Submitted:
07 December 2024
Posted:
09 December 2024
You are already at the latest version
Abstract

Keywords:
1. Introduction
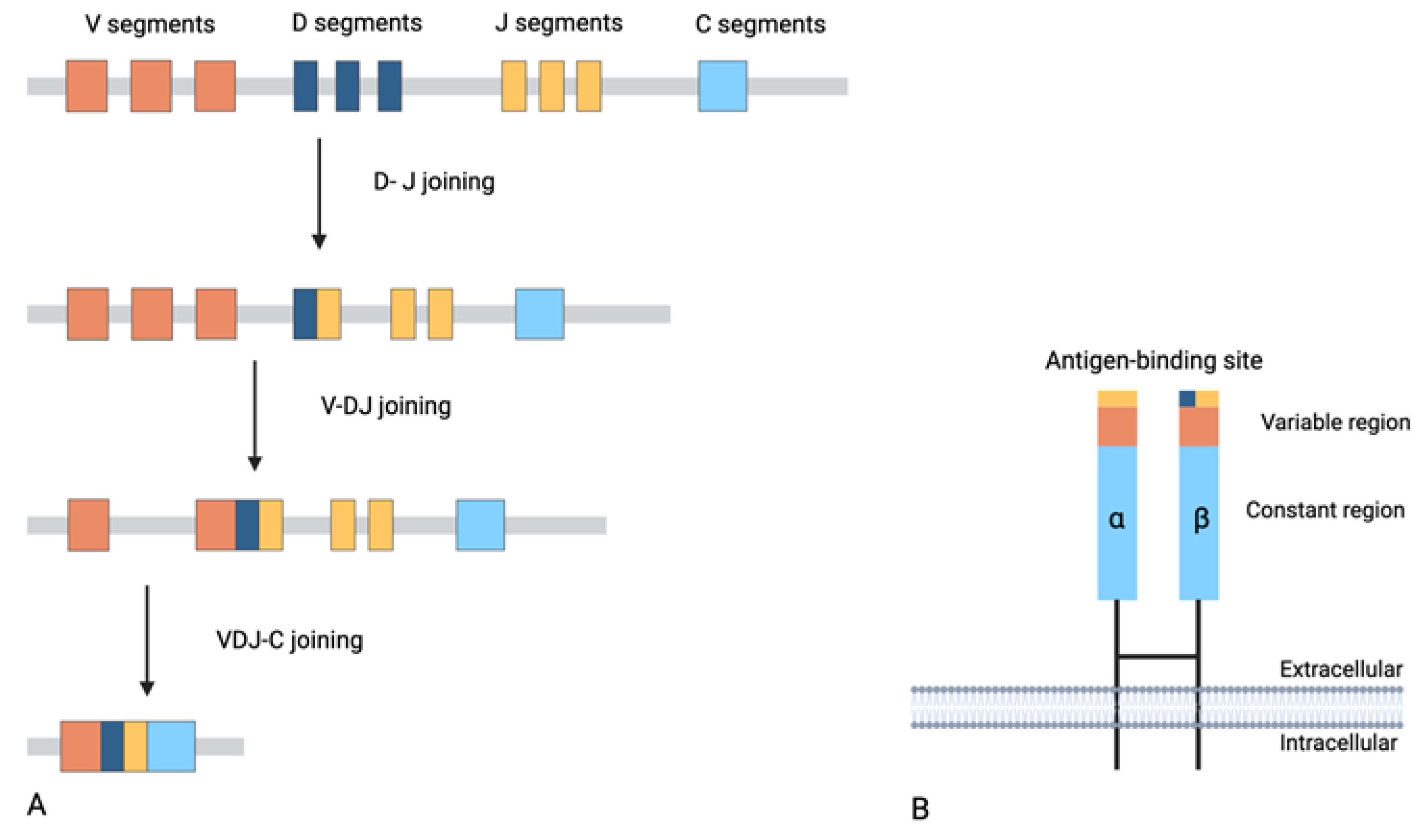
2. General Aspects of TCR Biology
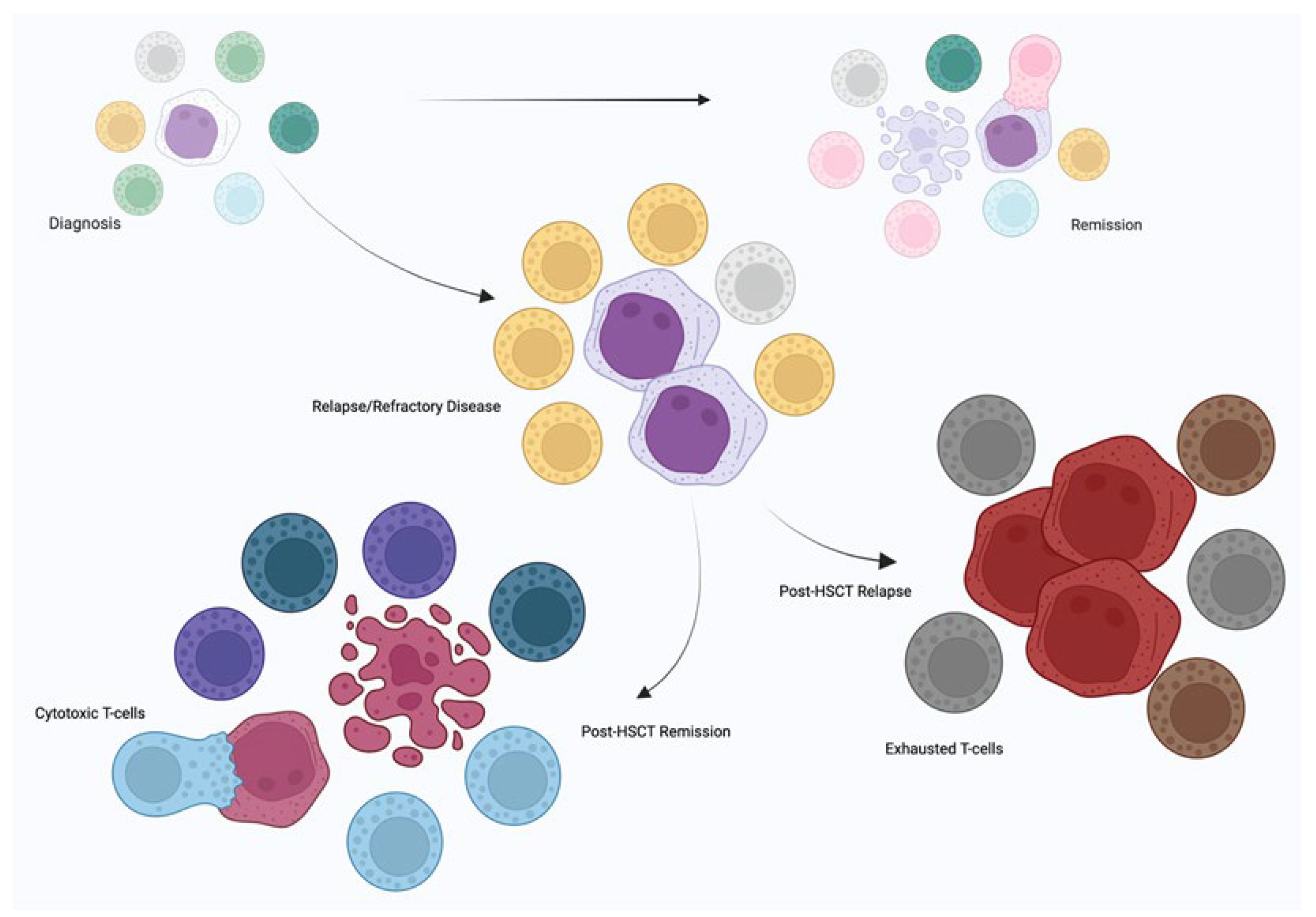
3. TCR Repertoire in MDS
3.1. Treatment effects on TCR repertoire
3.1.1. Immunotherapy and Hypomethylating Agents
3.1.2. Stem Cell Transplantation
4. TCR Repertoire in AML
4.1. Treatment effects on TCR repertoire
4.1.1. Hypomethylating Agents
4.1.2. Immune Checkpoint Inhibitors
4.1.3. Stem Cell Transplantation
5. Future Directions
6. Conclusions
| Reference | Population | Methods | Therapy | TCR/ Clonotype change | Prognostic/ Predictive correlations |
| Jin et al [46] | Newly diagnosed AML patients | RT-PCR, GeneScan | N/A | TRDV4, TRDV8 oligoclonality | Protective factor for CR |
| TRDV5, TRDV6 oligoclonality | Relapse | ||||
| Geng et al [47] | RAEB- MDS patients | RT-PCR, GeneScan | N/A | Reduced TRDV4 frequencies | Progression to AML |
| TRDV8 clonal expansion | |||||
| Kong et al [48] | Newly diagnosed- De novo AML patients |
RT-qPCR, GeneScan |
Various Chemotherapy regimens HSCT |
Higher TRGV expression levels | Improved OS |
| Increased TRGV9 usage Increased circulating Vγ9+Vδ2+ T cells |
Protective factor for CR | ||||
| Zhang et al [51] | TCGA and TARGET AML data | In silico, NGS | Various Chemotherapy regimens |
Specific TRD-CDR3 cluster associated with TRDV2, TRDJ3 usage | Superior OS |
| Pospiech et al [53] | TCGA AML data | In silico, NGS | Various Chemotherapy regimens |
Normalized unique TRA/TRB counts | No association with survival |
| Public clonotypes (CANTGELFF TRB CDR3 clone) | Superior OS | ||||
| Feng et al [56] | Newly diagnosed AML patients | NGS | DNR + AraC | CD8+ TRB clonotype expansion Decreased Shannon entropy Increased TRB clonotype evenness |
Disease relapse |
| Grimm et al [57] | De novo and secondary AML patients | NGS | 5’-AZA +/- Mitoxantrone + AraC |
Higher pre-treatment TRB diversity (Increased Shannon entropy) Increase in post-treatment TRB richness TRBV skewing (TRBV12-3, 5-7, 6-9) |
Superior EFS and OS |
| Beckford et al [58] | Newly diagnosed AML patients | NGS | 5’-AZA | TRBV16, TRBV12-5 increase post therapy TRBV12-2 increased pre therapy |
Response to 5’-AZA |
| TRBV7-4 increase post therapy | No response to 5’-AZA | ||||
| Abbas et al [60] | R/R AML patients | NGS | 5’-AZA + Nivolumab | αβ T- clonotype expansion | Response to therapy/ Stable disease |
| αβ T- clonotype contraction | Resistance to therapy | ||||
| Yew et al [64] | AML ( and 1 MDS) patients | NGS | allo-HSCT (MD+ Haplo-cord) | Early increase in TRA/ TRB diversity | Improved cord chimerism |
| Increased TCR diversity | Remission and no- GVHD occurrence | ||||
| Decreased TCR diversity | Remission and GVHD occurrence | ||||
| Schultze- Florey et al [69] | AML patients with relapse or increased host chimerism after allo-HSCT | NGS | DLI | Decreased CD8+ TRB diversity Clonotype expansion With pre- and post- treatment overlap |
GVL occurrence and durable remmision |
| Absence of clonotype expansion | Relapse | ||||
| van Bergen et al [70] | AML, CML, MDS patients with relapse after allo-HSCT | NGS | DLI | Reduced TRB diversity of MiHA- reactive CD8+ clones | GVL with CR and no GVHD emergence |
| Increased TRB diversity to a broader of MiHAs | GVL with concomitant GVHD occurrence | ||||
| Arruda et al [77] | AML patients | NGS | PBSC | TRG Clonotype expansion Lower usage of TRGV4-J2, TRGV5-J2, TRGV8-JP2 Higher usage of TRGV2-JP1, TRGV9-JP Increased public clonotypes |
Non- relapse |
| Overall TRG status | Not associated with acute GVHD development | ||||
| Lower TCR diversity More private clonotypes |
Donor CMV positivity | ||||
| Lee et al [29] | MDS and CMML patients | FC, PCR, in silico | IT + HMAs | Emergence of novel clonotypes vs TCRB repertoire contraction | Responders vs non-responders |
| HMAs | No change in TCR clonality | None | |||
| Fozza et al [31] | MDS and AML | FC, PCR | 5’-AZA | Restoration of CDR3 diversity | Response to 5’ AZA |
| Abbas et al [32] | MDS | PCR, in silico | HMAs | Emergence of novel clonotypes vs TRB repertoire contraction | Responders vs non-responders |
| Kochenderfer et al [33] | MDS | FC, PCR | ATG-based treatment | Regression of dominant TCR clonotypes | Response to ATG |
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Mazzotti, L.; Gaimari, A.; Bravaccini, S.; Maltoni, R.; Cerchione, C.; Juan, M.; Navarro, E.A.; Pasetto, A.; Nascimento Silva, D.; Ancarani, V.; et al. T cell Receptor Repertoire Sequencing and Its Applications: Focus on Infectious Diseases and Cancer. Int J Mol Sci 2022, 23. [Google Scholar] [CrossRef] [PubMed]
- Barakos, G.P.; Hatzimichael, E. Microenvironmental Features Driving Immune Evasion in Myelodysplastic Syndromes and Acute Myeloid Leukemia. Diseases 2022, 10. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Manero, G. Myelodysplastic syndromes: 2023 update on diagnosis, risk-stratification, and management. Am J Hematol 2023, 98, 1307–1325. [Google Scholar] [CrossRef] [PubMed]
- Fozza, C.; Longinotti, M. T cell receptor repertoire usage in hematologic malignancies. Crit Rev Oncol Hematol 2013, 86, 201–211. [Google Scholar] [CrossRef]
- Linder, K.; Lulla, P. Myelodysplastic syndrome and immunotherapy novel to next in-line treatments. Hum Vaccin Immunother 2021, 17, 2602–2616. [Google Scholar] [CrossRef]
- Shimony, S.; Stahl, M.; Stone, R.M. Acute myeloid leukemia: 2023 update on diagnosis, risk-stratification, and management. Am J Hematol 2023, 98, 502–526. [Google Scholar] [CrossRef]
- Chen, Y.; Wang, J.; Zhang, F.; Liu, P. A perspective of immunotherapy for acute myeloid leukemia: Current advances and challenges. Front Pharmacol 2023, 14, 1151032. [Google Scholar] [CrossRef]
- Bonilla, F.A.; Oettgen, H.C. Adaptive immunity. J Allergy Clin Immunol 2010, 125, S33–S40. [Google Scholar] [CrossRef]
- Nikolich-Zugich, J.; Slifka, M.K.; Messaoudi, I. The many important facets of T cell repertoire diversity. Nat Rev Immunol 2004, 4, 123–132. [Google Scholar] [CrossRef]
- Cardinale, A.; De Luca, C.D.; Locatelli, F.; Velardi, E. Thymic Function and T cell Receptor Repertoire Diversity: Implications for Patient Response to Checkpoint Blockade Immunotherapy. Front Immunol 2021, 12, 752042. [Google Scholar] [CrossRef]
- Takahama, Y.; Ohigashi, I.; Baik, S.; Anderson, G. Generation of diversity in thymic epithelial cells. Nat Rev Immunol 2017, 17, 295–305. [Google Scholar] [CrossRef] [PubMed]
- Frank, M.L.; Lu, K.; Erdogan, C.; Han, Y.; Hu, J.; Wang, T.; Heymach, J.V.; Zhang, J.; Reuben, A. T cell Receptor Repertoire Sequencing in the Era of Cancer Immunotherapy. Clin Cancer Res 2023, 29, 994–1008. [Google Scholar] [CrossRef] [PubMed]
- Lo Presti, E.; Dieli, F.; Meraviglia, S. Tumor-Infiltrating gammadelta T Lymphocytes: Pathogenic Role, Clinical Significance, and Differential Programing in the Tumor Microenvironment. Front Immunol 2014, 5, 607. [Google Scholar] [CrossRef] [PubMed]
- Rossjohn, J.; Gras, S.; Miles, J.J.; Turner, S.J.; Godfrey, D.I.; McCluskey, J. T cell antigen receptor recognition of antigen-presenting molecules. Annu Rev Immunol 2015, 33, 169–200. [Google Scholar] [CrossRef]
- Pauken, K.E.; Lagattuta, K.A.; Lu, B.Y.; Lucca, L.E.; Daud, A.I.; Hafler, D.A.; Kluger, H.M.; Raychaudhuri, S.; Sharpe, A.H. TCR-sequencing in cancer and autoimmunity: Barcodes and beyond. Trends Immunol 2022, 43, 180–194. [Google Scholar] [CrossRef]
- Aversa, I.; Malanga, D.; Fiume, G.; Palmieri, C. Molecular T cell Repertoire Analysis as Source of Prognostic and Predictive Biomarkers for Checkpoint Blockade Immunotherapy. Int J Mol Sci 2020, 21. [Google Scholar] [CrossRef]
- Chen, G.; Yang, X.; Ko, A.; Sun, X.; Gao, M.; Zhang, Y.; Shi, A.; Mariuzza, R.A.; Weng, N.P. Sequence and Structural Analyses Reveal Distinct and Highly Diverse Human CD8(+) TCR Repertoires to Immunodominant Viral Antigens. Cell Rep 2017, 19, 569–583. [Google Scholar] [CrossRef]
- Rodriguez-Sevilla, J.J.; Colla, S. T cell dysfunctions in myelodysplastic syndromes. Blood 2024, 143, 1329–1343. [Google Scholar] [CrossRef]
- Epperson, D.E.; Nakamura, R.; Saunthararajah, Y.; Melenhorst, J.; Barrett, A.J. Oligoclonal T cell expansion in myelodysplastic syndrome: Evidence for an autoimmune process. Leuk Res 2001, 25, 1075–1083. [Google Scholar] [CrossRef]
- Naylor, K.; Li, G.; Vallejo, A.N.; Lee, W.W.; Koetz, K.; Bryl, E.; Witkowski, J.; Fulbright, J.; Weyand, C.M.; Goronzy, J.J. The influence of age on T cell generation and TCR diversity. J Immunol 2005, 174, 7446–7452. [Google Scholar] [CrossRef]
- Melenhorst, J.J.; Eniafe, R.; Follmann, D.; Nakamura, R.; Kirby, M.; Barrett, A.J. Molecular and flow cytometric characterization of the CD4 and CD8 T cell repertoire in patients with myelodysplastic syndrome. Br J Haematol 2002, 119, 97–105. [Google Scholar] [CrossRef] [PubMed]
- Epling-Burnette, P.K.; Painter, J.S.; Rollison, D.E.; Ku, E.; Vendron, D.; Widen, R.; Boulware, D.; Zou, J.X.; Bai, F.; List, A.F. Prevalence and clinical association of clonal T cell expansions in Myelodysplastic Syndrome. Leukemia 2007, 21, 659–667. [Google Scholar] [CrossRef] [PubMed]
- Fozza, C.; Contini, S.; Galleu, A.; Simula, M.P.; Virdis, P.; Bonfigli, S.; Longinotti, M. Patients with myelodysplastic syndromes display several T cell expansions, which are mostly polyclonal in the CD4(+) subset and oligoclonal in the CD8(+) subset. Exp Hematol 2009, 37, 947–955. [Google Scholar] [CrossRef] [PubMed]
- Katsiki, K.; Tasis, A.; Filia, A.; Lamprianidou, E.; Liapis, K.; Kotsianidis, I.; Mitroulis, I. P742: TRANSCRIPTOMIC ANALYSIS OF ISOLATED CD4+ AND CD8+ T CELLS REVEALS DIFFERENCES IN TCR REPERTOIRE IN PATIENTS WITH HIGH RISK MDS COMPARED TO AML. HemaSphere 2022, 6, 637–638. [Google Scholar] [CrossRef]
- Campregher, P.V.; Srivastava, S.K.; Deeg, H.J.; Robins, H.S.; Warren, E.H. Abnormalities of the alphabeta T cell receptor repertoire in advanced myelodysplastic syndrome. Exp Hematol 2010, 38, 202–212. [Google Scholar] [CrossRef]
- Fonseca, S.; Pereira, V.; Lau, C.; Teixeira, M.D.A.; Bini-Antunes, M.; Lima, M. Human Peripheral Blood Gamma Delta T Cells: Report on a Series of Healthy Caucasian Portuguese Adults and Comprehensive Review of the Literature. Cells 2020, 9. [Google Scholar] [CrossRef]
- Hu, Y.; Hu, Q.; Li, Y.; Lu, L.; Xiang, Z.; Yin, Z.; Kabelitz, D.; Wu, Y. gammadelta T cells: Origin and fate, subsets, diseases and immunotherapy. Signal Transduct Target Ther 2023, 8, 434. [Google Scholar] [CrossRef]
- Kiladjian, J.-J.; Visentin, G.r.; Viey, E.; Ayari, S.; Bourhis, J.-H.; Chouaib, S.; Fenaux, P.; Caignard, A. Aberrant Repertoire and Deficient Proliferation of TCR γδ T Cells in Myelodysplastic Syndromes (MDS). Blood 2006, 108, 2639–2639. [Google Scholar] [CrossRef]
- Lee, S.E.; Wang, F.; Grefe, M.; Trujillo-Ocampo, A.; Ruiz-Vasquez, W.; Takahashi, K.; Abbas, H.A.; Borges, P.; Antunes, D.A.; Al-Atrash, G.; et al. Immunologic Predictors for Clinical Responses during Immune Checkpoint Blockade in Patients with Myelodysplastic Syndromes. Clin Cancer Res 2023, 29, 1938–1951. [Google Scholar] [CrossRef]
- Lindblad, K.E.; Goswami, M.; Hourigan, C.S.; Oetjen, K.A. Immunological effects of hypomethylating agents. Expert Rev Hematol 2017, 10, 745–752. [Google Scholar] [CrossRef]
- Fozza, C.; Corda, G.; Barraqueddu, F.; Virdis, P.; Contini, S.; Galleu, A.; Isoni, A.; Dore, F.; Angelucci, E.; Longinotti, M. Azacitidine improves the T cell repertoire in patients with myelodysplastic syndromes and acute myeloid leukemia with multilineage dysplasia. Leuk Res 2015, 39, 957–963. [Google Scholar] [CrossRef] [PubMed]
- Abbas, H.A.; Reville, P.K.; Jiang, X.; Yang, H.; Reuben, A.; Im, J.S.; Little, L.; Sinson, J.C.; Chen, K.; Futreal, A.; et al. Response to Hypomethylating Agents in Myelodysplastic Syndrome Is Associated With Emergence of Novel TCR Clonotypes. Front Immunol 2021, 12, 659625. [Google Scholar] [CrossRef] [PubMed]
- Kochenderfer, J.N.; Kobayashi, S.; Wieder, E.D.; Su, C.; Molldrem, J.J. Loss of T-lymphocyte clonal dominance in patients with myelodysplastic syndrome responsive to immunosuppression. Blood 2002, 100, 3639–3645. [Google Scholar] [CrossRef] [PubMed]
- Calderin Sollet, Z.; Schafer, A.; Ferrari-Lacraz, S.; Masouridi-Levrat, S.; Mamez, A.C.; Pradier, A.; Simonetta, F.; Chalandon, Y.; Villard, J.; Buhler, S. CMV serostatus and T cell repertoire diversity 5 years after allogeneic hematopoietic stem cell transplantation. Leukemia 2023, 37, 948–951. [Google Scholar] [CrossRef]
- Link-Rachner, C.S.; Eugster, A.; Rucker-Braun, E.; Heidenreich, F.; Oelschlagel, U.; Dahl, A.; Klesse, C.; Kuhn, M.; Middeke, J.M.; Bornhauser, M.; et al. T cell receptor-alpha repertoire of CD8+ T cells following allogeneic stem cell transplantation using next-generation sequencing. Haematologica 2019, 104, 622–631. [Google Scholar] [CrossRef]
- Lamble, A.J.; Lind, E.F. Targeting the Immune Microenvironment in Acute Myeloid Leukemia: A Focus on T Cell Immunity. Front Oncol 2018, 8, 213. [Google Scholar] [CrossRef]
- Vago, L.; Perna, S.K.; Zanussi, M.; Mazzi, B.; Barlassina, C.; Stanghellini, M.T.; Perrelli, N.F.; Cosentino, C.; Torri, F.; Angius, A.; et al. Loss of mismatched HLA in leukemia after stem-cell transplantation. N Engl J Med 2009, 361, 478–488. [Google Scholar] [CrossRef]
- Christopher, M.J.; Petti, A.A.; Rettig, M.P.; Miller, C.A.; Chendamarai, E.; Duncavage, E.J.; Klco, J.M.; Helton, N.M.; O'Laughlin, M.; Fronick, C.C.; et al. Immune Escape of Relapsed AML Cells after Allogeneic Transplantation. N Engl J Med 2018, 379, 2330–2341. [Google Scholar] [CrossRef]
- Vadakekolathu, J.; Rutella, S. Escape from T cell-targeting immunotherapies in acute myeloid leukemia. Blood 2024, 143, 2689–2700. [Google Scholar] [CrossRef]
- Le Dieu, R.; Taussig, D.C.; Ramsay, A.G.; Mitter, R.; Miraki-Moud, F.; Fatah, R.; Lee, A.M.; Lister, T.A.; Gribben, J.G. Peripheral blood T cells in acute myeloid leukemia (AML) patients at diagnosis have abnormal phenotype and genotype and form defective immune synapses with AML blasts. Blood 2009, 114, 3909–3916. [Google Scholar] [CrossRef]
- Behl, D.; Porrata, L.F.; Markovic, S.N.; Letendre, L.; Pruthi, R.K.; Hook, C.C.; Tefferi, A.; Elliot, M.A.; Kaufmann, S.H.; Mesa, R.A.; et al. Absolute lymphocyte count recovery after induction chemotherapy predicts superior survival in acute myelogenous leukemia. Leukemia 2006, 20, 29–34. [Google Scholar] [CrossRef] [PubMed]
- Ismail, M.M.; Abdulateef, N.A.B. Bone marrow T cell percentage: A novel prognostic indicator in acute myeloid leukemia. Int J Hematol 2017, 105, 453–464. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Yang, L.; Chen, S.; Li, R.; Zhang, Y.; Lu, Y.; Luo, G. Clonal expansion T cells identified in acute monoblastic leukemia by CDR3 size analysis of TCR V beta repertoire using RT-PCR and genescan. Chin Med J (Engl) 2002, 115, 69–71. [Google Scholar] [PubMed]
- Li, Y.; Chen, S.; Yang, L.; Zhou, Y.; Wu, X.; Huang, M.; Geng, S. Clonal expanded TCR Vbeta T cells in patients with APL. Hematology 2005, 10, 135–139. [Google Scholar] [CrossRef]
- Ou, Y.; Tong, C.; Zhang, Y.; Cai, P.; Gu, J.; Liu, Y.; Liu, H.; Wang, H.; Chu, B.; Zhu, P. An improved design of PCR primers for detection of human T cell receptor beta chain repertoire. Mol Biol Rep 2009, 36, 145–152. [Google Scholar] [CrossRef]
- Jin, Z.; Luo, Q.; Lu, S.; Wang, X.; He, Z.; Lai, J.; Chen, S.; Yang, L.; Wu, X.; Li, Y. Oligoclonal expansion of TCR Vdelta T cells may be a potential immune biomarker for clinical outcome of acute myeloid leukemia. J Hematol Oncol 2016, 9, 126. [Google Scholar] [CrossRef]
- Geng, S.; Weng, J.; Du, X.; Lai, P.; Huang, X.; Chen, S.; Yang, L.; Li, Y. Comparison of the distribution and clonal expansion features of the T cell gammadelta repertoire in myelodysplastic syndrome-RAEB and RAEB with progression to AML. DNA Cell Biol 2012, 31, 1563–1570. [Google Scholar] [CrossRef]
- Kong, X.; Zheng, J.; Liu, X.; Wang, W.; Jiang, X.; Chen, J.; Lai, J.; Jin, Z.; Wu, X. High TRGV 9 Subfamily Expression Marks an Improved Overall Survival in Patients With Acute Myeloid Leukemia. Front Immunol 2022, 13, 823352. [Google Scholar] [CrossRef]
- Gertner-Dardenne, J.; Castellano, R.; Mamessier, E.; Garbit, S.; Kochbati, E.; Etienne, A.; Charbonnier, A.; Collette, Y.; Vey, N.; Olive, D. Human Vgamma9Vdelta2 T cells specifically recognize and kill acute myeloid leukemic blasts. J Immunol 2012, 188, 4701–4708. [Google Scholar] [CrossRef]
- Di Lorenzo, B.; Simoes, A.E.; Caiado, F.; Tieppo, P.; Correia, D.V.; Carvalho, T.; da Silva, M.G.; Dechanet-Merville, J.; Schumacher, T.N.; Prinz, I.; et al. Broad Cytotoxic Targeting of Acute Myeloid Leukemia by Polyclonal Delta One T Cells. Cancer Immunol Res 2019, 7, 552–558. [Google Scholar] [CrossRef]
- Zhang, J.; Hu, X.; Wang, J.; Sahu, A.D.; Cohen, D.; Song, L.; Ouyang, Z.; Fan, J.; Wang, B.; Fu, J.; et al. Immune receptor repertoires in pediatric and adult acute myeloid leukemia. Genome Med 2019, 11, 73. [Google Scholar] [CrossRef] [PubMed]
- Peggs, K.S.; Verfuerth, S.; D'Sa, S.; Yong, K.; Mackinnon, S. Assessing diversity: Immune reconstitution and T cell receptor BV spectratype analysis following stem cell transplantation. Br J Haematol 2003, 120, 154–165. [Google Scholar] [CrossRef] [PubMed]
- Pospiech, M.; Beckford, J.; Kumar, A.M.S.; Tamizharasan, M.; Brito, J.; Liang, G.; Mangul, S.; Alachkar, H. The DNA methylation landscape across the TCR loci in patients with acute myeloid leukemia. Int Immunopharmacol 2024, 138, 112376. [Google Scholar] [CrossRef] [PubMed]
- Reuther, S.; Schmetzer, H.; Schuster, F.R.; Krell, P.; Grabrucker, C.; Liepert, A.; Kroell, T.; Kolb, H.J.; Borkhardt, A.; Buhmann, R. In vitro-induced response patterns of antileukemic T cells: Characterization by spectratyping and immunophenotyping. Clin Exp Med 2013, 13, 29–48. [Google Scholar] [CrossRef] [PubMed]
- Shi, L.; Chen, S.; Zha, X.; Xu, Y.; Xu, L.; Yang, L.; Lu, Y.; Zhu, K.; Li, Y. Enhancement of the TCRzeta expression, polyclonal expansion, and activation of t cells from patients with acute myeloid leukemia after IL-2, IL-7, and IL-12 induction. DNA Cell Biol 2015, 34, 481–488. [Google Scholar] [CrossRef]
- Feng, Z.; Fang, Q.; Kuang, X.; Liu, X.; Chen, Y.; Ma, D.; Wang, J. Clonal expansion of bone marrow CD8(+) T cells in acute myeloid leukemia patients at new diagnosis and after chemotherapy. Am J Cancer Res 2020, 10, 3973–3989. [Google Scholar]
- Grimm, J.; Simnica, D.; Jakel, N.; Paschold, L.; Willscher, E.; Schulze, S.; Dierks, C.; Al-Ali, H.K.; Binder, M. Azacitidine-induced reconstitution of the bone marrow T cell repertoire is associated with superior survival in AML patients. Blood Cancer J 2022, 12, 19. [Google Scholar] [CrossRef]
- Beckford, J.; Fulton, N.; Odenike, T.; Stock, W.; Alachkar, H. Abstract 4492: Characterization of the T cell receptor repertoire in patients with acute myeloid leukemia treated with 5-Azacytidine plus chemotherapy. Cancer Research 2020, 80, 4492–4492. [Google Scholar] [CrossRef]
- Desai, P.N.; Wang, B.; Fonseca, A.; Borges, P.; Jelloul, F.Z.; Reville, P.K.; Lee, E.; Ly, C.; Basi, A.; Root, J.; et al. Single-Cell Profiling of CD8+ T Cells in Acute Myeloid Leukemia Reveals a Continuous Spectrum of Differentiation and Clonal Hyperexpansion. Cancer Immunol Res 2023. [Google Scholar] [CrossRef]
- Abbas, H.A.; Hao, D.; Tomczak, K.; Barrodia, P.; Im, J.S.; Reville, P.K.; Alaniz, Z.; Wang, W.; Wang, R.; Wang, F.; et al. Single cell T cell landscape and T cell receptor repertoire profiling of AML in context of PD-1 blockade therapy. Nat Commun 2021, 12, 6071. [Google Scholar] [CrossRef]
- Hino, C.; Xu, Y.; Xiao, J.; Baylink, D.J.; Reeves, M.E.; Cao, H. The potential role of the thymus in immunotherapies for acute myeloid leukemia. Front Immunol 2023, 14, 1102517. [Google Scholar] [CrossRef] [PubMed]
- Matsutani, T.; Yoshioka, T.; Tsuruta, Y.; Iwagami, S.; Toyosaki-Maeda, T.; Horiuchi, T.; Miura, A.B.; Watanabe, A.; Takada, G.; Suzuki, R.; et al. Restricted usage of T cell receptor alpha-chain variable region (TCRAV) and T cell receptor beta-chain variable region (TCRBV) repertoires after human allogeneic haematopoietic transplantation. Br J Haematol 2000, 109, 759–769. [Google Scholar] [CrossRef] [PubMed]
- Epperson, D.E.; Margolis, D.A.; McOlash, L.; Janczak, T.; Barrett, A.J. In vitro T cell receptor V beta repertoire analysis may identify which T cell V beta families mediate graft-versus-leukaemia and graft-versus-host responses after human leucocyte antigen-matched sibling stem cell transplantation. Br J Haematol 2001, 114, 57–62. [Google Scholar] [CrossRef] [PubMed]
- Yew, P.Y.; Alachkar, H.; Yamaguchi, R.; Kiyotani, K.; Fang, H.; Yap, K.L.; Liu, H.T.; Wickrema, A.; Artz, A.; van Besien, K.; et al. Quantitative characterization of T cell repertoire in allogeneic hematopoietic stem cell transplant recipients. Bone Marrow Transplant 2015, 50, 1227–1234. [Google Scholar] [CrossRef]
- Chen, X.; Barfield, R.; Benaim, E.; Leung, W.; Knowles, J.; Lawrence, D.; Otto, M.; Shurtleff, S.A.; Neale, G.A.; Behm, F.G.; et al. Prediction of T cell reconstitution by assessment of T cell receptor excision circle before allogeneic hematopoietic stem cell transplantation in pediatric patients. Blood 2005, 105, 886–893. [Google Scholar] [CrossRef]
- Paranal, R.M.; Kantarjian, H.M.; Reuben, A.; Kerros, C.; Koppikar, P.; Little, L.; Gumbs, C.; Short, N.J.; Cortes, J.E.; Jabbour, E.; et al. Characterization of Changes in the T cell Receptor Repertoire in Patients with Acute Myeloid Leukemia with Durable Remission Following Allogeneic Stem Cell Transplant. Blood 2019, 134, 5186–5186. [Google Scholar] [CrossRef]
- Hu, K.X.; Guo, M.; Yu, C.L.; Qiao, J.H.; Sun, Q.Y.; Cai, B.; Zhan, X.R.; Shen, X.L.; Fan, C.B.; Ai, H.S.; et al. Hematopoietic stem cell microtransplantation in patients aged over 70 with acute myeloid leukemia: A multicenter study. Am J Cancer Res 2023, 13, 1509–1521. [Google Scholar]
- Noviello, M.; Manfredi, F.; Ruggiero, E.; Perini, T.; Oliveira, G.; Cortesi, F.; De Simone, P.; Toffalori, C.; Gambacorta, V.; Greco, R.; et al. Bone marrow central memory and memory stem T cell exhaustion in AML patients relapsing after HSCT. Nat Commun 2019, 10, 1065. [Google Scholar] [CrossRef]
- Schultze-Florey, C.R.; Kuhlmann, L.; Raha, S.; Barros-Martins, J.; Odak, I.; Tan, L.; Xiao, Y.; Ravens, S.; Hambach, L.; Venturini, L.; et al. Clonal expansion of CD8+ T cells reflects graft-versus-leukemia activity and precedes durable remission following DLI. Blood Adv 2021, 5, 4485–4499. [Google Scholar] [CrossRef]
- van Bergen, C.A.; van Luxemburg-Heijs, S.A.; de Wreede, L.C.; Eefting, M.; von dem Borne, P.A.; van Balen, P.; Heemskerk, M.H.; Mulder, A.; Claas, F.H.; Navarrete, M.A.; et al. Selective graft-versus-leukemia depends on magnitude and diversity of the alloreactive T cell response. J Clin Invest 2017, 127, 517–529. [Google Scholar] [CrossRef]
- Casorati, G.; Locatelli, F.; Pagani, S.; Garavaglia, C.; Montini, E.; Lisini, D.; Turin, I.; Rossi, F.; Dellabona, P.; Maccario, R.; et al. Bone marrow-resident memory T cells survive pretransplant chemotherapy and contribute to early immune reconstitution of patients with acute myeloid leukemia given mafosfamide-purged autologous bone marrow transplantation. Exp Hematol 2005, 33, 212–218. [Google Scholar] [CrossRef] [PubMed]
- Montagna, D.; Maccario, R.; Locatelli, F.; Montini, E.; Pagani, S.; Bonetti, F.; Daudt, L.; Turin, I.; Lisini, D.; Garavaglia, C.; et al. Emergence of antitumor cytolytic T cells is associated with maintenance of hematologic remission in children with acute myeloid leukemia. Blood 2006, 108, 3843–3850. [Google Scholar] [CrossRef] [PubMed]
- Suessmuth, Y.; Mukherjee, R.; Watkins, B.; Koura, D.T.; Finstermeier, K.; Desmarais, C.; Stempora, L.; Horan, J.T.; Langston, A.; Qayed, M.; et al. CMV reactivation drives posttransplant T cell reconstitution and results in defects in the underlying TCRbeta repertoire. Blood 2015, 125, 3835–3850. [Google Scholar] [CrossRef] [PubMed]
- Link, C.S.; Eugster, A.; Heidenreich, F.; Rucker-Braun, E.; Schmiedgen, M.; Oelschlagel, U.; Kuhn, D.; Dietz, S.; Fuchs, Y.; Dahl, A.; et al. Abundant cytomegalovirus (CMV) reactive clonotypes in the CD8(+) T cell receptor alpha repertoire following allogeneic transplantation. Clin Exp Immunol 2016, 184, 389–402. [Google Scholar] [CrossRef]
- Godder, K.T.; Henslee-Downey, P.J.; Mehta, J.; Park, B.S.; Chiang, K.Y.; Abhyankar, S.; Lamb, L.S. Long term disease-free survival in acute leukemia patients recovering with increased gammadelta T cells after partially mismatched related donor bone marrow transplantation. Bone Marrow Transplant 2007, 39, 751–757. [Google Scholar] [CrossRef]
- Perko, R.; Kang, G.; Sunkara, A.; Leung, W.; Thomas, P.G.; Dallas, M.H. Gamma delta T cell reconstitution is associated with fewer infections and improved event-free survival after hematopoietic stem cell transplantation for pediatric leukemia. Biol Blood Marrow Transplant 2015, 21, 130–136. [Google Scholar] [CrossRef]
- Arruda, L.C.M.; Gaballa, A.; Uhlin, M. Graft gammadelta TCR Sequencing Identifies Public Clonotypes Associated with Hematopoietic Stem Cell Transplantation Efficacy in Acute Myeloid Leukemia Patients and Unravels Cytomegalovirus Impact on Repertoire Distribution. J Immunol 2019, 202, 1859–1870. [Google Scholar] [CrossRef]
- Chiffelle, J.; Genolet, R.; Perez, M.A.; Coukos, G.; Zoete, V.; Harari, A. T cell repertoire analysis and metrics of diversity and clonality. Curr Opin Biotechnol 2020, 65, 284–295. [Google Scholar] [CrossRef]
| Metric | Definition |
| Density | Proportion of T cells within a specific area |
| Richness | Number of unique TCR sequences in a sample |
| Evenness | Distribution spectrum of TCR sequences |
| Diversity | Richness and evenness of clonotypes in a sample |
| Clonality | Proliferation of specific clonotypes within a sample |
| Shannon entropy | A measure of TCR diversity that accounts for both the richness and evenness of clonotypes within a sample (higher values indicate greater diversity) |
| (Gini-) Simpson index | A measure of TCR diversity reflecting the probability that two randomly selected clonotypes from a sample are different (higher values represent greater diversity) |
| Jaccard index | TCR overlap between samples |
| Morisita index | Frequency of shared TCRs between samples |
| Public clonotype | Clonotypes shared among samples |
| Private clonotype | Clonotypes unique to specific samples |
| CPK (CDR3s per kilo of TCR reads) | An estimation of TCR diversity in “bulk” NGS data (higher values represent greater diversity) |
| Normalized unique clone count | An estimation of TCR diversity in “bulk” NGS data (higher values represent greater diversity) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





