1. Introduction
Skin lesions, whether benign or malignant, are common on or beneath the skin’s surface. While benign lesions, such as moles, are generally harmless, malignant lesions, including melanoma, pose significant health risks [
1,
2]. Accurate differentiation of malignant tumors from dermoscopic images remains a challenging task, underscoring the need for effective deep learning methodologies. Skin cancer is a major public health concern, with incidence rates rising globally [
3,
4]. Early detection is crucial for successful treatment, as delayed diagnosis can have severe consequences.
Over the past decade, various automated approaches for classifying skin lesion images have been developed, with Convolutional Neural Networks (CNNs) and machine learning classifiers playing a predominant role. The following studies represent a sample of the extensive research in this domain. Tschandl et al. [5] introduced a hybrid approach to skin lesion classification by extracting deep features from three pre-trained models (AlexNet, VGG16, and ResNet-18) and classifying them using multiple support vector machine (SVM) classifiers. Murugan et al. [6] explored three distinct methods for feature extraction from skin cancer images, focusing on shape, the ABCD rule, and Gray Level Co-occurrence Matrix (GLCM) features, which were then classified using KNN, Random Forest, and SVM classifiers. Patil et al. [7] compared three classifiers—K-Nearest Neighbors (KNN), Naive Bayes, and SVM—finding that SVM achieved the highest precision when combined with de-duplication techniques. Melbin et al. [8] proposed an integrated approach for skin lesion classification using modified ABCD features with SVM as the classifier, achieving high precision for melanoma, seborrheic keratosis, and lupus erythematosus, though other skin diseases were not considered. Perez et al. [9] evaluated the factors influencing the selection of the optimal CNN architecture for skin lesion analysis, comparing the performance of simple ensemble models against single models across 13 factors and nine architectures. Mporas et al. [10] presented an architecture for classifying pigmented skin lesions from dermatoscopic images by extracting color-based features and utilizing an AdaBoost classifier in conjunction with Random Forest. Lastly, Hosny et al. [11] proposed fine-tuning the AlexNet architecture by replacing its final layer with a softmax layer to classify lesions into three distinct classes. The integration of deep learning (DL) and computer vision in skin cancer classification presents a transformative approach to medical technology, offering significant potential for early detection and effective disease management. However, several challenges persist due to the task’s complexity and the limitations of current technologies. The quality of the images used for training and analysis is critical to classification accuracy. Variations in lighting, background, resolution, and the presence of occlusions such as hair or skin folds can hinder the accurate identification of cancerous lesions [12,13,14,15]. Furthermore, the subtlety of skin cancer symptoms can make it difficult for deep learning models to distinguish malignant lesions from benign ones. Numerous studies have leveraged the combination of deep learning and machine learning techniques, demonstrating the effectiveness and versatility of this hybrid approach across various fields and applications [15,16,17,18,19,20,21,22,23,24,25]. To address these challenges, this study proposes an integrated approach that combines DL with ML to enhance the efficiency and effectiveness of skin cancer classification. This approach accelerates the training process and addresses the limitations posed by the limited availability of annotated data in medical contexts. In particular, DL is used to extract meaningful features from deep representations, improving the accuracy and precision of distinguishing between different types of skin lesions. This study employs several CNN models for feature extraction, paired with a robust ML algorithm, specifically Random Forest (RF), to enhance the system’s discriminatory power.
The remainder of this paper is structured as follows:
Section 2 outlines the proposed methodology, including preprocessing, feature extraction using pretrained CNN models, and classification using RF.
Section 3 presents the experimental analysis, discusses the results, and provides a comparison with recent studies. Finally,
Section 4 offers concluding remarks and suggests directions for future research.
2. Methodology
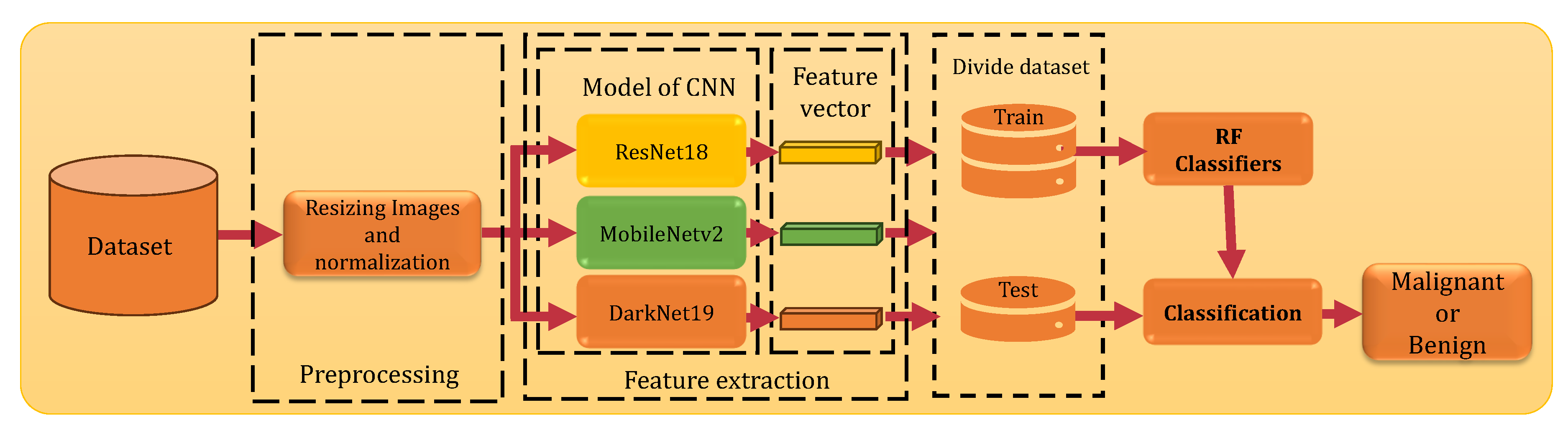
The main objective of this research is to develop a smart system that can accurately classify and detect skin cancer based on images of skin moles. The system specifically distinguishing between malignant and benign lesions using CNN features extraction. This section provides an overview of the proposed skin cancer classification system.
Figure 1 illustrates the proposed skin cancer classification system which is designed to train and deploy a smart system capable of differentiate between two different skin cancer malignant and benign using images of the skin moles. The proposed system first based on the extraction of deep CNN features and then describes the classification method employed in the final stage.
2.1. Preprocessing
Data preprocessing includes two essential processing steps that are important to ensure high quality input to the model, enabling effective learning and enhancing performance. Initially, resizing the images to dimension compatible with the input requirements of the CNN model. Secondly, normalization which adjusts pixel values to a common scale, typically using min-max scaling, for prevents attributes with larger ranges from dominating the learning process, reduces data inconsistencies, and enhances model performance through faster convergence during feature extraction. By employing these preprocessing steps, features extraction convergence is improved, effectively ensuring that all features contribute proportionally regardless of their original scales [
26,
27].
2.2. Feature Extraction Using CNNs
Deep learning has revolutionized the field of computer vision, especially in the area of features extraction from images a critical step in various applications, including skin cancer classification. In this study, we employ three CNN models to extract features crucial for the accurate classification of skin cancer. The selected architectures are: ResNet18, Darknet19, and MobileNetv2 were chosen based on their proven efficacy in the literature and their ability to achieve high accuracy across diverse datasets.
2.2.1. ResNet18
ResNet18 [
28] is a Deep CNN (DCNN) architecture known for introducing the concept of residual learning, a technique that substantially simplifies the training of very deep networks. Comprising 18 layers, ResNet18 utilizes residual connections, also known as skip connections, to bypass one or more layers. This architecture mitigates the vanishing gradient problem, enabling for the training of deeper networks without performance degradation.
2.2.2. Darknet19
Darknet19 [
29] is a lightweight and streamlined yet powerful CNN architecture initially developed as the backbone for the YOLO (You Only Look Once) object detection system. It consists of 19 convolutional layers and 5 maxpooling layers, optimized for real-time object detection tasks. Darknet19 architecture emphasizes both speed and efficiency while maintaining high levels of accuracy.
2.2.3. MobileNetv2
MobileNetv2 [
30] is a highly efficient CNN architecture specifically designed for mobile and embedded devices, emphasizing computational efficiency. It introduces the concepts of inverted residuals and linear bottlenecks, which significantly reduce computational costs and memory usage without compromising accuracy.
2.3. Classification
In the final stage of our system, following the features extraction using CNN models, we employ the Random Forest (RF) classifier [
31], a well-established and highly effective ML algorithm. RF is an ensemble learning method capable of performing both regression and classification tasks. It functions by constructing a multiple decision trees during the training process and then aggregating their outputs to determine the final classification. The key advantage of RF is its ability to improve prediction accuracy and mitigate overfitting by leveraging the combined results of multiple decision trees. This ensemble approach enhances the model’s robustness and reliability, making it a suitable choice for our classification task.
3. Experimentation and Results
3.1. Dataset Description
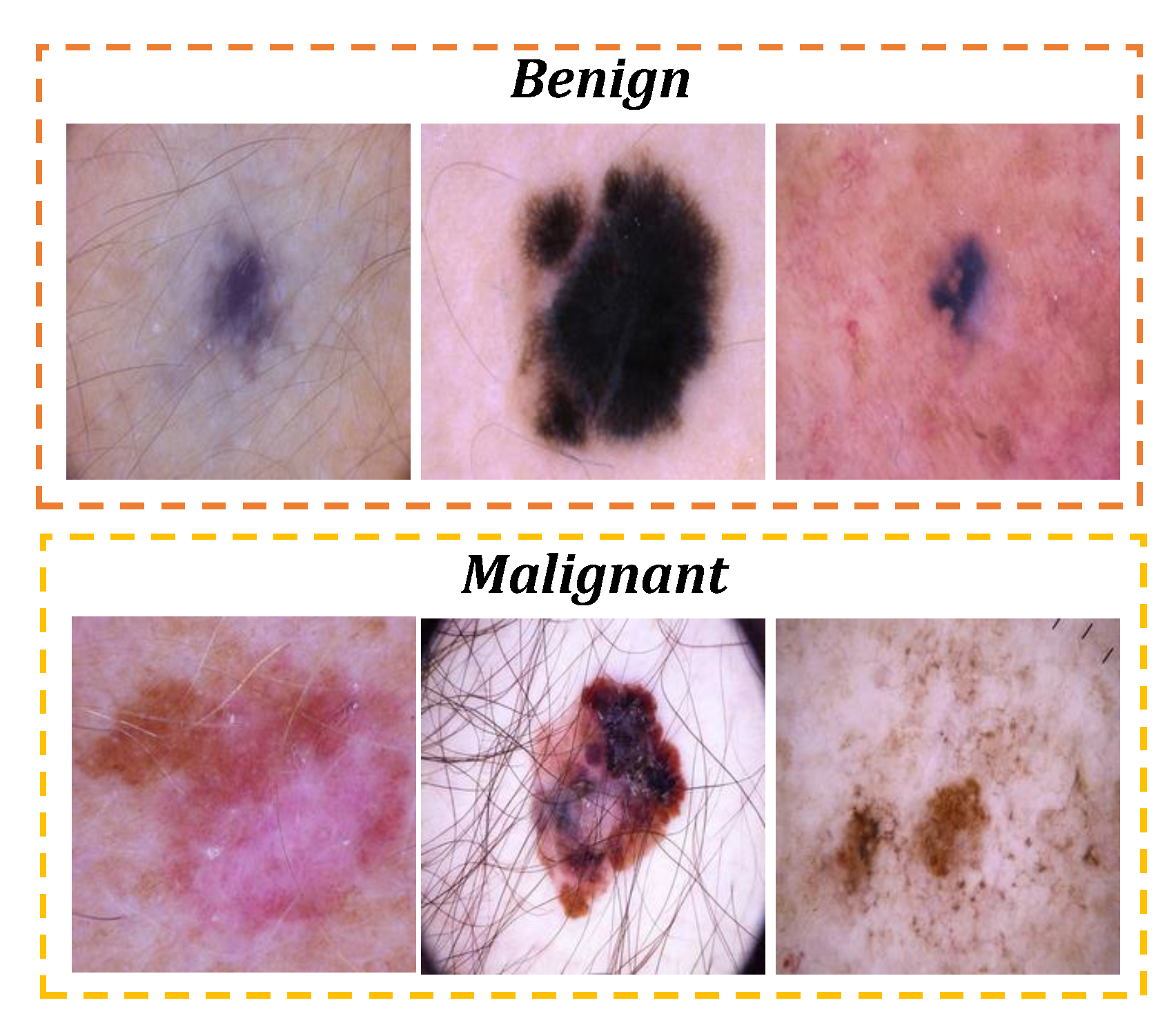
This study leverages the "Skin Cancer: Malignant vs. Benign" dataset available at [
32], comprising 3,297 high resolution images (224x224 pixels) of skin lesions. The dataset includes 1,497 images of malignant and 1,800 images of benign lesions, which are divided into two subset, training set contains 2,637 images and a testing set encopasse 660 images. The training set is employed for model development, while the testing set is used to evaluate the model’s performance. Representative examples of these lesions are shown in
Figure 2. The dataset is governed by the ISIC-Archive rights.
3.2. Evaluation Metrics
The performance of the employed skin cancer classification system is evaluated using five critical evaluation metrics: Accuracy (
), Precision (
), Sensitivity (
), Specificity (
), and F1-score (
). These metrics are defined as follows:
Where represents True Positives, denotes True Negatives, stands for False Positives, and indicates False Negatives.
3.3. Parameter Settings
In this study, we employ three pre-trained CNN models: ResNet18, Darknet19, and MobileNetv2. The input image dimensions are configured as 224x224x3 for ResNet18 and MobileNetv2, and 256x256x3 for Darknet19. Features extraction is performed using the penultimate layers of each model model specifically, the ’conv19’ layer for Darknet19, the ’Logits’ layer for MobileNetv2, and the ’fc1000’ layer for ResNet18, resulting in features vectors of size (1 × 1000). For the RF classifier, the model was configured with 50 decision trees.
3.4. Results and Discussion
Table 1 presents the performance metrics of the proposed model, which utilizes various pre-trained CNN architectures for feature extraction in conjunction with a Random Forest (RF) classifier. To identify the most effective feature extraction method, we systematically evaluated multiple pre-trained models.
Our experiments demonstrate that the ResNet18 model, paired with the RF classifier, achieved the highest accuracy of 82.85%. In comparison, the Darknet19 and MobileNetv2 models, both integrated with the RF classifier, attained accuracies of 81.18% and 82.39%, respectively. These findings highlight the substantial impact of the selected pre-trained model architecture on the overall performance of our skin cancer classification system. The superior performance of the ResNet18-based model across all metrics underscores the critical importance of model architecture in optimizing accuracy and reliability.
Convolutional Neural Networks (CNNs) have revolutionized computer vision by enabling the automatic learning and extraction of hierarchical features from raw data. These networks capture both low-level details, such as edges and textures, and high-level abstractions, like shapes and objects, enhancing their generalization across diverse datasets and tasks. This capability is particularly advantageous in complex classification challenges, such as skin cancer detection. In this study, the features extracted from the penultimate layers of pre-trained CNN models ResNet18, Darknet19, and MobileNetv2 were crucial in achieving high classification accuracy. When combined with the RF classifier, these features effectively distinguished between malignant and benign skin lesions, emphasizing the significance of CNN-derived features in medical image analysis. Furthermore, the adaptability of CNNs to different architectures allows for fine-tuning and transfer learning, further enhancing their utility across various applications. The results of our experiments underscore the critical role of CNN model features in driving classification performance, reinforcing the importance of leveraging these powerful representations in the design of advanced image-based diagnostic tools.
3.5. Comparison with Recent Works
The performance of our proposed CNN-RF model is comprehensively compared with recent related studies in Table 2. This comparison underscores the enhanced effectiveness and reliability of our approach, particularly emphasizing its accuracy and methodological strengths. Notably, our CNN-RF model, built on ResNet18, achieved an accuracy of 82.85%, representing a substantial improvement over several existing methods in the field.
In previous studies, Mijwil et al. reached an accuracy of 81.64% through a combination of ResNet50 and DenseNet, while Ismail et al. reported an accuracy of 75.31% using a standalone ResNet model. Although these approaches demonstrated effectiveness, our proposed CNN-RF model surpassed them, underscoring its significant potential for robust and reliable skin cancer classification. This enhancement in accuracy and model efficiency suggests that the CNN-RF model could serve as a valuable tool in advancing early and accurate detection in medical diagnostics, potentially informing clinical decision-making with greater precision.
This comparison underscores the competitiveness of our model, particularly given its simpler architecture compared to other methods. The results indicate that our CNN-RF approach offers a promising balance between accuracy and model complexity in the context of skin cancer classification.
4. Conclusion and Future Works
In this paper, we have explored the application of a CNN based features extraction combined with RF classifier for skin cancer detection, utilizing the "Skin Cancer: Malignant vs. Benign" dataset. Our objective was to develop a robust system capable of accurately classifying the skin cancer types. By leveraging the power of the CNNs pre-trained model, we effectively utilized the spatial features extracted from data. Through extensive experimentation, we identified the most accurate model, which achieved an accuracy of 82.85%, a precision of 82.69%, sensitivity of 82.86%, a specificity of 82.86%, and a F1-Score of 82.75%. These results underscore the effectiveness of the proposed approach in distinguishing between malignant and benign skin lesions. Looking ahead, promising avenues for future research include the integration of additional skin cancer classes, which could further enhance the robustness and versatility of the detection system. Additionally, the implementation of federated learning could bolster the system’s security, enabling the development of a more secure and decentralized approach to skin cancer detection.
References
- A. Adegun and S. Viriri, “Deep learning techniques for skin lesion analysis and melanoma cancer detection: a survey of state-of-the-art,” Artificial Intelligence Review, vol. 54, no. 2, pp. 811–841, 2021. [CrossRef]
- Y. Wu, B. Chen, A. Zeng, D. Pan, R. Wang, and S. Zhao, “Skin cancer classification with deep learning: a systematic review,” Frontiers in Oncology, vol. 12, p. 893972, 2022. [CrossRef]
- R. L. Siegel, K. D. Miller, H. E. Fuchs, and A. Jemal, “Cancer statistics, 2022.” CA: a cancer journal for clinicians, vol. 72, no. 1, 2022.
- H. Sung, J. Ferlay, R. L. Siegel, M. Laversanne, I. Soerjomataram, A. Jemal, and F. Bray, “Global cancer statistics 2020: Globocan estimates of incidence and mortality worldwide for 36 cancers in 185 countries,” CA: a cancer journal for clinicians, vol. 71, no. 3, pp. 209–249, 2021. [CrossRef]
- P. Tschandl, C. Rosendahl, B. N. Akay, G. Argenziano, A. Blum, R. P. Braun, H. Cabo, J.-Y. Gourhant, J. Kreusch, A. Lallas et al., “Expert-level diagnosis of nonpigmented skin cancer by combined convolutional neural networks,” JAMA dermatology, vol. 155, no. 1, pp. 58–65, 2019. [CrossRef]
- A. Murugan, S. A. H. Nair, and K. S. Kumar, “Detection of skin cancer using svm, random forest and knn classifiers,” Journal of medical systems, vol. 43, no. 8, p. 269, 2019.
- M. Patil and N. Dongre, “Melanoma detection using hsv with svm classifier and de-duplication technique to increase efficiency,” in Computing Science, Communication and Security: First International Conference, COMS2 2020, Gujarat, India, 26 March–27, 2020, Revised Selected Papers 1. Springer, 2020, pp. 109–120.
- K. Melbin and Y. J. V. Raj, “Integration of modified abcd features and support vector machine for skin lesion types classification,” Multimedia Tools and Applications, vol. 80, no. 6, pp. 8909–8929, 2021. [CrossRef]
- F. Perez, S. Avila, and E. Valle, “Solo or ensemble? choosing a cnn architecture for melanoma classification,” in Proceedings of the IEEE/CVF conference on computer vision and pattern recognition workshops, 2019, pp. 0–0.
- I. Mporas, I. Perikos, and M. Paraskevas, “Color models for skin lesion classification from dermatoscopic images,” in Advances in Integrations of Intelligent Methods: Post-workshop volume of the 8th International Workshop CIMA 2018, Volos, Greece, November 2018 (in conjunction with IEEE ICTAI 2018). Springer, 2020, pp. 85–98.
- K. M. Hosny, M. A. Kassem, and M. M. Foaud, “Skin cancer classification using deep learning and transfer learning,” in 2018 9th Cairo international biomedical engineering conference (CIBEC). IEEE, 2018, pp. 90–93.
- V. Anand, S. Gupta, and D. Koundal, “Skin disease diagnosis: challenges and opportunities,” in Proceedings of Second Doctoral Symposium on Computational Intelligence: DoSCI 2021. Springer, 2022, pp. 449–459.
- B. Zhang, X. Zhou, Y. Luo, H. Zhang, H. Yang, J. Ma, and L. Ma, “Opportunities and challenges: Classification of skin disease based on deep learning,” Chinese Journal of Mechanical Engineering, vol. 34, pp. 1–14, 2021. [CrossRef]
- M. Goyal, T. Knackstedt, S. Yan, and S. Hassanpour, “Artificial intelligence-based image classification methods for diagnosis of skin cancer: Challenges and opportunities,” Computers in biology and medicine, vol. 127, p. 104065, 2020. [CrossRef]
- M. R. A. Berkani, A. Chouchane, Y. Himeur, A. Ouamane, and A. Amira, “An intelligent edge-deployable indoor air quality monitoring and activity recognition approach,” in 2023 6th International Conference on Signal Processing and Information Security (ICSPIS). IEEE, 2023, pp. 184–189.
- Y. Habchi, Y. Himeur, H. Kheddar, A. Boukabou, S. Atalla, A. Chouchane, A. Ouamane, and W. Mansoor, “Ai in thyroid cancer diagnosis: Techniques, trends, and future directions,” Systems, vol. 11, no. 10, p. 519, 2023.
- M. Bessaoudi, A. Chouchane, A. Ouamane, and E. Boutellaa, “Multilinear subspace learning using handcrafted and deep features for face kinship verification in the wild,” Applied Intelligence, vol. 51, pp. 3534–3547, 2021. [CrossRef]
- A. A. Gharbi, A. Chouchane, A. Ouamane, Y. Himeur, S. Bourennane et al., “A hybrid multilinear-linear subspace learning approach for enhanced person re-identification in camera networks,” Expert Systems with Applications, vol. 257, p. 125044, 2024. [CrossRef]
- A. Ouamane, A. Chouchane, Y. Himeur, A. Debilou, S. Nadji, N. Boubakeur, and A. Amira, “Enhancing plant disease detection: a novel cnn-based approach with tensor subspace learning and howsvd-mda,” Neural Computing and Applications, pp. 1–25, 2024. [CrossRef]
- M. Khammari, A. Chouchane, A. Ouamane, M. Bessaoudi et al., “Kinship verification using multiscale retinex preprocessing and integrated 2dswt-cnn features,” in 2024 8th International Conference on Image and Signal Processing and their Applications (ISPA). IEEE, 2024, pp. 1–8.
- A. A. Gharbi, A. Chouchane, A. Ouamane, and M. Bessaoudi, “Advancing person re-identification: Tensor-based feature fusion and multilinear subspace learning,” in 2024 8th International Conference on Image and Signal Processing and their Applications (ISPA). IEEE, 2024, pp. 1–7.
- M. Khammari, A. Chouchane, M. Bessaoudi, A. Ouamane, Y. Himeur, S. Alallal, W. Mansoor et al., “Enhancing kinship verification through multiscale retinex and combined deep-shallow features,” in 2023 6th International Conference on Signal Processing and Information Security (ICSPIS). IEEE, 2023, pp. 178–183.
- B. Bengherbia, M. R. A. Berkani, Z. Achir, A. Tobbal, M. Rebiai, and M. Maazouz, “Real-time smart system for ecg monitoring using a one-dimensional convolutional neural network,” in 2022 International Conference on Theoretical and Applied Computer Science and Engineering (ICTASCE). IEEE, 2022, pp. 32–37.
- M. Khammari, A. Chouchane, A. Ouamane, M. Bessaoudi, Y. Himeur, M. Hassaballah et al., “High-order knowledge-based discriminant features for kinship verification,” Pattern Recognition Letters, vol. 175, pp. 30–37, 2023. [CrossRef]
- A. Chouchane, A. Ouamanea, Y. Himeur, A. Amira et al., “Deep learning-based leaf image analysis for tomato plant disease detection and classification,” in 2024 IEEE International Conference on Image Processing (ICIP). IEEE, 2024, pp. 2923–2929.
- I. Manole, A.-I. Butacu, R. N. Bejan, and G.-S. Tiplica, “Enhancing dermatological diagnostics with efficientnet: A deep learning approach,” Bioengineering, vol. 11, no. 8, p. 810, 2024. [CrossRef]
- M. Rebiai, M. R. A. Berkani, M. Y. Bentegri, and M. T. Seghir, “Mathematical morphology-based envelope detection for cardiac disease classification from phonocardiogram signals,” in 2024 8th International Conference on Image and Signal Processing and their Applications (ISPA). IEEE, 2024, pp. 1–5.
- K. He, X. Zhang, S. Ren, and J. Sun, “Deep residual learning for image recognition,” in Proceedings of the IEEE conference on computer vision and pattern recognition, 2016, pp. 770–778.
- J. Redmon and A. Farhadi, “Yolo9000: better, faster, stronger,” in Proceedings of the IEEE conference on computer vision and pattern recognition, 2017, pp. 7263–7271.
- M. Sandler, A. Howard, M. Zhu, A. Zhmoginov, and L.-C. Chen, “Mobilenetv2: Inverted residuals and linear bottlenecks,” in Proceedings of the IEEE conference on computer vision and pattern recognition, 2018, pp. 4510–4520.
- L. Breiman, “Random forests,” Machine learning, vol. 45, pp. 5–32, 2001.
- C. Fanconi, “Skin cancer: Malignant vs. benign,” https://www.kaggle.com/fanconic/skin-cancer-malignant-vs-benign, Available: [Access date], processed skin cancer pictures of the ISIC Archive Kaggle dataset.
- M. M. Mijwil, “Skin cancer disease images classification using deep learning solutions,” Multimedia Tools and Applications, vol. 80, no. 17, pp. 26 255–26 271, 2021. [CrossRef]
- M. A. Ismail, N. Hameed, and J. Clos, “Deep learning-based algorithm for skin cancer classification,” in Proceedings of International Conference on Trends in Computational and Cognitive Engineering: Proceedings of TCCE 2020. Springer, 2021, pp. 709–719.
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).