Submitted:
04 December 2024
Posted:
06 December 2024
You are already at the latest version
Abstract
Keywords:
Graphical Abstract

1. Introduction
2. Results and Discussion
2.1. The Performance of the Extraction Systems Tested
2.2. Bioactivities of the E. punicifolia Leaf Extracts
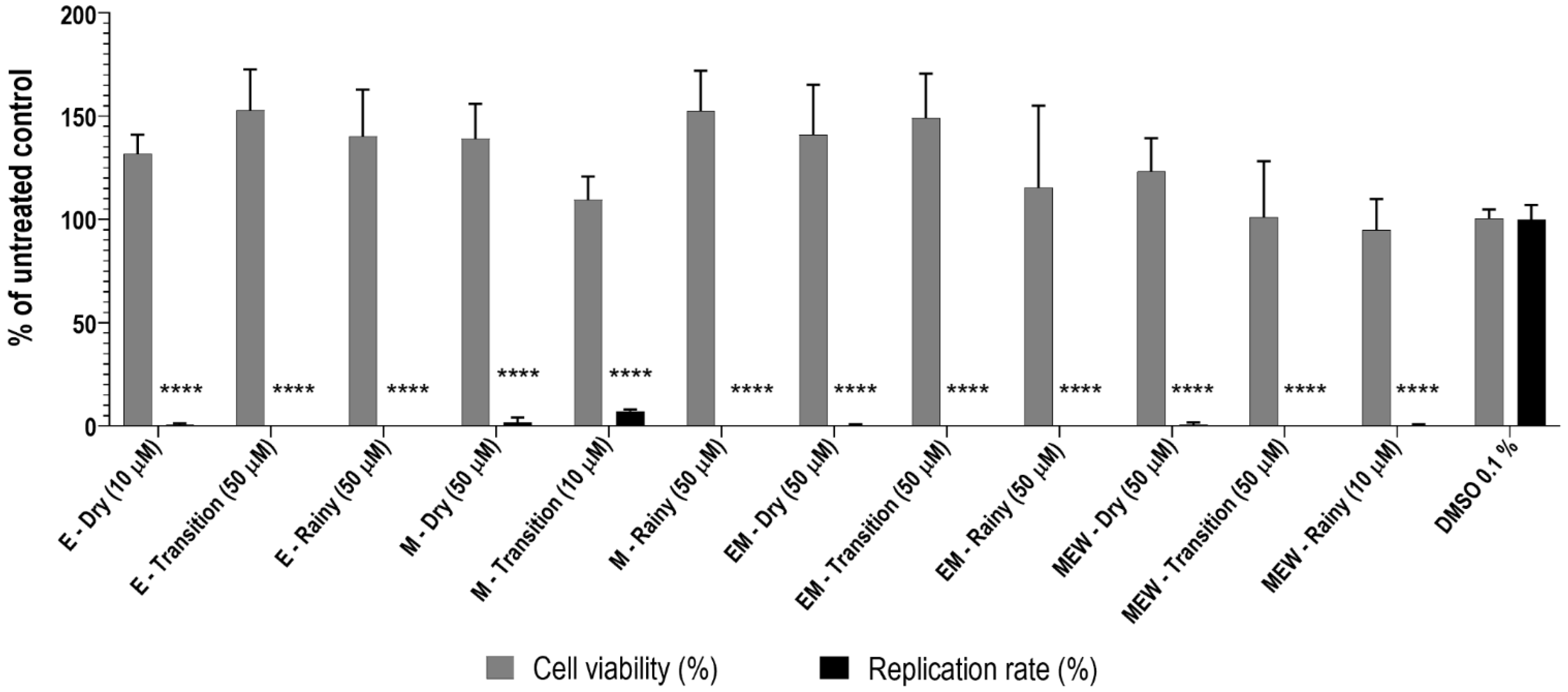
2.2.1. Cytotoxicity of Eugenia punicifolia Leaf Extracts
2.2.2. Anti-ZIKV Activity
2.2.3. Antioxidant Activity via DPPH and ABTS Assays
2.2.4. Antiglycation Activity: Non-Oxidative Pathway
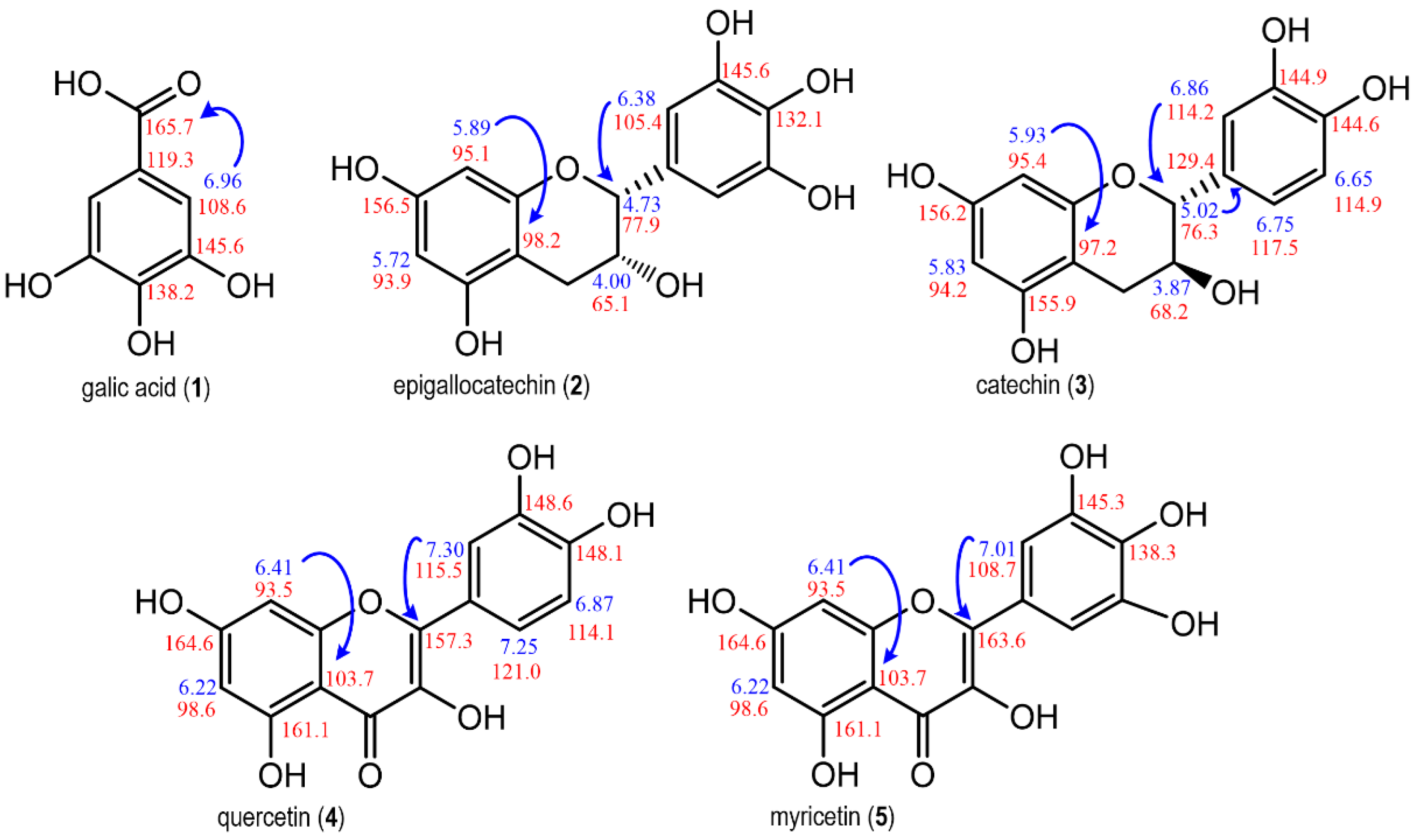
2.3. Identification of Phenolic Compounds in E. punicifolia Leaf Extracts
2.4. qNMR of Phenolic Compounds by PULCON
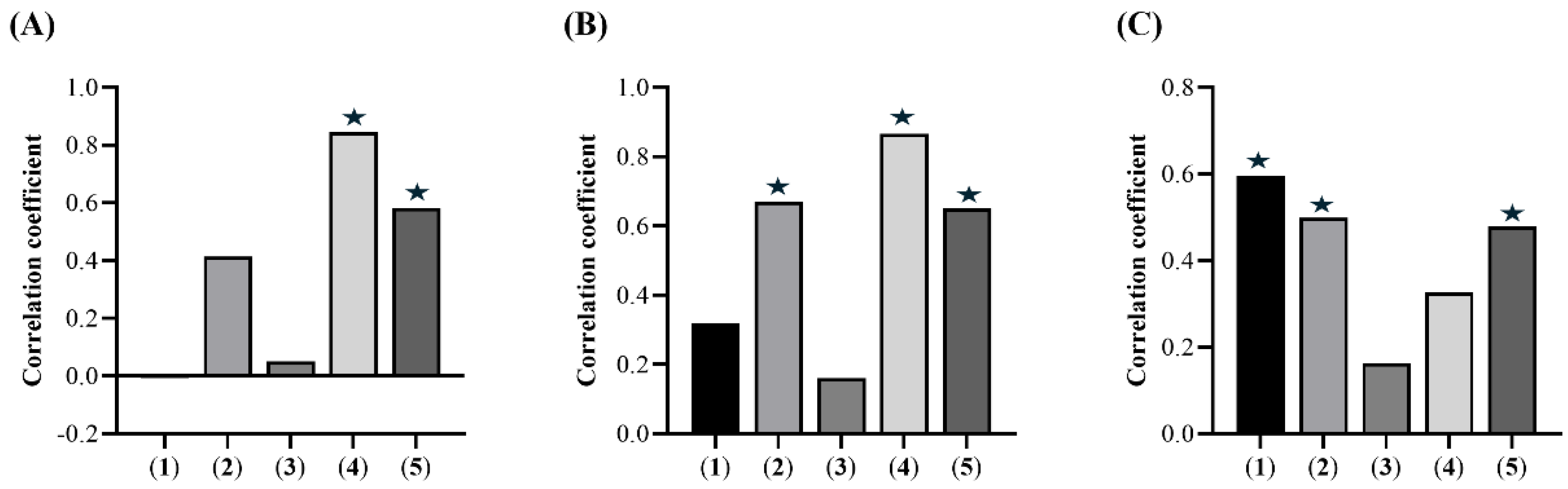
2.5. Chemical Composition and Bioactivities of the E. punicifolia Leaf Extracts: Searching for Correlations
3. Materials and Methods
3.1. Materials
3.2. Plant Material
3.3. Extraction Procedure
3.4. Acquisition and Processing of NMR Data
3.5. Cell Culture
3.6. Cell Viability Assay
3.7. Antiviral Assay - ZIKV
3.8. Determination of Antioxidant Potential
3.8.1. DPPH Radical Scavenging Capacity
3.8.2. ABTS Radical Cation Scavenging Capacity
3.9. Antiglycation Activity: Non-Oxidative Pathway
3.10. Canonical Correlation Analysis (CCA)
4. Conclusions
Supplementary Materials
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Jorge, U.; Aguiar, J.; Silva, M. Anatomia foliar de pedra-hume-caá (Myrcia sphaerocarpa, Myrcia guianensis, Eugenia punicifolia - Myrtaceae). Acta Amaz. 2000, 30, 49-49.
- Oliveira, E. S. C.; Pontes, F. L. D.; Acho, L. D. R.; Silva, B. J. P.; Rosário, A. S.; Chaves, F. C. M.; Campos, F. R.; Bezerra, J. A.; Lima, E. S.; Machado, M. B. NMR and multivariate methods: Identification of chemical markers in extracts of pedra-ume-caá and their antiglycation, antioxidant, and enzymatic inhibition activities. Phytochem. Anal. 2024, 35, 552-566.
- Amorim, M.; Verdan, M.; Oliveira, C.; Santos, A. D. C. Essential Oils of Neotropical Myrtaceae Species from 2011 until 2023: An Update. Chem. Biodivers. 2024, e202401503. [CrossRef]
- Teixeira, R. G. S.; Pascual, R.; Araújo, K. G. L.; Brito, M. A.; Rios, C. L.; Carmo, A. F.; Gandía, L.; Silva, C. L. M.; Machado, T. B.; Santos, W. C. In vitro and in silico studies for barbinervic acid, a triterpene isolated from Eugenia punicifolia that inhibits vasopressor tone. Nat. Prod. Res. 2021, 35, 4870-4875.
- Basting, R. T.; Nishijima, C. M.; Lopes, J. A.; Santos, R. C.; Lucena Périco, L.; Laufer, S.; Bauer, S.; Costa, M. F.; Santos, L. C.; Rocha, L. R. M.; et al. Antinociceptive, anti-inflammatory and gastroprotective effects of a hydroalcoholic extract from the leaves of Eugenia punicifolia (Kunth) DC. in rodents. J. Ethnopharmacol. 2014, 157, 257-267. [CrossRef]
- Sales D. S.; Carmona F.; Azevedo B. C.; Taleb-Contini S. H.; Bartolomeu A. C. D.; Honorato F. B.; Martinez E. Z.; Pereira A. M. S. Eugenia punicifolia (Kunth) DC. as an Adjuvant Treatment for Type-2 Diabetes Mellitus: A non-Controlled, Pilot Study. Phytother. Res. 2014, 28, 1816–1821.
- Ramos, A.; Mar, J.; Silva, L.; Acho, L. D.; Silva, B.; Lima, E.; Campelo, P.; Sanches, E.; Bezerra, A. J.; Chaves, F.; et al. Pedra-ume caá fruit: An Amazon cherry rich in phenolic compounds with antiglycant and antioxidant properties. Food Res. Int. 2019, 123. [CrossRef]
- Souza, A.; Oliveira, C.; Oliveira, V.; Betim, F.; Miguel, M. Traditional Uses, Phytochemistry, and Antimicrobial Activities of Eugenia Species - A Review. Planta Med. 2018, 84.
- Santos, C.; Mizobucchi, A. L.; Escaramboni, B.; Lopes, B. P.; Angolini, C. F. F.; Eberlin, M. N.; de Toledo, K. A.; Núñez, E. G. F. Optimization of Eugenia punicifolia (Kunth) DC. leaf extraction using a simplex centroid design focused on extracting phenolics with antioxidant and antiproliferative activities. BMC Chem. 2020, 14, 34. [CrossRef]
- Zhang, Q. W.; Lin, L. G.; Ye, W. C. Techniques for extraction and isolation of natural products: a comprehensive review. Chin. Med. 2018, 13, 20. [CrossRef]
- Neves, K. O. G.; Santos, M. F. C.; Mar, J. M.; Pontes, F. L. D.; Tormena, C. F.; Chaves, F. C. M.; Campos, F. R.; Sanches, E. A.; Bezerra, J. A.; Machado, M. B.; et al. 1H NMR Chemical Profile and Antioxidant Activity of Eugenia punicifolia Extracts Over Seasons: A Metabolomic Pilot Study. J. Braz. Chem. Soc. 2024, 35, e-20240010. [CrossRef]
- Brunetti, I. L.; Vendramini, R. C.; Januário, A. H.; França, S. C.; Pepato, M. T. Effects and Toxicity of Eugenia punicifolia. Extracts in Streptozotocin-Diabetic Rats. Pharm. Biol. 2006, 44, 35.
- Dogara, A. M.; Ibrahim, M. T.; Mahmud, A. A.; Danladi, M. D.; Lema, A. A.; Usman, M.; Tahir, A. S.; Tabti, K. Biological activity, chemical composition, and molecular docking of Eugenia punicifolia (Kunth) DC. J.Umm Al-Qura Univ. Appll. Sci. 2024. [CrossRef]
- Sehlakgwe P. F.; Lall N.; Prinsloo G. 1H-NMR Metabolomics and LC-MS Analysis to Determine Seasonal Variation in a Cosmeceutical Plant Leucosidea sericea. Front. Pharmacol. 2020, 11, 219.
- Ravaglia, L. M.; Oliveira, P. D.; Holzgrabe, U.; Alcantara, G. B. qNMR in natural products: practical approaches. What nobody tells you before starting your qNMR study! Front. Nat. Prod. 2024, 3, Review.
- Fátima C. S. M.; Rech, K. S.; Dutra, L. M.; Menezes, L. R. A.; Santos, A. D. C.; Nagata, N.; Stefanello, M. É. A.; Barison, A. 1H HR-MAS NMR chemical profile and chemometric analysis as a tool for quality control of different cultivars of green tea (Camellia sinensis). Food Chem. 2023, 408, 135016. [CrossRef]
- Delgado-Altamirano, R.; García-Aguilera, M. E.; Delgado-Domínguez, J.; Becker, I.; Rodríguez de San Miguel, E.; Rojas-Molina, A.; Esturau-Escofet, N. 1H NMR profiling and chemometric analysis as an approach to predict the leishmanicidal activity of dichloromethane extracts from Lantana camara (L.). J. Pharm. Biomed. Anal. 2021, 199, 114060. [CrossRef]
- Bailey, N. J.; Wang, Y.; Sampson, J.; Davis, W.; Whitcombe, I.; Hylands, P. J.; Croft, S. L.; Holmes, E. Prediction of anti-plasmodial activity of Artemisia annua extracts: application of 1H NMR spectroscopy and chemometrics. J. Pharm. Biomed. Anal. 2004, 35, 117-126.
- Souza , C. A; Oliveira, A. P.; Santos, A. D. C.; Guimarães, A. L.; Silva, N. D. S.; Queiroz, M. A. Á.; Araújo, E. C. C.; Almeida, J. R. G. S. Total content of kaurene diterpenes in Annona vepretorum stems via 1H qNMR: A method for speeding the identification of bioactive extracts. Phytochem. Anal. 2019, 30, 83-88. [CrossRef]
- Khalef, L.; Lydia, R.; Filicia, K.; Moussa, B. Cell viability and cytotoxicity assays: Biochemical elements and cellular compartments. Cell Biochem. Funct. 2024, 42, e4007. [CrossRef]
- Huang, D. J.; Ou, B. X.; Prior, R. L. The chemistry behind antioxidant capacity assays. J. Agric. Food Chem. 2005, 53, 1841-1856. [CrossRef]
- Dudonne, S.; Vitrac, X.; Coutiere, P.; Woillez, M.; Merillon, J.M. Comparative Study of Antioxidant Properties and Total Phenolic Content of 30 Plant Extracts of Industrial Interest Using DPPH, ABTS, FRAP, SOD, and ORAC Assays. J. Agric. Food Chem. 2009, 57, 1768-1774. [CrossRef]
- Zargoosh, Z.; Ghavam, M.; Bacchetta, G.; Tavili, A. Effects of ecological factors on the antioxidant potential and total phenol content of Scrophularia striata Boiss. Sci. Rep. 2019, 9, 16021. [CrossRef]
- Li, H.; Tsao, R.; Deng, Z. Factors affecting the antioxidant potential and health benefits of plant foods. Can. J. Plant. Sci. 2012, 92, 1101-1111. [CrossRef]
- Khalid, M.; Petroianu, G.; Adem, A. Advanced Glycation End Products and Diabetes Mellitus: Mechanisms and Perspectives. Biomolecules 2022, 12, 542. [CrossRef]
- López-Martínez, L. M.; Santacruz-Ortega, H.; Navarro, R. E.; Sotelo-Mundo, R. R.; González-Aguilar, G. A. A ¹H NMR Investigation of the Interaction between Phenolic Acids Found in Mango (Manguifera indica cv Ataulfo) and Papaya (Carica papaya cv Maradol) and 1,1-diphenyl-2-picrylhydrazyl (DPPH) Free Radicals. PLoS One 2015, 10, e0140242.
- Napolitano, J.; Gödecke, T.; Lankin, D.; Jaki, B.; McAlpine, J.; Chen, S.N.; Pauli, G. Orthogonal analytical methods for botanical standardization: Determination of green tea catechins by qNMR and LC–MS/MS. J. Pharm. Biomed. Anal. 2013, 93, 59-67. [CrossRef]
- Oliveira, E. S. C.; Acho, L. D. R.; Silva, B. J. P.; Morales-Gamba, R. D.; Pontes, F. L. D.; Rosário, A. S.; Bezerra, J. A.; Campos, F. R.; Barcellos, J. F. M.; Lima, E. S.; Machado, M. B. Hypoglycemic effect and toxicity of the dry extract of Eugenia biflora (L.) DC. leaves. J. Ethnopharmacol. 2022, 293, 115276. [CrossRef]
- Oliveira, E. S. C.; Pontes, F. L. D.; Acho, L. D. R.; Rosário, A. S.; Silva, B. J. P.; Araújo, B. J.; Campos, F. R.; Lima, E. S.; Machado, M. B. qNMR quantification of phenolic compounds in dry extract of Myrcia multiflora leaves and its antioxidant, anti-AGE, and enzymatic inhibition activities. J. Pharm. Biomed. Anal. 2021, 201, 114109. [CrossRef]
- Truzzi, E.; Marchetti, L.; Benvenuti, S.; Righi, V.; Rossi, M. C.; Gallo, V.; Bertelli, D. A Novel qNMR Application for the Quantification of Vegetable Oils Used as Adulterants in Essential Oils. Molecules 2021, 26, 5439. [CrossRef]
- Mak, J. Y. W. Determination of Sample Concentrations by PULCON NMR Spectroscopy. Aust. J. Chem. 2022, 75, 160-164. [CrossRef]
- Watanabe, R.; Sugai, C.; Yamazaki, T.; Matsushima, R.; Uchida, H.; Matsumiya, M.; Takatsu, A.; Suzuki, T. Quantitative Nuclear Magnetic Resonance Spectroscopy Based on PULCON Methodology: Application to Quantification of Invaluable Marine Toxin, Okadaic Acid. Toxins (Basel). 2016, 8, 294. [CrossRef]
- Yang, X.; Liu, W.; Tao, D. A Survey on Canonical Correlation Analysis. EEE Trans. Knowl. Data Eng. 2021, 33, 2349-2368. [CrossRef]
- Zhou, X.; Liu, H.; Zhang, M.; Li, C.; Li, G. Spectrum-effect relationship between UPLC fingerprints and anti-lung cancer effect of Panax ginseng. Phytochem. Anal. 2021, 32, 339-346.
- Chen, Y.; Pan, G.; Xu, W.; Sun, Q.; Wang, B.; Zhang, Y.; Yang, T. Spectrum-effect relationship study between HPLC fingerprints and antioxidant activity of Sabia parviflora. J. Chromatogr. B. 2020, 1140, 121970. [CrossRef]
- Ugur, S.; kılınc, O.; Selamoglu, Z. Antioxidant Activity of Quercetin: A Mechanistic Review. Turk. J. Agric. For. 2016, 4, 1134–1138. [CrossRef]
- Hassanpour, S. H.; Doroudi, A. Review of the antioxidant potential of flavonoids as a subgroup of polyphenols and partial substitute for synthetic antioxidants. Avicenna J. Phytomed. 2023, 13, 354-376. [CrossRef]
- Figueiroa, E. O.; Silva, L. C. N.; Melo, C. M. L.; Neves, J. K. A. L.; Silva, N. H.; Pereira, V. R. A.; Correia, M. T. S. Evaluation of Antioxidant, Immunomodulatory, and Cytotoxic Action of Fractions from Eugenia uniflora L. and Eugenia malaccensis L.: Correlation with Polyphenol and Flavanoid Content. Sci. World J. 2013, 125027, 1.
- Yeh, W. J.; Hsia, S. M.; Lee, W. H.; Wu, C. H. Polyphenols with antiglycation activity and mechanisms of action: A review of recent findings. J. Food Drug. Anal. 2017, 25, 84-92. [CrossRef]
- Wang, Y.; Li, S.; Zhang, T.; Wang, J.; Zhang, X.; Li, M.; Gao, Y.; Zhang, M.; Chen, H. Effects of myricetin and its derivatives on nonenzymatic glycation: A mechanism study based on proteomic modification and fluorescence spectroscopy analysis. Food Chem. 2024, 455, 139880. [CrossRef]
- Cataneo, A. H. D.; Ávila, E. P.; Mendes, L. A. d. O.; de Oliveira, V. G.; Ferraz, C. R.; Almeida, M. V.; Frabasile, S.; Santos, C. N. D.; Verri, W. A.; Bordignon, J.; Wowk, P. F. Flavonoids as Molecules With Anti-Zika virus Activity. Front. Microbiol. 2021, 12, 710359. [CrossRef]
- Pereira, R. S.; Santos, F. C.; Campana, P. R.; Costa, V. V.; de Pádua, R. M.; Souza, D. G.; Teixeira, M. M.; Braga, F. C. Natural Products and Derivatives as Potential Zika virus Inhibitors: A Comprehensive Review. Viruses, 2023, 15, 1211. [CrossRef]
- Liga, S.; Paul, C.; Péter, F. Flavonoids: Overview of Biosynthesis, Biological Activity, and Current Extraction Techniques. Plants 2023, 12, 2732. [CrossRef]
- Rahmani, A. H.; Almatroudi, A.; Allemailem, K. S.; Alwanian, W. M.; Alharbi, B. F.; Alrumaihi, F.; Khan, A. A.; Almatroodi, S. A. Myricetin: A Significant Emphasis on Its Anticancer Potential via the Modulation of Inflammation and Signal Transduction Pathways. Int. J. Mol. Sci. 2023, 24, 9665. [CrossRef]
- Oliveira M. R.; Nabavi S. M.; Braidy N., Setzer W. N.; Ahmed T., Nabavi S. F. Quercetin and the mitochondria: A mechanistic view. Biotechnol. Adv. 2016, 34, 532-549. [CrossRef]
- Lim, H. J.; Nguyen, T. T. H.; Kim, N. M.; Park, J.S.; Jang, T.S.; Kim, D. Inhibitory effect of flavonoids against NS2B-NS3 protease of ZIKA virus and their structure activity relationship. Biotechnol. Lett. 2017, 39, 415-421. [CrossRef]
- Ramos, P. R.; Mottin, M.; Lima, C. S.; Assis, L. R.; de Oliveira, K. Z.; Mesquita, N. C.; Cassani, N. M.; Santos, I. A.; Borba, J. V.; Fiaia Costa, V. A.; Neves, B. J.; Guido, R. V. C.; Oliva, G.; Jardim, A. C. G.; Regasini, L. O.; Andrade, C. H. Natural Compounds as Non-Nucleoside Inhibitors of Zika Virus Polymerase through Integration of In Silico and In Vitro Approaches. Pharmaceuticals (Basel), 2022, 15, 1493. [CrossRef]
- Zou, M.; Liu, H.; Li, J.; Yao, X.; Chen, Y.; Ke, C.; Liu, S. Structure-activity relationship of flavonoid bifunctional inhibitors against Zika virus infection. Biochem. Pharmacol. 2020, 177, 113962. [CrossRef]
- Lum, F. M.; Lin, C.; Susova, O. Y.; Teo, T. H.; Fong, S. W.; Mak, T. M.; Lee, L. K.; Chong, C. Y.; Lye, D. C. B.; Lin, R. T. P.; Merits, A.; Leo, Y. L.; Ng, L. F. P. A Sensitive Method for Detecting Zika Virus Antigen in Patients’ Whole-Blood Specimens as an Alternative Diagnostic Approach. J. Infect. Dis. 2017, 216, 182-190.
- Minitab® 18.1; Minitab Inc., State College: PA, USA, 2017.
- TopSpin 3.6.3; Academia License, Bruker Optics GmbH & Co. KG: Ettlingen, Germany, 2021.
- Hong, R.; Hwang, K.; Kim, S.; Cho, H.; Lee, H.; Hong, J.; Moon, D. C. Survey of ERETIC2 NMR for quantification. J. Korean Soc. Magn. Reson. Med. 2013, 17, 98-104. [CrossRef]
- Donald, C. L.; Brennan, B.; Cumberworth, S. L.; Rezelj, V. V.; Clark, J. J.; Cordeiro, M. T.; França, R. F. O., Pena, L. J.; Wilkie, G. S.; Silva, F. A.; Davis, C, Hughes, J.; Varjak, M.; Selinger, M.; Zuvanov, L.; Owsianka, A. M.; Patel, A. H., McLauchlan, J.; Lindenbach, B. D.; Fall, G.; Sall, A. A.; Biek, R.; Rehwinkel, J.; Schnettler, E.; Kohl, A. Full Genome Sequence and sfRNA Interferon Antagonist Activity of Zika Virus from Recife, Brazil. PLoS Negl. Trop. Dis. 2016, 10, e0005048. [CrossRef]
- Cassani, N. M.; Santos, I. A.; Grosche, V. R.; Ferreira, G. M.; Guevara-Vega, M.; Rosa, R. B.; Pena, L. J.; Nicolau-Junior, N.; Cintra, A. C. O.; Mineo, T. P. Roles of Bothrops jararacussu toxins I and II: Antiviral findings against Zika virus. Int. J. Biol. Macromol. 2023, 227, 630-640.
- One-way ANOVA followed by Dunnett’s multiple comparisons test was performed using GraphPad Prism version 10.3.0 for Windows; GraphPad Software: Boston, MA, USA, 2024.
- Mar, J. M.; Silva, L. S.; Moreira, W. P.; Biondo, M. M.; Pontes, F. L. D.; Campos, F. R.; Kinupp, V. F.; Campelo, P. H.; Sanches, E. A.; Bezerra, J. A. Edible flowers from Theobroma speciosum: Aqueous extract rich in antioxidant compounds. Food Chem. 2021, 356, 129723. [CrossRef]
- Kiho, T.; Usui, S.; Hirano, K.; Aizawa, K.; Inakuma, T. Tomato paste fraction inhibiting the formation of advanced glycation end-products. Biosci. Biotechnol. Biochem. 2004, 68, 200-205. [CrossRef]
| Sample | MEW (mg g−1 dry extract) |
M (mg g−1 dry extract) |
EM (mg g−1 dry extract) |
E (mg g−1 dry extract) |
|---|---|---|---|---|
| Dry | 312.2 ± 13.5ab | 275.8 ± 5.9c | 207.4 ± 4.7e | 117.0 ± 8.9f |
| Transition | 334.1 ± 7.3a | 298.9 ± 8.6b | 224.9 ± 1.9ed | 118.9 ± 7.8f |
| Rainy | 328.2 ± 8.1a | 304.2 ± 1.9b | 242.9 ± 9.1d | 124.4 ± 7.1f |
| Sample | Dry | Transition | Rainy | |||
|---|---|---|---|---|---|---|
| DPPH• | ABTS•+ | DPPH• | ABTS•+ | DPPH• | ABTS•+ | |
| MEW | 1317.5 ± 6.6a | 1848.8 ± 6.9a | 1449.2 ± 8.8a | 2008.8 ± 8.4a | 1530.8 ± 5.2a | 2121.0 ± 6.7a |
| EM | 1139.2 ± 10.1b | 1702.1 ± 10.7b | 1213.3 ± 8.0b | 1766.5 ± 6.9b | 1343.33 ± 8.0b | 1919.9 ± 8.4b |
| E | 1115.0 ± 9.0c | 1685.4 ± 6.9b | 1189.2 ± 8.8c | 1751.0 ± 6.7b | 1318.3 ± 6.3c | 1827.7 ± 6.7d |
| M | 1025.8 ± 5.2d | 1645.4 ± 8.4c | 1085.8 ± 3.8d | 1703.2 ± 10.2a | 1140.8 ± 7.6d | 1878.8 ± 8.4c |
| Sample | MEW (%inhibition of AGEs) |
M (%inhibition ofAGEs) |
EM (% inhibition of AGEs) |
E (% inhibition ofAGEs) |
|---|---|---|---|---|
| Dry | 80.5 ± 1.4b | 90.1 ± 2.0a | 80.4 ± 3.0b | 76.0 ± 1.8b |
| Transition | 94.3 ± 1.5a | 82.4 ± 2.4b | 83.4 ± 1.6b | 94.0 ± 3.0a |
| Rainy | 93.1 ± 3.7a | 88.2 ± 3.8ab | 94.8 ± 1.4a | 87.5 ± 3.0a |
| Sample | Quercetin (mg g−1 dry extract) |
Myricetin (mg g−1 dry extract) |
Gallic acid (mg g−1 dry extract) |
Catechin (mg g−1 dry extract) |
Epigallocatechin (mg g−1 dry extract) |
Sum of total phenolics (mg g−1 dry extract) |
|---|---|---|---|---|---|---|
| E - Dry | 1.94 ± 0.01 | 1.40 ± 0.00 | 3.49 ± 0.01 | 5.02 ± 0.01 | 3.34 ± 0.01 | 15.24 ± 0.20 |
| E - Transition | 2.23 ± 0.01 | 1.62 ± 0.00 | 3.54 ± 0.01 | 4.65 ± 0.02 | 3.63 ± 0.02 | 15.75 ± 0.31 |
| E - Rainy | 2.23 ± 0.00 | 1.69 ± 0.00 | 3.50 ± 0.01 | 4.54 ± 0.01 | 3.55 ± 0.01 | 15.60 ± 0.17 |
| EM - Dry | 1.24 ± 0.01 | 1.45 ± 0.01 | 3.23 ± 0.01 | 5.50 ± 0.02 | 3.60 ± 0.02 | 15.09 ± 0.37 |
| EM - Transition | 1.73 ± 0.00 | 1.71 ± 0.00 | 4.04 ± 0.01 | 6.55 ± 0.01 | 5.01 ± 0.00 | 19.13 ± 0.18 |
| EM - Rainy | 2.44 ± 0.00 | 1.87 ± 0.00 | 3.61 ± 0.01 | 5.78 ± 0.01 | 4.47 ± 0.00 | 18.27 ± 0.07 |
| M - Dry | 1.47 ± 0.00 | 1.75 ± 0.01 | 3.85 ± 0.02 | 6.54 ± 0.03 | 4.28 ± 0.02 | 17.98 ± 0.43 |
| M - Transition | 2.07 ± 0.00 | 1.60 ± 0.00 | 4.00 ± 0.02 | 6.35 ± 0.01 | 5.55 ± 0.01 | 19.65 ± 0.18 |
| M - Rainy | 2.29 ± 0.01 | 1.83 ± 0.01 | 3.91 ± 0.00 | 5.89 ± 0.02 | 5.08 ± 0.01 | 19.11 ± 0.43 |
| MEW - Dry | 2.89 ± 0.01 | 2.13 ± 0.00 | 3.38 ± 0.03 | 6.97 ± 0.03 | 4.87 ± 0.03 | 20.35 ± 0.35 |
| MEW - Transition | 2.70 ± 0.01 | 1.56 ± 0.01 | 3.91 ± 0.01 | 6.67 ± 0.03 | 5.85 ± 0.02 | 20.94 ± 0.56 |
| MEW - Rainy | 3.16 ± 0.00 | 1.98 ± 0.00 | 3.79 ± 0.02 | 6.12 ± 0.01 | 5.07 ± 0.01 | 20.22 ± 0.27 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





