Submitted:
02 November 2024
Posted:
04 November 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results
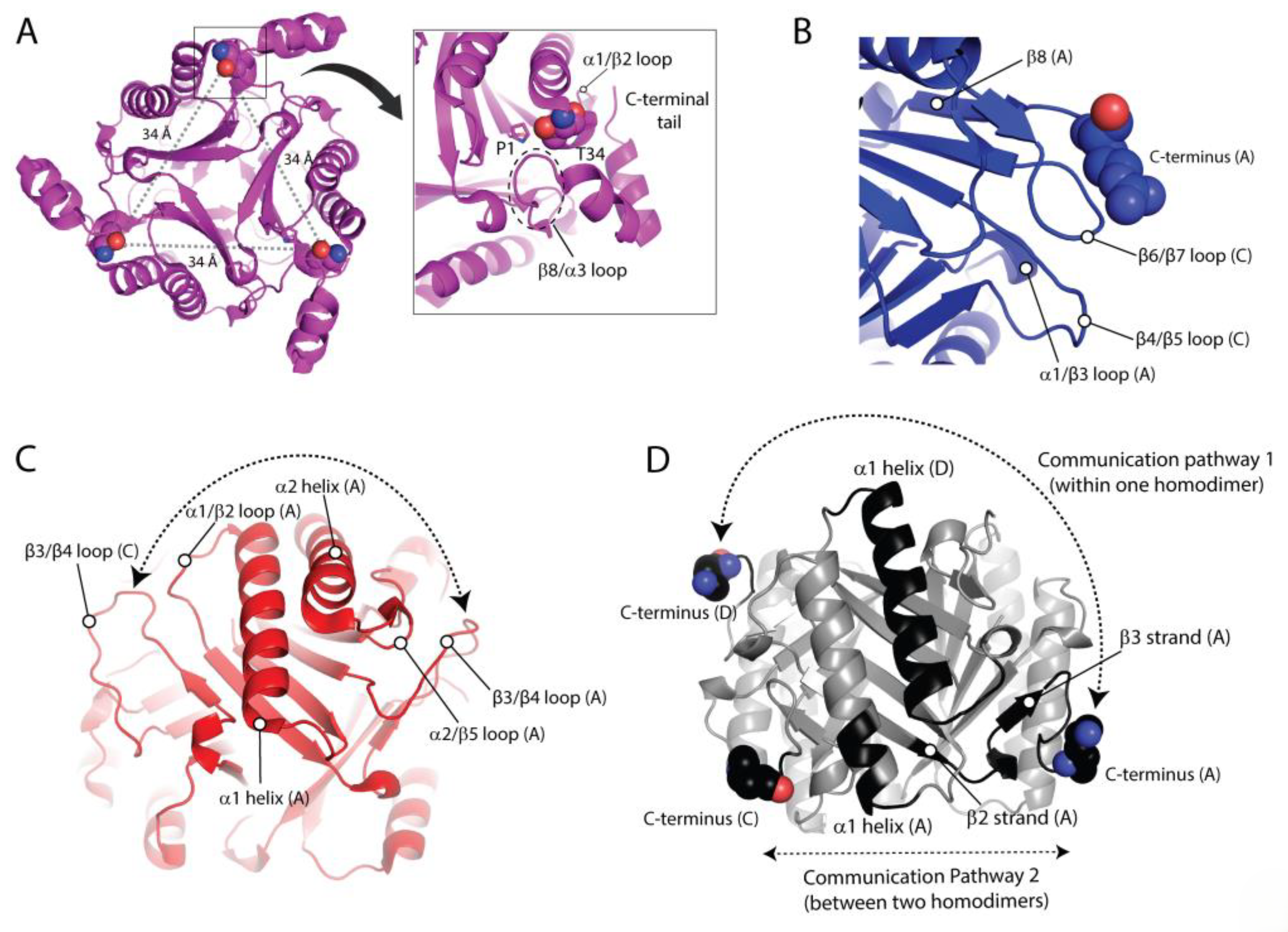
2.1. Crystallographic Analysis of TSF Representative Members Reveals Noticeable Differences in the C-Terminal Region
2.2. RMSF Analysis Across the Target Proteins Demonstrates Diverse Dynamic Profiles
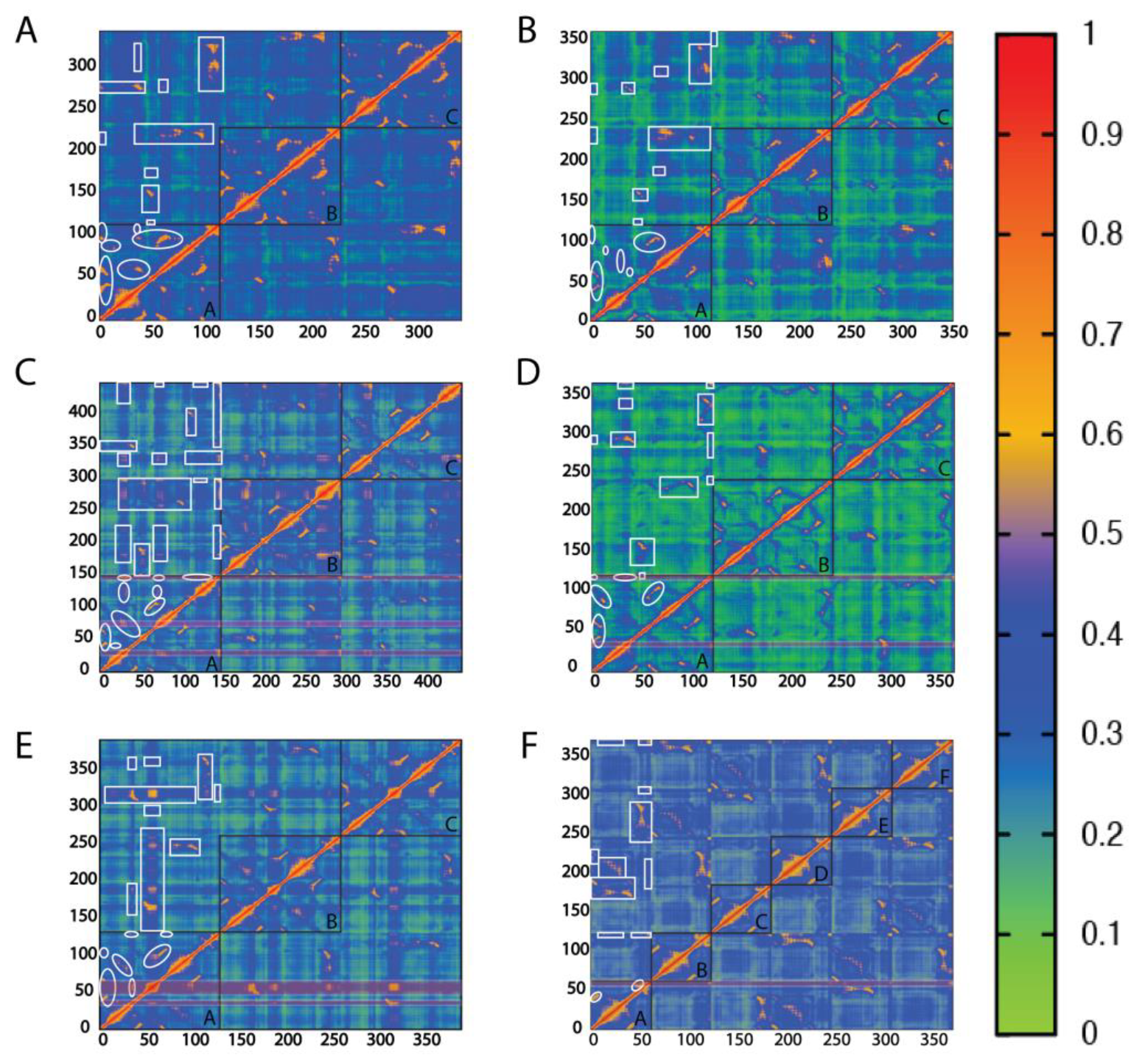
2.3. Correlation Plots Expose Communication Pathways with Mechanistic Interest
2.4. Correlation Analyses Reveals Communications Between the C-Terminal Region and Active Site Residues
3. Discussion and Conclusions
4. Materials and Methods
4.1. Multiple Sequence Alignments
4.2. MD Simulations
4.3. Correlation Analysis
4.4. RMSF Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Davidson, R.; Baas, B. J.; Akiva, E.; Holliday, G. L.; Polacco, B. J.; LeVieux, J. A.; Pullara, C. R.; Zhang, Y. J.; Whitman, C. P.; Babbitt, P. C., A global view of structure-function relationships in the tautomerase superfamily. J Biol Chem 2018, 293, (7), 2342-2357. [CrossRef]
- Poelarends, G. J.; Veetil, V. P.; Whitman, C. P., The chemical versatility of the beta-alpha-beta fold: catalytic promiscuity and divergent evolution in the tautomerase superfamily. Cell Mol Life Sci 2008, 65, (22), 3606-18.
- Whitman, C. P., The 4-oxalocrotonate tautomerase family of enzymes: how nature makes new enzymes using a beta-alpha-beta structural motif. Arch Biochem Biophys 2002, 402, (1), 1-13.
- Sun, H. W.; Bernhagen, J.; Bucala, R.; Lolis, E., Crystal structure at 2.6-A resolution of human macrophage migration inhibitory factor. Proc Natl Acad Sci U S A 1996, 93, (11), 5191-6. [CrossRef]
- Sugimoto, H.; Taniguchi, M.; Nakagawa, A.; Tanaka, I.; Suzuki, M.; Nishihira, J., Crystal structure of human D-dopachrome tautomerase, a homologue of macrophage migration inhibitory factor, at 1.54 A resolution. Biochemistry 1999, 38, (11), 3268-79. [CrossRef]
- Taylor, A. B.; Czerwinski, R. M.; Johnson, W. H., Jr.; Whitman, C. P.; Hackert, M. L., Crystal structure of 4-oxalocrotonate tautomerase inactivated by 2-oxo-3-pentynoate at 2.4 A resolution: analysis and implications for the mechanism of inactivation and catalysis. Biochemistry 1998, 37, (42), 14692-700.
- Pantouris, G.; Khurana, L.; Ma, A.; Skeens, E.; Reiss, K.; Batista, V. S.; Lisi, G. P.; Lolis, E. J., Regulation of MIF Enzymatic Activity by an Allosteric Site at the Central Solvent Channel. Cell Chem Biol 2020, 27, (6), 740-750 e5. [CrossRef]
- Pantouris, G.; Bucala, R.; Lolis, E. J., Structural Plasticity in the C-Terminal Region of Macrophage Migration Inhibitory Factor-2 Is Associated with an Induced Fit Mechanism for a Selective Inhibitor. Biochemistry 2018, 57, (26), 3599-3605. [CrossRef]
- Parkins, A.; Skeens, E.; McCallum, C. M.; Lisi, G. P.; Pantouris, G., The N-terminus of MIF regulates the dynamic profile of residues involved in CD74 activation. Biophys J 2021, 120, (18), 3893-3900. [CrossRef]
- Parkins, A.; Chen, E.; Rangel, V. M.; Singh, M.; Xue, L.; Lisi, G. P.; Pantouris, G., Ligand-induced conformational changes enable intersubunit communications in D-dopachrome tautomerase. Biophys J 2023. [CrossRef]
- Parkins, A.; Pilien, A. V. R.; Wolff, A. M.; Argueta, C.; Vargas, J.; Sadeghi, S.; Franz, A. H.; Thompson, M. C.; Pantouris, G., The C-terminal Region of D-DT Regulates Molecular Recognition for Protein-Ligand Complexes. J Med Chem 2024, 67, (9), 7359-7372. [CrossRef]
- Parkins, A.; Das, P.; Prahaladan, V.; Rangel, V. M.; Xue, L.; Sankaran, B.; Bhandari, V.; Pantouris, G., 2,5-Pyridinedicarboxylic acid is a bioactive and highly selective inhibitor of D-dopachrome tautomerase. Structure 2023, 31, (3), 355-367 e4. [CrossRef]
- Pantouris, G.; Syed, M. A.; Fan, C.; Rajasekaran, D.; Cho, T. Y.; Rosenberg, E. M., Jr.; Bucala, R.; Bhandari, V.; Lolis, E. J., An Analysis of MIF Structural Features that Control Functional Activation of CD74. Chem Biol 2015, 22, (9), 1197-205. [CrossRef]
- Singh, A. K.; Pantouris, G.; Borosch, S.; Rojanasthien, S.; Cho, T. Y., Structural basis for decreased induction of class IB PI3-kinases expression by MIF inhibitors. J Cell Mol Med 2017, 21, (1), 142-153. [CrossRef]
- Tilstam, P. V.; Pantouris, G.; Corman, M.; Andreoli, M.; Mahboubi, K.; Davis, G.; Du, X.; Leng, L.; Lolis, E.; Bucala, R., A selective small-molecule inhibitor of macrophage migration inhibitory factor-2 (MIF-2), a MIF cytokine superfamily member, inhibits MIF-2 biological activity. J Biol Chem 2019, 294, (49), 18522-18531. [CrossRef]
- Parkins, A.; Sandin, S. I.; Knittel, J.; Franz, A. H.; Ren, J.; de Alba, E.; Pantouris, G., Underrepresented Impurities in 4-Hydroxyphenylpyruvate Affect the Catalytic Activity of Multiple Enzymes. Anal Chem 2023, 95, (11), 4957-4965. [CrossRef]
- Bloom, J.; Pantouris, G.; He, M.; Aljabari, B.; Mishra, L.; Manjula, R.; Parkins, A.; Lolis, E. J.; Al-Abed, Y., Iguratimod, an allosteric inhibitor of macrophage migration inhibitory factor (MIF), prevents mortality and oxidative stress in a murine model of acetaminophen overdose. Mol Med 2024, 30, (1), 43. [CrossRef]
- Li, J.; Leng, L.; Pantouris, G.; Manjula, R.; Piecychna, M.; Abriola, L.; Hu, B.; Lolis, E.; Armstrong, M. E.; Donnelly, S. C.; Bucala, R., A small-molecule allele-selective transcriptional inhibitor of the MIF immune susceptibility locus. J Biol Chem 2024, 300, (7), 107443. [CrossRef]
- Lubetsky, J. B.; Swope, M.; Dealwis, C.; Blake, P.; Lolis, E., Pro-1 of macrophage migration inhibitory factor functions as a catalytic base in the phenylpyruvate tautomerase activity. Biochemistry 1999, 38, (22), 7346-54. [CrossRef]
- Odh, G.; Hindemith, A.; Rosengren, A. M.; Rosengren, E.; Rorsman, H., Isolation of a new tautomerase monitored by the conversion of D-dopachrome to 5,6-dihydroxyindole. Biochem Biophys Res Commun 1993, 197, (2), 619-24. [CrossRef]
- Chen, L. H.; Kenyon, G. L.; Curtin, F.; Harayama, S.; Bembenek, M. E.; Hajipour, G.; Whitman, C. P., 4-Oxalocrotonate tautomerase, an enzyme composed of 62 amino acid residues per monomer. J Biol Chem 1992, 267, (25), 17716-21. [CrossRef]
- Stivers, J. T.; Abeygunawardana, C.; Whitman, C. P.; Mildvan, A. S., 4-Oxalocrotonate tautomerase, a 41-kDa homohexamer: backbone and side-chain resonance assignments, solution secondary structure, and location of active site residues by heteronuclear NMR spectroscopy. Protein Sci 1996, 5, (4), 729-41. [CrossRef]
- Subramanya, H. S.; Roper, D. I.; Dauter, Z.; Dodson, E. J.; Davies, G. J.; Wilson, K. S.; Wigley, D. B., Enzymatic ketonization of 2-hydroxymuconate: specificity and mechanism investigated by the crystal structures of two isomerases. Biochemistry 1996, 35, (3), 792-802. [CrossRef]
- de Jong, R. M.; Bazzacco, P.; Poelarends, G. J.; Johnson, W. H., Jr.; Kim, Y. J.; Burks, E. A.; Serrano, H.; Thunnissen, A. M.; Whitman, C. P.; Dijkstra, B. W., Crystal structures of native and inactivated cis-3-chloroacrylic acid dehalogenase. Structural basis for substrate specificity and inactivation by (R)-oxirane-2-carboxylate. J Biol Chem 2007, 282, (4), 2440-9.
- Almrud, J. J.; Poelarends, G. J.; Johnson, W. H., Jr.; Serrano, H.; Hackert, M. L.; Whitman, C. P., Crystal structures of the wild-type, P1A mutant, and inactivated malonate semialdehyde decarboxylase: a structural basis for the decarboxylase and hydratase activities. Biochemistry 2005, 44, (45), 14818-27.
- Johnson, W. H., Jr.; Hajipour, G.; Whitman, C. P., Stereochemical Studies of 5-(Carboxymethyl)-2-hydroxymuconate Isomerase and 5-(Carboxymethyl)-2-oxo-3-hexene-1,6-dioate Decarboxylase from Escherichia coli C: Mechanistic and Evolutionary Implications. Journal of the American Chemical Society 1995, 117, (34), 8719-8726. [CrossRef]
- Poelarends, G. J.; Whitman, C. P., Evolution of enzymatic activity in the tautomerase superfamily: mechanistic and structural studies of the 1,3-dichloropropene catabolic enzymes. Bioorg Chem 2004, 32, (5), 376-92. [CrossRef]
- Poelarends, G. J.; Johnson, W. H., Jr.; Murzin, A. G.; Whitman, C. P., Mechanistic characterization of a bacterial malonate semialdehyde decarboxylase: identification of a new activity on the tautomerase superfamily. J Biol Chem 2003, 278, (49), 48674-83.
- Poelarends, G. J.; Serrano, H.; Johnson, W. H., Jr.; Hoffman, D. W.; Whitman, C. P., The hydratase activity of malonate semialdehyde decarboxylase: mechanistic and evolutionary implications. J Am Chem Soc 2004, 126, (48), 15658-9. [CrossRef]
- Poelarends, G. J.; Serrano, H.; Johnson, W. H., Jr.; Whitman, C. P., Inactivation of malonate semialdehyde decarboxylase by 3-halopropiolates: evidence for hydratase activity. Biochemistry 2005, 44, (26), 9375-81. [CrossRef]
- Pantouris, G.; Ho, J.; Shah, D.; Syed, M. A.; Leng, L.; Bhandari, V.; Bucala, R.; Batista, V. S.; Loria, J. P.; Lolis, E. J., Nanosecond Dynamics Regulate the MIF-Induced Activity of CD74. Angew Chem Int Ed Engl 2018, 57, (24), 7116-7119.
- Mischke, R.; Gessner, A.; Kapurniotu, A.; Juttner, S.; Kleemann, R.; Brunner, H.; Bernhagen, J., Structure activity studies of the cytokine macrophage migration inhibitory factor (MIF) reveal a critical role for its carboxy terminus. FEBS Lett 1997, 414, (2), 226-32.
- El-Turk, F.; Cascella, M.; Ouertatani-Sakouhi, H.; Narayanan, R. L.; Leng, L.; Bucala, R.; Zweckstetter, M.; Rothlisberger, U.; Lashuel, H. A., The conformational flexibility of the carboxy terminal residues 105-114 is a key modulator of the catalytic activity and stability of macrophage migration inhibitory factor. Biochemistry 2008, 47, (40), 10740-56. [CrossRef]
- Henzler-Wildman, K.; Kern, D., Dynamic personalities of proteins. Nature 2007, 450, (7172), 964-72. [CrossRef]
- McLean, L. R.; Zhang, Y.; Li, H.; Choi, Y. M.; Han, Z.; Vaz, R. J.; Li, Y., Fragment screening of inhibitors for MIF tautomerase reveals a cryptic surface binding site. Bioorg Med Chem Lett 2010, 20, (6), 1821-4. [CrossRef]
- Fenwick, R. B.; Orellana, L.; Esteban-Martin, S.; Orozco, M.; Salvatella, X., Correlated motions are a fundamental property of beta-sheets. Nat Commun 2014, 5, 4070.
- Bouvignies, G.; Bernadó, P.; Meier, S.; Cho, K.; Grzesiek, S.; Brüschweiler, R.; Blackledge, M., Identification of slow correlated motions in proteins using residual dipolar and hydrogen-bond scalar couplings. Proc Natl Acad Sci U S A 2005, 102, (39), 13885-90. [CrossRef]
- Chen, E.; Widjaja, V.; Kyro, G.; Allen, B.; Das, P.; Prahaladan, V. M.; Bhandari, V.; Lolis, E. J.; Batista, V. S.; Lisi, G. P., Mapping N- to C-terminal allosteric coupling through disruption of a putative CD74 activation site in D-dopachrome tautomerase. J Biol Chem 2023, 299, (6), 104729. [CrossRef]
- Skeens, E.; Pantouris, G.; Shah, D.; Manjula, R.; Ombrello, M. J.; Maluf, N. K.; Bhandari, V.; Lisi, G. P.; Lolis, E. J., A Cysteine Variant at an Allosteric Site Alters MIF Dynamics and Biological Function in Homo- and Heterotrimeric Assemblies. Front Mol Biosci 2022, 9, 783669. [CrossRef]
- Robert, X.; Gouet, P., Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res 2014, 42, (Web Server issue), W320-4. [CrossRef]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T. J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Soding, J.; Thompson, J. D.; Higgins, D. G., Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 2011, 7, 539. [CrossRef]
- DeLano, W. L., The PyMOL Molecular Graphics System. Delano Scientific, San Carlos. 2002.
- Pettersen, E. F.; Goddard, T. D.; Huang, C. C.; Couch, G. S.; Greenblatt, D. M.; Meng, E. C.; Ferrin, T. E., UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem 2004, 25, (13), 1605-12. [CrossRef]
- Humphrey, W.; Dalke, A.; Schulten, K., VMD: visual molecular dynamics. J Mol Graph 1996, 14, (1), 33-8, 27-8. [CrossRef]
- Huang, J.; Rauscher, S.; Nawrocki, G.; Ran, T.; Feig, M.; de Groot, B. L.; Grubmuller, H.; MacKerell, A. D., Jr., CHARMM36m: an improved force field for folded and intrinsically disordered proteins. Nat Methods 2017, 14, (1), 71-73. [CrossRef]
- Phillips, J. C.; Hardy, D. J.; Maia, J. D. C.; Stone, J. E.; Ribeiro, J. V.; Bernardi, R. C.; Buch, R.; Fiorin, G.; Henin, J.; Jiang, W.; McGreevy, R.; Melo, M. C. R.; Radak, B. K.; Skeel, R. D.; Singharoy, A.; Wang, Y.; Roux, B.; Aksimentiev, A.; Luthey-Schulten, Z.; Kale, L. V.; Schulten, K.; Chipot, C.; Tajkhorshid, E., Scalable molecular dynamics on CPU and GPU architectures with NAMD. J Chem Phys 2020, 153, (4), 044130. [CrossRef]
- Lange, O. F.; Grubmuller, H., Generalized correlation for biomolecular dynamics. Proteins 2006, 62, (4), 1053-61. [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




