prediction, and the predicted probability. Across these cases, the model accurately identifies both regression and progression with high confidence (probabilities close to or equal to 1.00).
These examples emphasize the model’s robust capability to differentiate between treatment response types, reinforcing its clinical relevance. Consistently high confidence levels indicate that the Vision Transformer model effectively recognizes patterns associated with each response type, thereby supporting its potential utility in real-world clinical settings for monitoring brain metastasis.
4. Discussion
This study demonstrates the potential of vision transformers in predicting early treatment responses in brain metastases after SRS using minimal MRI pre-processing [
21]. The promising results indicate that ViTs can be integrated into clinical workflows to aid in personalized treatment planning and improve patient outcomes. By leveraging minimal pre-processing, this approach addresses the need for efficient, scalable solutions in clinical practice, especially in resource-limited settings.
The findings align with previous studies highlighting the efficacy of deep learning models in radiological applications [
22,
23,
24,
25,
26]. The high accuracy and AUC-ROC underscore the robustness of ViTs in handling the complexity of medical images. Moreover, the use of minimal pre-processing simplifies the pipeline, making it feasible for implementation in busy clinical environments where rapid decision-making is essential.
The interpretability of the model, demonstrated through attention maps, presents an additional advantage, allowing clinicians to understand the rationale behind predictions and potentially enhance the trustworthiness of AI applications in medical settings [
27,
28]. Future research should explore the integration of additional clinical and radiological features to further enhance predictive accuracy and generalizability across diverse patient populations.
Limitations
Sample Size: The study is based on a relatively small cohort of 19 patients. While the initial results are promising, a larger and more diverse dataset is necessary to validate the model's performance across different populations and to ensure its generalizability. A limited sample size may also increase the risk of overfitting, making it crucial to confirm these findings with additional data. It may limit also the generalizability of our findings, particularly in diverse populations. A larger cohort with varied demographics and tumor characteristics will be crucial for validating the model's robustness. Furthermore, the retrospective nature of this study could introduce biases, necessitating future prospective studies to mitigate these concerns.
Imaging Modalities: The model's effectiveness was evaluated using specific MRI sequences (FLAIR and CE T1w). While these modalities are common in clinical practice, the performance of the model on other imaging types or combinations (e.g., diffusion-weighted imaging or functional MRI) remains untested. Future research should explore the integration of multiple imaging modalities to enhance predictive accuracy.
Retrospective Nature: The study utilized a retrospective design, relying on pre-existing medical data. This approach may introduce biases related to patient selection and data quality. Prospective studies would help address these biases and provide a clearer understanding of the model's performance in real-time clinical settings.
Class Imbalance: Although the model demonstrated good performance metrics, there is a slight imbalance in the number of cases between progression and regression classes. This imbalance can affect the model's learning and may lead to biased predictions. Techniques such as data augmentation or synthetic data generation could be employed to mitigate this issue in future studies.
Clinical Context: The model's predictions are based solely on imaging data and do not incorporate other potentially influential clinical factors, such as genetic markers, histopathological features, or patient comorbidities. Integrating these additional variables could enhance the model's predictive power and its applicability in clinical decision-making.
Future Directions
Larger Multi-Center Studies: Future research should focus on conducting larger, multi-center studies to validate the model's performance across diverse patient populations. This would help establish robustness and generalizability, ensuring that the model is applicable in various clinical settings.
Integration of Additional Data Types: Expanding the model to incorporate various data types, including genomic information, histopathological data, and clinical features, could improve predictive accuracy. Developing a multimodal approach would allow for a more comprehensive assessment of patient responses to treatment.
Real-Time Predictive Systems: Efforts should be made to develop real-time predictive systems that can be integrated into clinical workflows. Such systems would assist clinicians in making timely decisions about patient management based on live imaging data and model predictions.
Model Refinement: Continuous refinement of the Vision Transformer architecture and training methodologies should be pursued to enhance model performance. Experimentation with hyperparameter tuning, different model architectures, and advanced training techniques (such as transfer learning) could lead to improved outcomes.
Longitudinal Studies: Future studies should consider a longitudinal approach to assess how the model's predictions correlate with long-term patient outcomes. This could provide valuable insights into the model's utility in guiding treatment and monitoring disease progression over time.
User-Friendly Interfaces: Developing user-friendly interfaces that facilitate the use of the model in clinical practice is essential. These tools should be designed to allow clinicians to input MRI data and obtain predictions with minimal complexity, ultimately aiding in the decision-making process.
In conclusion, while the Vision Transformer model shows great promise in predicting early responses of brain metastases to radiosurgery, addressing its limitations and pursuing these future directions will be critical for enhancing its utility in clinical practice and improving patient outcomes.
5. Conclusions
The Vision Transformer model has demonstrated exceptional performance in predicting early responses of brain metastases to radiosurgery, achieving an overall accuracy of 97%. With high precision (96% for progression and 97% for regression) and strong recall rates (92% for progression and 99% for regression), the model effectively distinguishes between treatment outcomes. The confusion matrix analysis further supports its reliability, showing minimal misclassifications. The almost perfect area under the ROC curve (AUC = 0.99) indicates that the model can accurately differentiate between responders and non-responders across various thresholds. These findings suggest that the Vision Transformer model is not only robust but also holds significant promise for clinical applications in oncology, enhancing decision-making processes and ultimately improving patient outcomes. By improving early response predictions for brain metastases, this model not only has the potential to refine clinical decision-making but also to contribute to the development of personalized treatment strategies in oncology.
Future research should focus on validating these results in larger, diverse cohorts, integrating additional data types, and refining the model to further enhance its utility in clinical practice.
Author Contributions
Conceptualization: S.R.V., D.I.B., L.O.; data curation: M.R.O., C.C.V., D.I.R.; investigation: D.I.B., M.A., M.I.U., D.V.; software: C.G.B.; supervision: S.R.V., D.I.B., D.I.R.; validation: S.R.V., M.A., D.V.; visualization: C.G.B., C.C.V., L.O.;writing—original draft: S.R.V., D.I.B., M.R.O., D.I.R.; writing—review and editing: M.A., M.I.U., C.C.V., D.V.; funding acquisition: S.R.V., M.I.U., D.V. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding
Institutional Review Board Statement
Since the study was retrospective, there was no need for approval from the Ethics Committee.
Informed Consent Statement
The study used only pre-existing medical data, therefore, patient consent was not required.
Data Availability Statement:
.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Kuksis M, Gao Y, Tran W, Hoey C, Kiss A, Komorowski AS, Dhaliwal AJ, Sahgal A, Das S, Chan KK, Jerzak KJ. The incidence of brain metastases among patients with metastatic breast cancer: a systematic review and meta-analysis. Neuro Oncol. 2021 Jun 1;23(6):894-904. [CrossRef]
- Gavrilovic IT, Posner JB. Brain metastases: epidemiology and pathophysiology. J Neurooncol. 2005 Oct;75(1):5-14. [CrossRef]
- Lehrer EJ, Jones BM, Sindhu KK, Dickstein DR, Cohen M, Lazarev S, Quiñones-Hinojosa A, Green S, Trifiletti DM. A Review of the Role of Stereotactic Radiosurgery and Immunotherapy in the Management of Primary Central Nervous System Tumors. Biomedicines. 2022 Nov 19;10(11):2977. [CrossRef]
- Mangesius J, Seppi T, Arnold CR, Mangesius S, Kerschbaumer J, Demetz M, Minasch D, Vorbach SM, Sarcletti M, Lukas P, Nevinny-Stickel M, Ganswindt U. Prognosis versus Actual Outcomes in Stereotactic Radiosurgery of Brain Metastases: Reliability of Common Prognostic Parameters and Indices. Curr Oncol. 2024 Mar 26;31(4):1739-1751. [CrossRef]
- Kondziolka, D.; Patel, A.; Lunsford, L.; Kassam, A.; Flickinger, J.C. Stereotactic radiosurgery plus whole brain radiotherapy versus radiotherapy alone for patients with multiple brain metastases. Int. J. Radiat. Oncol. Biol. Phys. 1999, 45, 427–434. [CrossRef]
- Patchell, R.A.; Tibbs, P.A.; Walsh, J.W.; Dempsey, R.J.; Maruyama, Y.; Kryscio, R.J.; Markesbery, W.R.; Macdonald, J.S.; Young, B. A Randomized trial of surgery in the treatment of single metastases to the brain. N. Engl. J. Med. 1990, 322, 494–500. [CrossRef]
- Park, Y.G.; Choi, J.Y.; Chang, J.W.; Chung, S.S. Gamma knife radiosurgery for metastatic brain tumors. Ster. Funct. Neurosurg. 2001, 76, 201–203. [CrossRef]
- Kocher, M.; Soffietti, R.; Abacioglu, U.; Villà, S.; Fauchon, F.; Baumert, B.G.; Fariselli, L.; Tzuk-Shina, T.; Kortmann, R.-D.; Carrie, C.; et al. Adjuvant whole-brain radiotherapy versus observation after radiosurgery or surgical resection of one to three cerebral metastases: Results of the EORTC 22952-26001 study. J. Clin. Oncol. 2011, 29, 134–141. [CrossRef]
- Kim, Y.-J.; Cho, K.H.; Kim, J.-Y.; Lim, Y.K.; Min, H.S.; Lee, S.H.; Kim, H.J.; Gwak, H.S.; Yoo, H.; Lee, S.H. Single-dose versus fractionated stereotactic radiotherapy for brain metastases. Int. J. Radiat. Oncol. Biol. Phys. 2011, 81, 483–489. [CrossRef]
- Jee, T.K.; Seol, H.J.; Im, Y.-S.; Kong, D.-S.; Nam, D.-H.; Park, K.; Shin, H.J.; Lee, J.-I. Fractionated gamma knife radiosurgery for benign perioptic tumors: outcomes of 38 patients in a single institute. Brain Tumor Res. Treat. 2014, 2, 56–61. [CrossRef]
- Ernst-Stecken, A.; Ganslandt, O.; Lambrecht, U.; Sauer, R.; Grabenbauer, G. Phase II trial of hypofractionated stereotactic radiotherapy for brain metastases: Results and toxicity. Radiother. Oncol. 2006, 81, 18–24. [CrossRef]
- Kim, J.W.; Park, H.R.; Lee, J.M.; Kim, J.W.; Chung, H.T.; Kim, D.G.; Paek, S.H. Fractionated stereotactic gamma knife radiosurgery for large brain metastases: A retrospective, single center study. PLoS ONE 2016, 11, e0163304. [CrossRef]
- Ewend, M.G.; Elbabaa, S.; Carey, L.A. Current treatment paradigms for the management of patients with brain metastases. Neurosurgery 2005, 57 (Suppl. S5), S66–77, Discusssion S1. [CrossRef]
- Cho, K.R.; Lee, M.H.; Kong, D.-S.; Seol, H.J.; Nam, D.-H.; Sun, J.-M.; Ahn, J.S.; Ahn, M.-J.; Park, K.; Kim, S.T.; et al. Outcome of gamma knife radiosurgery for metastatic brain tumors derived from non-small cell lung cancer. J. Neuro-Oncol. 2015, 125, 331– 338. [CrossRef]
- Dosovitskiy, Alexey. "An image is worth 16x16 words: Transformers for image recognition at scale." arXiv preprint arXiv:2010.11929 (2020).
- Christos Matsoukas, Johan Fredin Haslum, Magnus Söderberg,Kevin Smith, Is it Time to Replace CNNs with Transformers for Medical Images? https://ar5iv.labs.arxiv.org/html/2108.09038.
- Brenner, D.J. the linear-quadratic model is an appropriate methodology for determining isoeffective doses at large doses perfraction. Semin. Radiat. Oncol. 2008, 18, 234–239. [CrossRef]
- Fowler, J.F. The linear-quadratic formula and progress in fractionated radiotherapy. Br. J. Radiol. 1989, 62, 679–694. [CrossRef]
- Higuchi, Y.; Serizawa, T.; Nagano, O.; Matsuda, S.; Ono, J.; Sato, M.; Iwadate, Y.; Saeki, N. Three-staged stereotactic radiotherapywithout whole brain irradiation for large metastatic brain tumors. Int. J. Radiat. Oncol. Biol. Phys. 2009, 74, 1543–1548. [CrossRef]
- Lin NU, Lee EQ, Aoyama H, Barani IJ, Barboriak DP, Baumert BG, Bendszus M, Brown PD, Camidge DR, Chang SM, Dancey J, de Vries EG, Gaspar LE, Harris GJ, Hodi FS, Kalkanis SN, Linskey ME, Macdonald DR, Margolin K, Mehta MP, Schiff D, Soffietti R, Suh JH, van den Bent MJ, Vogelbaum MA, Wen PY; Response Assessment in Neuro-Oncology (RANO) group. Response assessment criteria for brain metastases: proposal from the RANO group. Lancet Oncol. 2015 Jun;16(6):e270-8. [CrossRef]
- Ching T, Himmelstein DS, Beaulieu-Jones BK, Kalinin AA, Do BT, Way GP, Ferrero E, Agapow PM, Zietz M, Hoffman MM, Xie W, Rosen GL, Lengerich BJ, Israeli J, Lanchantin J, Woloszynek S, Carpenter AE, Shrikumar A, Xu J, Cofer EM, Lavender CA, Turaga SC, Alexandari AM, Lu Z, Harris DJ, DeCaprio D, Qi Y, Kundaje A, Peng Y, Wiley LK, Segler MHS, Boca SM, Swamidass SJ, Huang A, Gitter A, Greene CS. Opportunities and obstacles for deep learning in biology and medicine. J R Soc Interface. 2018 Apr;15(141):20170387. [CrossRef]
- Topol, E. J. (2019). "High-Performance Medicine: The convergence of human and artificial intelligence." Nature Medicine, 25, 44–56.
- Litjens G, Kooi T, Bejnordi BE, Setio AAA, Ciompi F, Ghafoorian M, van der Laak JAWM, van Ginneken B, Sánchez CI. A survey on deep learning in medical image analysis. Med Image Anal. 2017 Dec;42:60-88. [CrossRef]
- Trofin, A.-M.; Buzea, C.G.; Buga, R.; Agop, M.; Ochiuz, L.; Iancu, D.T.; Eva, L. Predicting Tumor Dynamics Post-Staged GKRS: Machine Learning Models in Brain Metastases Prognosis. Diagnostics 2024, 14, 1268. [CrossRef]
- Buzea CG, Buga R, Paun MA, Albu M, Iancu DT, Dobrovat B, Agop M, Paun VP, Eva L. AI Evaluation of Imaging Factors in the Evolution of Stage-Treated Metastases Using Gamma Knife. Diagnostics (Basel). 2023 Sep 4;13(17):2853. [CrossRef]
- Buzea, CG, Mirestean, CC, Agop, M, Paun, VP, Iancu, DT. Classification of good and bad responders in locally advanced rectal cancer after neoadjuvant radio-chemotherapy using radiomics signature. University Politehnica of Bucharest Scientific Bulletin-series A-Applied Mathematics and Physics, Volume 81, Issue 2, Page 265-278 (2019).
- Caruana, R., & Niculescu-Mizil, A. (2006). "An Empirical Comparison of Supervised Learning Algorithms." In Proceedings of the 23rd International Conference on Machine Learning (ICML), 161-168.
- Rajkomar A, Dean J, Kohane I. Machine Learning in Medicine. N Engl J Med. 2019 Apr 4;380(14):1347-1358. [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).