Submitted:
05 November 2024
Posted:
06 November 2024
You are already at the latest version
Abstract
The pursuit of understanding the brain’s complex mechanisms is at the forefront of neuroscience, especially in developing early diagnostic tools for neurological diseases. Advances in neural fields, brain dynamics, and the integration of applied physics have opened new possibilities for identifying biomarkers indicative of neurological conditions such as Alzheimer’s disease, schizophrenia, and ADHD. Each of these fields contributes uniquely: neural fields offer a spatially distributed model of brain activity, brain dynamics provide insights into temporal patterns and oscillations, and applied surface physics enhances the precision of neural interface technologies and imaging. Combined, they form a multidisciplinary foundation that allows researchers to study the brain’s intricate structures and functions with unprecedented clarity, revealing potential pathways for diagnosing and treating neurological disorders.
Keywords:
1. Introduction
1.1. Neural Fields: Mapping Brain Activity in Space
1.2. Mathematical Models in Neural Field Theory
1.3. Brain Dynamics: Temporal Patterns and Oscillations
1.3.1. Oscillatory Activity as a Diagnostic Tool
1.3.2. Noise and Stochasticity in Brain Dynamics
1.4. Neurological Disease Biomarkers: From Theory to Clinical Application
1.4.1. EEG Biomarkers in Neurological Disorders
1.4.2. Biomarkers in Alzheimer’s Disease
1.5. Applied Surface Physics in Neurology
1.5.1. Enhancing Neural Data Accuracy with Surface Physics
1.6. Topological Data Analysis (TDA) in Brain Mapping
1.7.1. Conclusion
2. Methodology
2.1. Neural Field Theory
- : neural activity at position and time ,
- : synaptic weight or connectivity function describing the strength of interaction between neurons at and ,
- : nonlinear activation function, often chosen as a sigmoid or threshold function to model neural firing responses,
- : external input to the network, which could represent sensory input or stimulation.
2.1.1. Connectivity Function
- : amplitude parameters for excitation and inhibition,
- : spatial scales for excitation and inhibition, respectively.
2.2. Stochastic Dynamics for Brain Oscillations
- : amplitude of the noise, which can vary spatially and temporally,
- : Wiener process (or white noise), representing random fluctuations in neural activity.
2.2.1. Mean Field Analysis of Oscillatory Activity
2.3. Signal Processing and Surface Physics for Neural Interfaces
2.3.1. Laplace Surface Filtering for EEG Signals
- : potential at the primary electrode location,
- : potentials at neighbouring electrodes surrounding .
2.4. Topological Data Analysis (TDA) on Surface Maps
2.5. Model Validation and Biomarker Extraction
2.6. Electrodynamics and Maxwell's Equations in Neural Modelling
2.7. Quasi-Static Approximation for EEG Signal Modelling
2.8. Cross-Correlation for Synchrony Detection
Conclusion
2.9. Computational Methods
-
Neural Field Dynamics:
- o
- A Gaussian connectivity function models localized neural interactions, promoting nearby excitation and distant inhibition.
- o
- External input III is added at specific locations to simulate stimulus.
- o
- Neural field dynamics are updated iteratively, including a tanh nonlinearity to simulate neuron firing and Gaussian noise to model stochastic effects.
-
EEG Signal Calculation:
- o
- EEG signals are approximated by summing local neural activity around specific electrode positions, capturing nearby electric potentials in the field.
-
Visualization:
- o
- Neural Field Activity Over Time: A heatmap showing the spatial-temporal evolution of neural activity.
- o
- Simulated EEG Signals: EEG signals at each electrode position over time.
- o
- Power Spectral Density (PSD): Frequency-domain representation of EEG signals, showing power distribution across different frequencies.
3. Results and Discusion
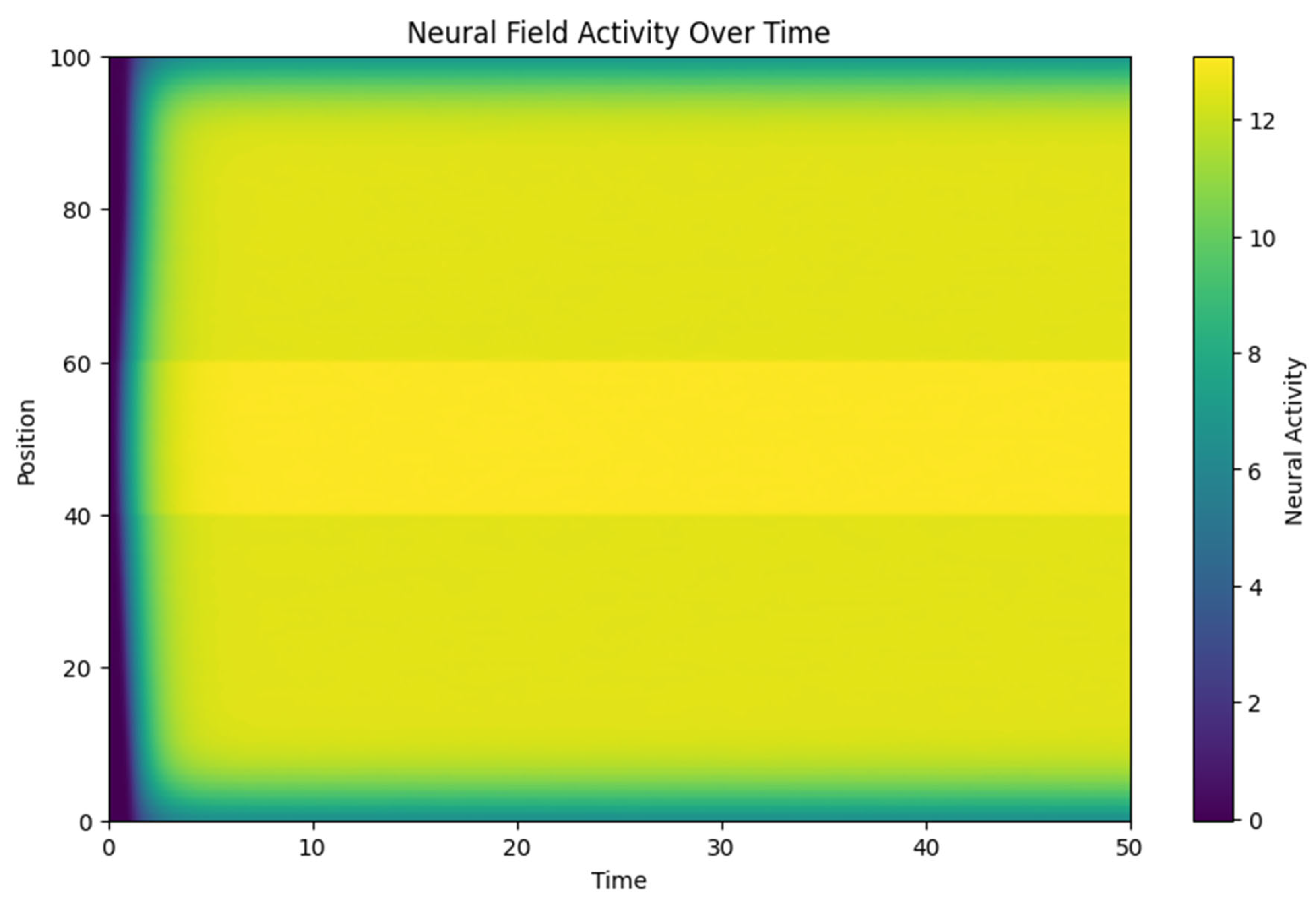
3.1. Neural Activity Propagation:
- o The heatmap of neural field activity illustrates how an initial external stimulus (applied between positions 40 and 60) propagates through the field. The spatial spread is influenced by the connectivity function w(x,y), with neighboring regions showing synchronized activity.
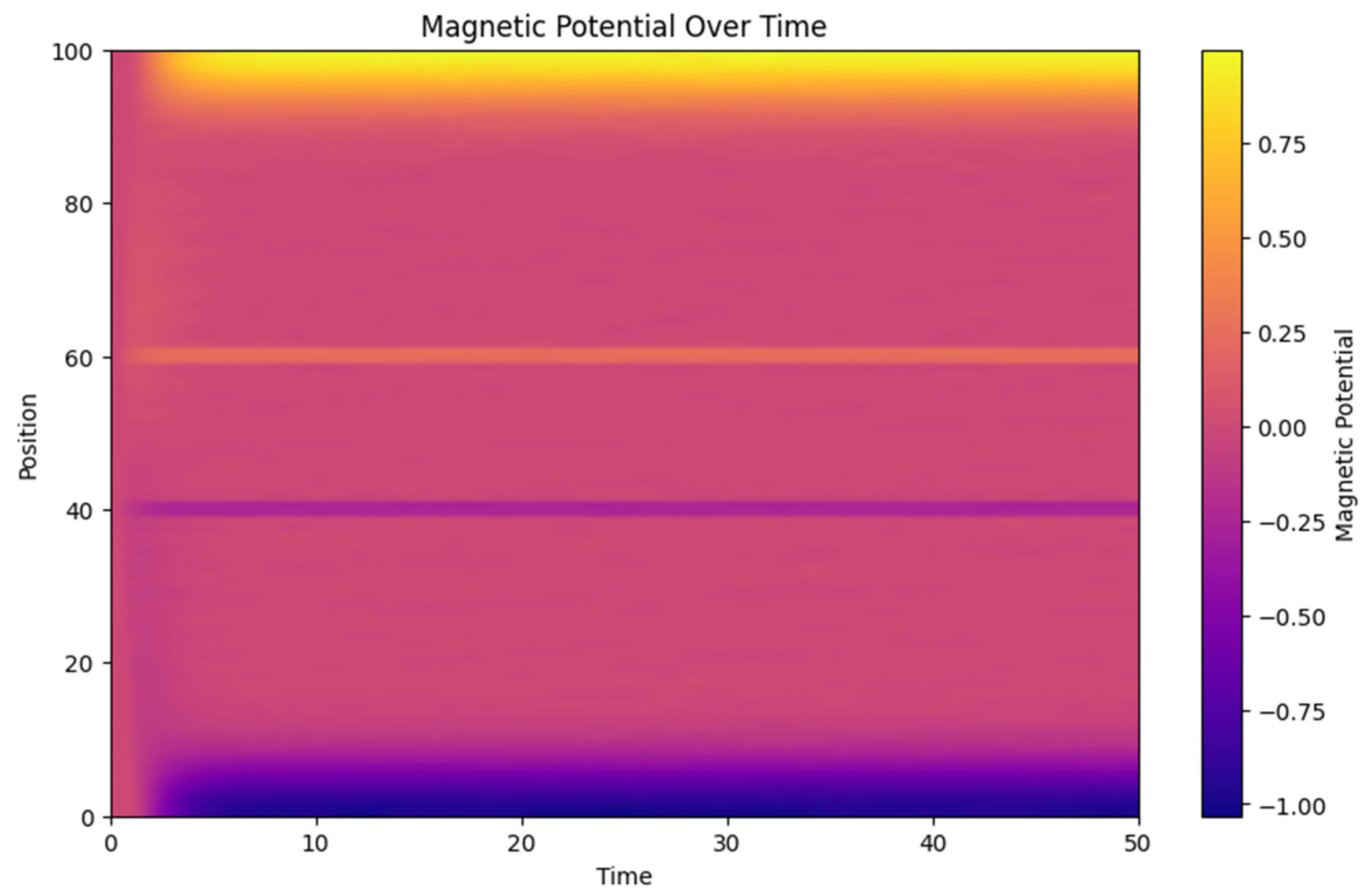
3.2. Magnetic Potential Influence
- o The magnetic potential heatmap (Figure 2) shows a clear interaction between neural activity and the generated magnetic fields. The magnetic field acts as feedback, altering the neural activity in subsequent time steps, which reflects the bidirectional coupling between electrical and magnetic components in the model.
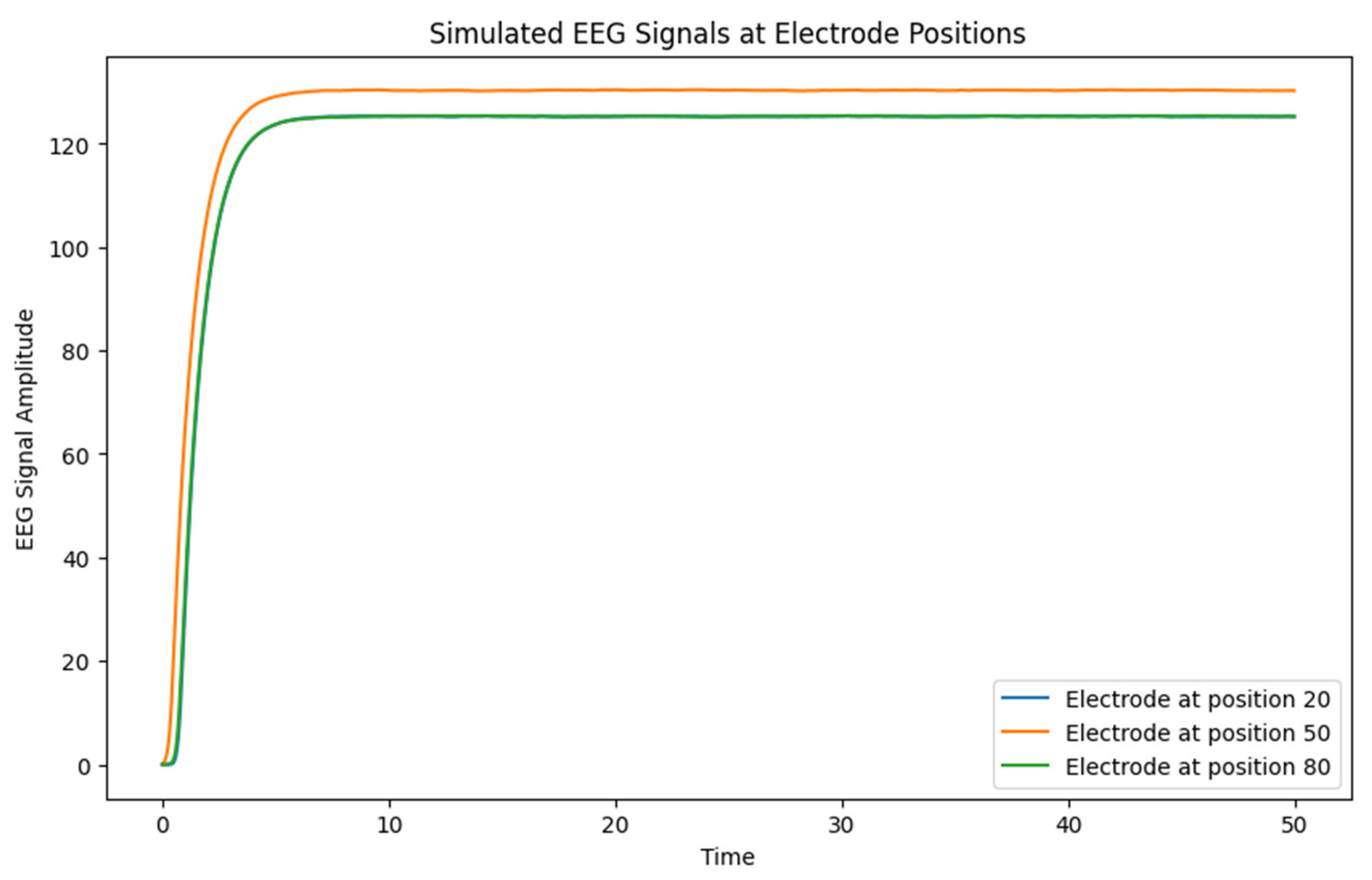
3.3. EEG Signals
- o The simulated EEG signals reflect the local neural dynamics around each electrode. Differences between signals at different electrode positions highlight how spatial variations in connectivity and external inputs lead to different patterns of neural activity.
3.4. Frequency Analysis (PSD):
- o The PSD show the frequency components present in the EEG signals. Peaks at certain frequencies indicate rhythmic neural activity, which could be indicative of underlying brain states such as relaxation (alpha waves) or cognitive engagement (beta waves).
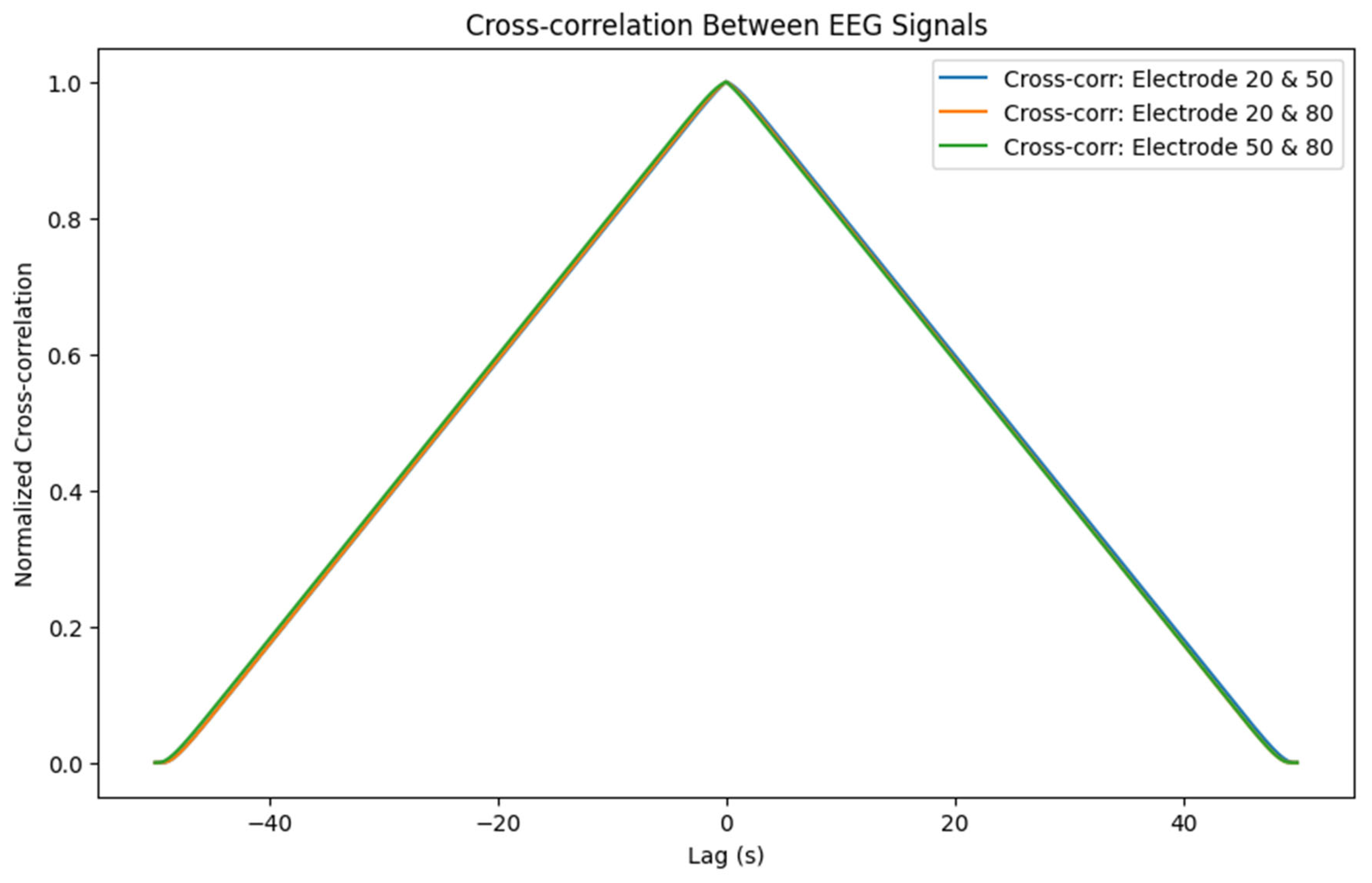
3.4.1. Functional Connectivity (Cross-Correlation)
- o Cross-correlation between EEG signals provides insights into the temporal relationships between different parts of the neural field. High correlations indicate synchronized activity, potentially pointing to functional connectivity. Synchronization between electrodes might signify coordinated neural processes or pathological synchrony (as seen in epileptic seizures).
Conclusion
3.5. Implications for Biomarker Identification
- Amplitude of Neural Activity: The level of activity in response to stimuli may indicate neural tissue excitability, a characteristic associated with epilepsy and other hyper-excitability conditions (Crunelli & Leresche, 2002).
- Oscillatory Patterns: Distinct frequency components of EEG signals, as shown in PSD plots, can help identify specific cognitive or pathological states. For instance, reduced beta and elevated theta power often correlate with attentional deficits, suggesting potential biomarkers for ADHD (Barry et al., 2003).
- Functional Connectivity: Cross-correlation provides insights into synchronisation patterns, which are altered in various neurological and psychiatric disorders. For example, functional disconnection in schizophrenia is marked by decreased synchronisation in gamma oscillations (Uhlhaas & Singer, 2010).
Limitations and Future Directions
4. Conclusion
References
- Amari, S. Dynamics of pattern formation in lateral-inhibition type neural fields. Biological Cybernetics 1977, 27, 77–87. [Google Scholar] [CrossRef] [PubMed]
- Barry, R.J.; Clarke, A.R.; McCarthy, R. ; Selikowitz MEEG differences in children with attention-deficit/hyperactivity disorder inattentive type without, A. D.H.D. Psychiatry Research 2003, 93, 141–153. [Google Scholar]
- Breakspear, M. Dynamic models of large-scale brain activity. Nature Neuroscience 2017, 20, 340–352. [Google Scholar] [CrossRef] [PubMed]
- Bullmore, E.; Sporns, O. Complex brain networks: graph theoretical analysis of structural and functional systems. Nature Reviews Neuroscience 2009, 10, 186–198. [Google Scholar] [CrossRef] [PubMed]
- Buzsáki, G. (2006). Rhythms of the Brain. Oxford University Press.
- Buzsáki, G.; Draguhn, A. Neuronal oscillations in cortical networks. Science 2004, 304, 1926–1929. [Google Scholar] [CrossRef]
- Coombes, S. Waves, bumps, and patterns in neural field theories. Biological Cybernetics 2005, 93, 91–108. [Google Scholar] [CrossRef]
- Crunelli, V.; Leresche, N. Childhood absence epilepsy: genes, channels, neurons, and networks. Nature Reviews Neuroscience 2002, 3, 371–382. [Google Scholar] [CrossRef]
- Deco, G.; Jirsa, V.K.; McIntosh, A.R. Emerging concepts for the dynamical organization of resting-state activity in the brain. Nature Reviews Neuroscience 2011, 12, 43–56. [Google Scholar] [CrossRef]
- Engel, A.K.; Singer, W. Temporal binding and the neural correlates of sensory awareness. Trends in Cognitive Sciences 2001, 5, 16–25. [Google Scholar] [CrossRef]
- Faisal, A.A.; Selen, L.P.; Wolpert, D.M. Noise in the nervous system. Nature Reviews Neuroscience 2008, 9, 292–303. [Google Scholar] [CrossRef]
- Fries, P. A mechanism for cognitive dynamics: neuronal communication through neuronal coherence. Trends in Cognitive Sciences 2005, 9, 474–480. [Google Scholar] [CrossRef] [PubMed]
- Froemke, R.C.; Schreiner, C.E. Synaptic plasticity in auditory cortical neurons is constrained by the natural sound statistics. Nature Neuroscience 2015, 18, 383–392. [Google Scholar]
- Hebb, D.O. (1949). The Organization of Behavior: A Neuropsychological Theory. Wiley.
- Jack, C.R. , et al. Hypothetical model of dynamic biomarkers of the Alzheimer's pathological cascade. The Lancet Neurology 2010, 9, 119–128. [Google Scholar]
- Jirsa, V.K.; Stacey, W.C.; Quilichini, P.P.; Ivanov, E.N.; Bernard, C. On the nature of seizure dynamics. Brain 2014, 137, 2210–2230. [Google Scholar] [CrossRef] [PubMed]
- Jiruska, P.; de Curtis, M.; Jefferys, J.G.; Schevon, C.A.; Schiff, S.J.; Schindler, K. Synchronization and desynchronization in epilepsy: controversies and hypotheses. The Journal of Physiology 2013, 591, 787–797. [Google Scholar] [CrossRef]
- Kassanos, P.; Kosmas, P.; Nader, C. Nanotechnology-enabled neural interfaces for high-resolution mapping of brain activity. Nano Research 2016, 9, 1778–1796. [Google Scholar]
- Klimesch, W. EEG alpha and theta oscillations reflect cognitive and memory performance: a review and analysis. Brain Research Reviews 1999, 29(2-3), 169-195.
- Maxwell, J.C. A dynamical theory of the electromagnetic field. Philosophical Transactions of the Royal Society of London 1865, 155, 459–512. [Google Scholar]
- Montgomery, R M. , Evaluating EEG-Based Parameters for Bipolar Disorder Diagnosis Using a Synthetic Dataset. Int Internal Med J 2024, 2, 01–08.
- Montgomery, R M. Topological Dynamics in Ecological Biomes and Toroidal Structures: Mathematical Models of Stability, Bifurcation, and Structural Failure. J Gene Engg Bio Res 2024, 6, 1–9.
- Montgomery, R.M. Exploring Altered Neural Connectivity in Autism Spectrum Disorder: A Simulation of Spatial and Topological Proximities. Int J Psychiatry 2024, 9, 01–08. [Google Scholar] [CrossRef]
- Montgomery, R.M. (2023). Possible Influence of Cortical Neuronal Circuits on Each Other: A Model. 10.20944/preprints202405.1224.
- Montgomery, R.M. ; (2024)a. The Intersection of ADHD and Autism: Neurodevelopmental Disorders with Shared Origins and Distinct Trajectories. Preprints. [CrossRef]
- Murthy, V.N.; Fetz, E.E. Coherent 25- to 35-Hz oscillations in the sensorimotor cortex of awake behaving monkeys. Proceedings of the National Academy of Sciences 1992, 89, 5670–5674. [Google Scholar] [CrossRef] [PubMed]
- Niedermeyer, E. , & da Silva, F.L. (2004). Electroencephalography: Basic Principles, Clinical Applications, and Related Fields. Lippincott Williams & Wilkins.
- Nunez, P.L. ; Srinivasan R (2006) Electric Fields of the Brain: The Neurophysics of, E.E.G. Oxford University Press.
- Pietrobon, D.; Moskowitz, M.A. Pathophysiology of migraine. Annual Review of Physiology 2014, 75, 365–391. [Google Scholar] [CrossRef] [PubMed]
- Plonsey, R.; Heppner, D.B. Considerations of quasi-stationarity in electrophysiological systems. The Bulletin of Mathematical Biophysics 1967, 29, 657–664. [Google Scholar] [CrossRef] [PubMed]
- Uhlhaas, P.J.; Singer, W. Abnormal neural oscillations and synchrony in schizophrenia. Nature Reviews Neuroscience 2010, 11, 100–113. [Google Scholar] [CrossRef] [PubMed]
- Wilson, H.R.; Cowan, J.D. Excitatory and inhibitory interactions in localized populations of model neurons. Biophysical Journal 1972, 12, 1–24. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




