Submitted:
13 January 2025
Posted:
14 January 2025
You are already at the latest version
Abstract
Keywords:
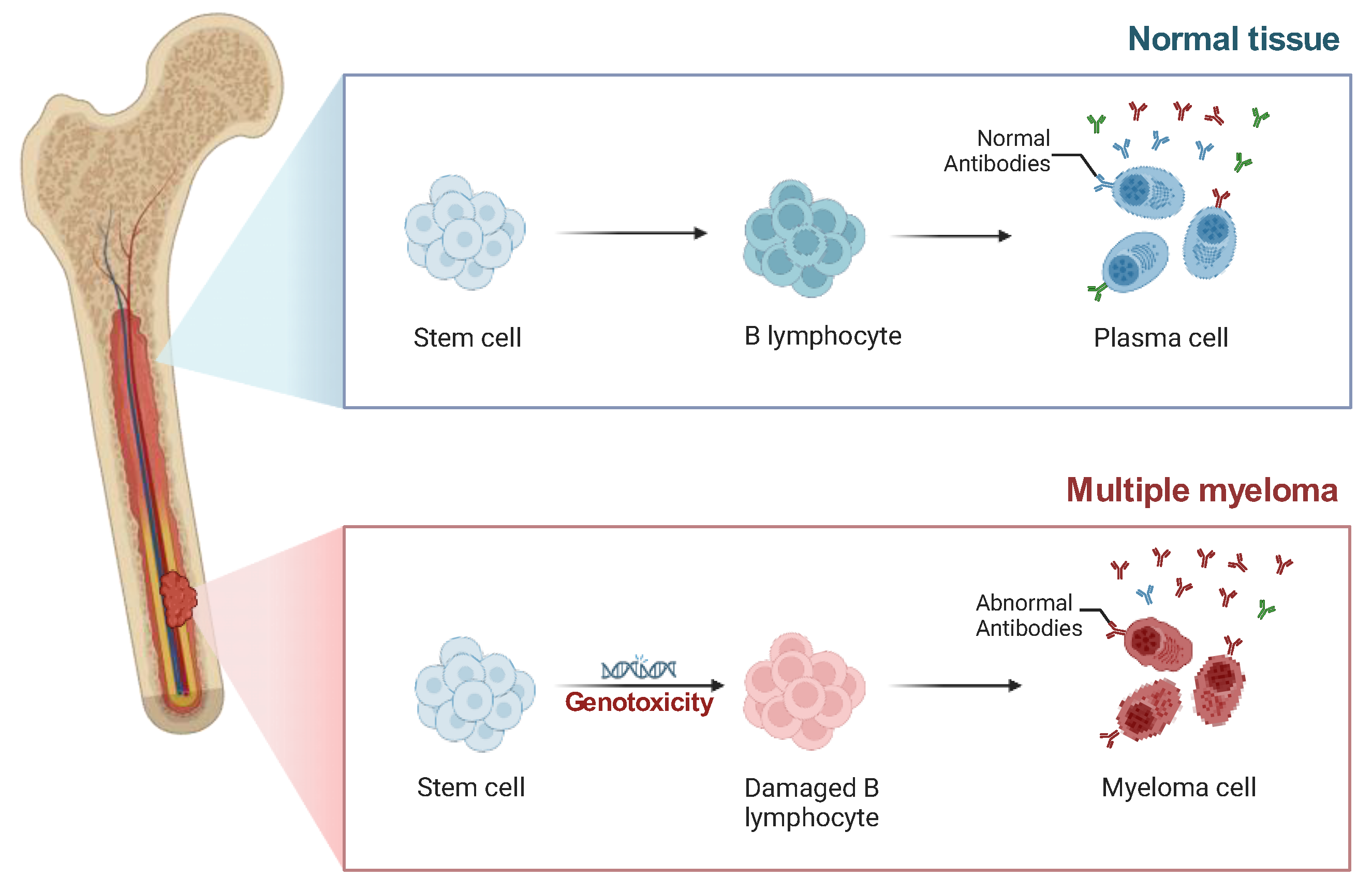
Introduction
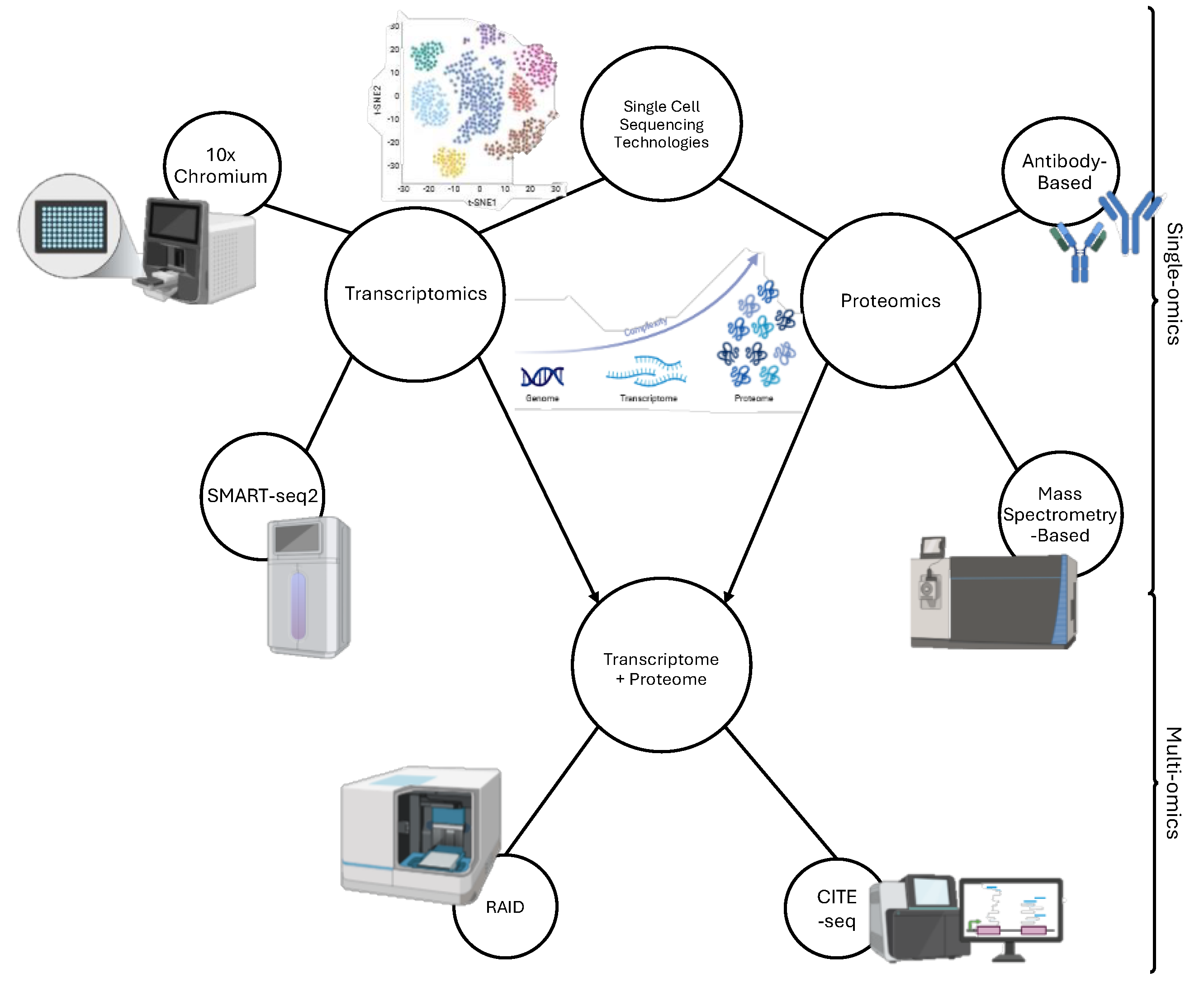
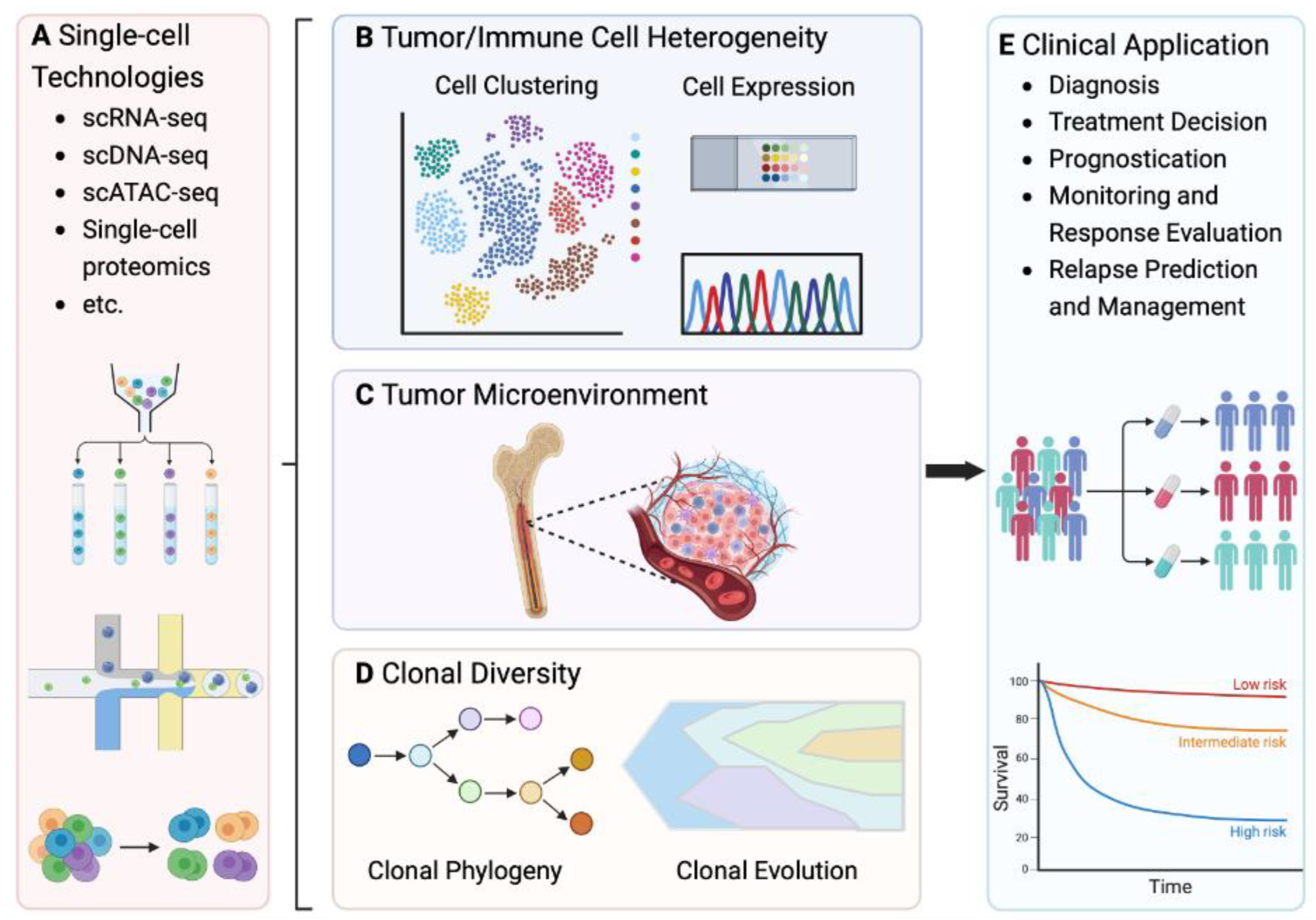
Single Cell Analysis Technologies in Mm
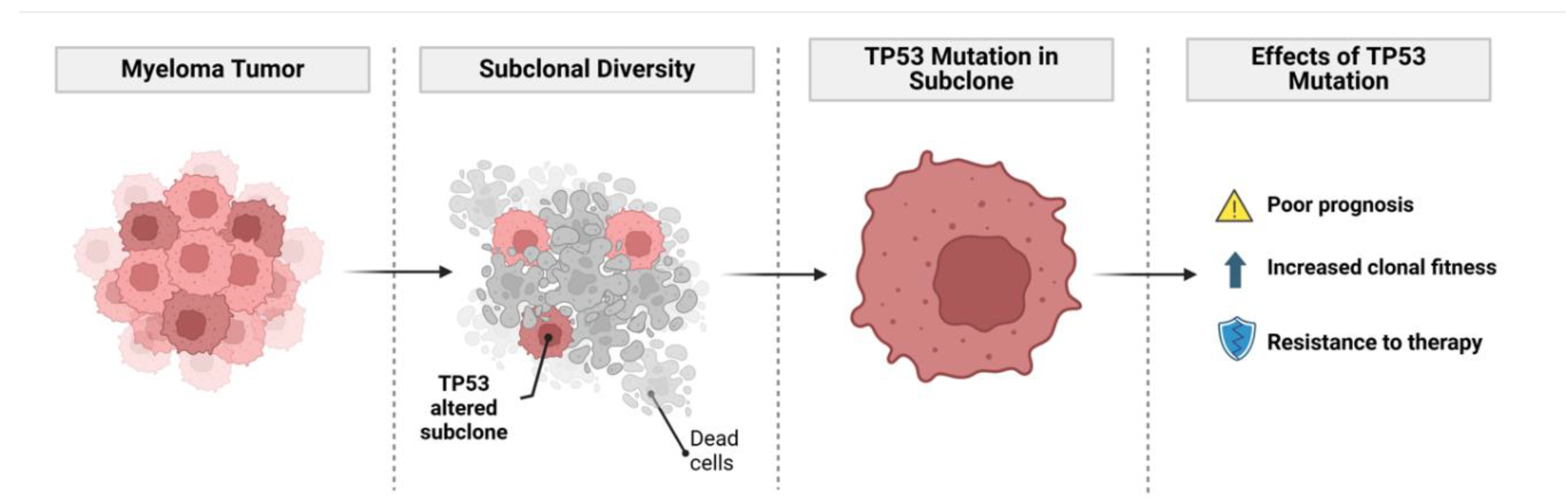
Subclonality and Tumor Evolution
Genomic Instability Drives Subclonal Diversity in MM
Tumor Evolution in Myeloma
Interplay Between Subclones and the Microenviroment
Subclone Competition
Subclone Cooperation
Inter-Subclone Communication
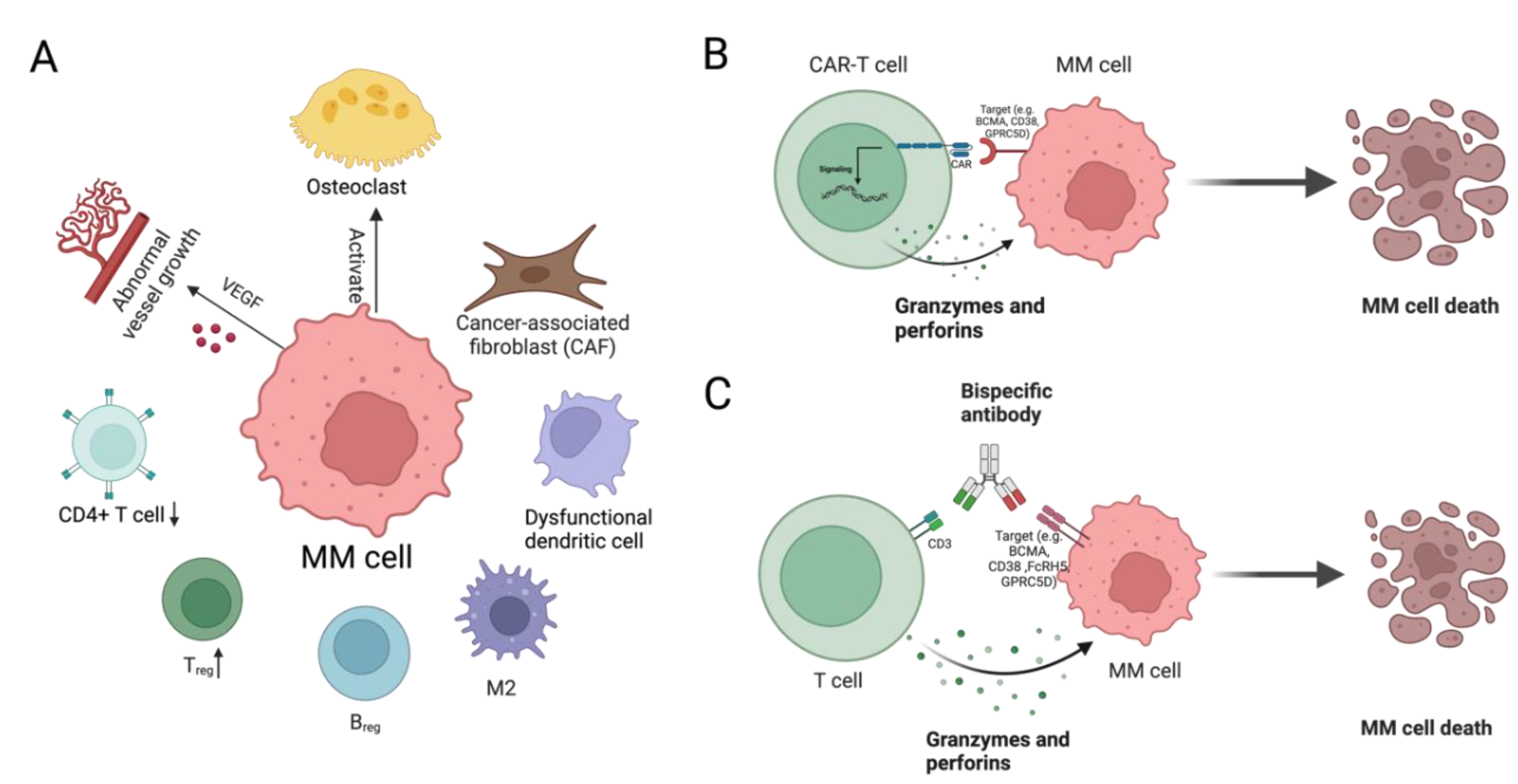
Subclone and Microenvironment Interactions
Immunotherapies
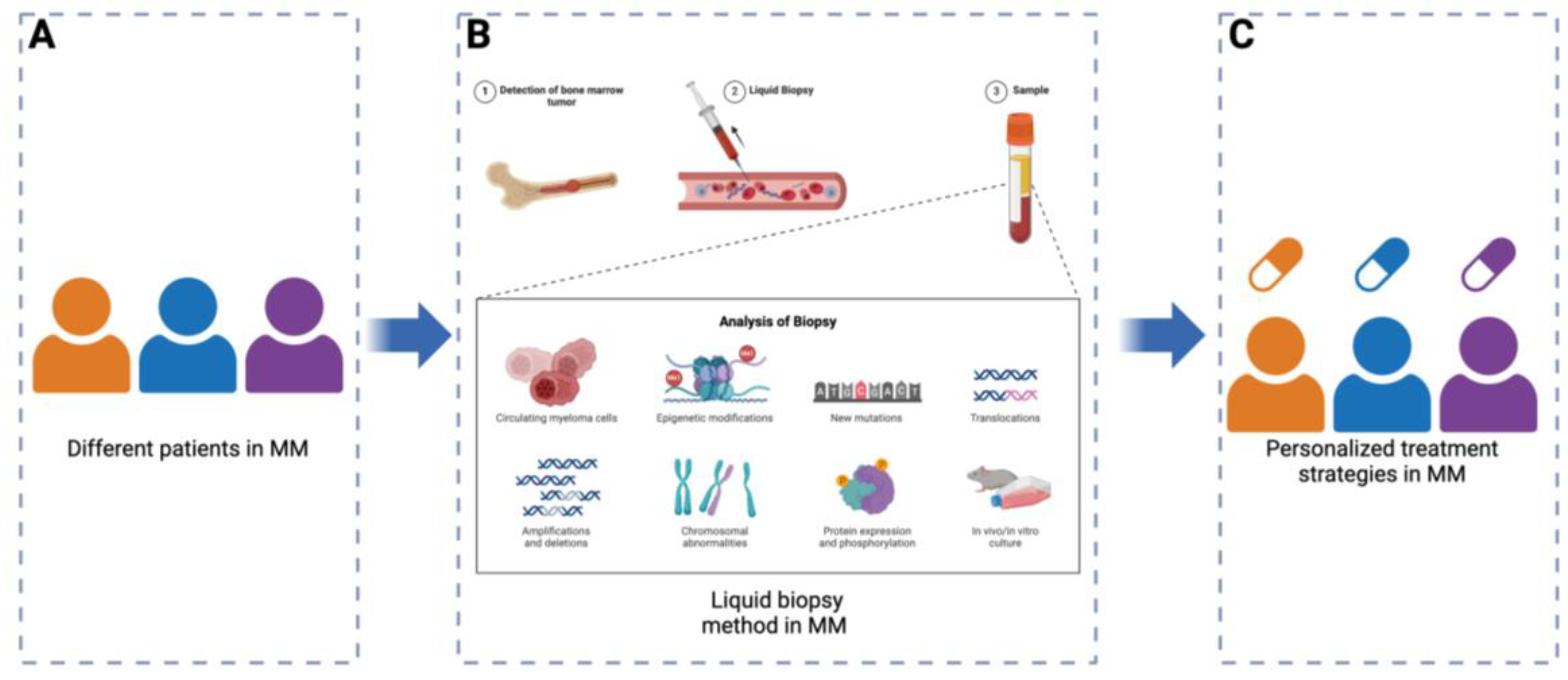
Clinical Utility of Single Cell Technologies in the Myeloma
Discussion
References
- Lannes, R.; Samur, M.; Perrot, A.; Mazzotti, C.; Divoux, M.; Cazaubiel, T.; Leleu, X.; Schavgoulidze, A.; Chretien, M.-L.; Manier, S.; et al. In Multiple Myeloma, High-Risk Secondary Genetic Events Observed at Relapse Are Present From Diagnosis in Tiny, Undetectable Subclonal Populations. Journal of Clinical Oncology 2023, 41, 1695–1702. [Google Scholar] [CrossRef] [PubMed]
- Cowan, A.J.; Green, D.J.; Kwok, M.; Lee, S.; Coffey, D.G.; Holmberg, L.A.; Tuazon, S.; Gopal, A.K.; Libby, E.N. Diagnosis and Management of Multiple Myeloma: A Review. JAMA 2022, 327, 464–477. [Google Scholar] [CrossRef] [PubMed]
- Alexander, D.D.; Mink, P.J.; Adami, H.-O.; Cole, P.; Mandel, J.S.; Oken, M.M.; Trichopoulos, D. Multiple myeloma: A review of the epidemiologic literature. International Journal of Cancer 2007, 120, 40–61. [Google Scholar] [CrossRef]
- Bergsagel, P.L.; Kuehl, W.M. Chromosome translocations in multiple myeloma. Oncogene 2001, 20, 5611–5622. [Google Scholar] [CrossRef]
- Institute, N.C. Cancer Stat Facts: Myeloma. 2024.
- Nightingale, B.; Decker, M.; Ryan, R.; Kaczmarczyk, K.; Jandir, P.; Waykole, T.; Ashkar, R.; Harmon, G.; Mathur, A.; Levitt, M. Multiple Myeloma: A Review of the Literature and a Case Report Highlighting the Immunocompromised State of Myeloma Patients; 2024.
- Mohty, M.; Facon, T.; Malard, F.; Harousseau, J.L. A roadmap towards improving outcomes in multiple myeloma. Blood Cancer J 2024, 14, 135. [Google Scholar] [CrossRef]
- Dima, D.; Jiang, D.; Singh, D.J.; Hasipek, M.; Shah, H.S.; Ullah, F.; Khouri, J.; Maciejewski, J.P.; Jha, B.K. Multiple Myeloma Therapy: Emerging Trends and Challenges. Cancers (Basel) 2022, 14. [Google Scholar] [CrossRef]
- Gohil, S.H.; Iorgulescu, J.B.; Braun, D.A.; Keskin, D.B.; Livak, K.J. Applying high-dimensional single-cell technologies to the analysis of cancer immunotherapy. Nature Reviews Clinical Oncology 2021, 18, 244–256. [Google Scholar] [CrossRef]
- Ramsköld, D.; Luo, S.; Wang, Y.C.; Li, R.; Deng, Q.; Faridani, O.R.; Daniels, G.A.; Khrebtukova, I.; Loring, J.F.; Laurent, L.C.; et al. Full-length mRNA-Seq from single-cell levels of RNA and individual circulating tumor cells. Nat Biotechnol 2012, 30, 777–782. [Google Scholar] [CrossRef]
- Picelli, S.; Faridani, O.R.; Björklund, A.K.; Winberg, G.; Sagasser, S.; Sandberg, R. Full-length RNA-seq from single cells using Smart-seq2. Nat Protoc 2014, 9, 171–181. [Google Scholar] [CrossRef]
- Jang, J.S.; Li, Y.; Mitra, A.K.; Bi, L.; Abyzov, A.; van Wijnen, A.J.; Baughn, L.B.; Van Ness, B.; Rajkumar, V.; Kumar, S.; et al. Molecular signatures of multiple myeloma progression through single cell RNA-Seq. Blood Cancer J 2019, 9, 2. [Google Scholar] [CrossRef]
- Melchor, L.; Brioli, A.; Wardell, C.P.; Murison, A.; Potter, N.E.; Kaiser, M.F.; Fryer, R.A.; Johnson, D.C.; Begum, D.B.; Hulkki Wilson, S.; et al. Single-cell genetic analysis reveals the composition of initiating clones and phylogenetic patterns of branching and parallel evolution in myeloma. Leukemia 2014, 28, 1705–1715. [Google Scholar] [CrossRef] [PubMed]
- Cohen, Y.C.; Zada, M.; Wang, S.-Y.; Bornstein, C.; David, E.; Moshe, A.; Li, B.; Shlomi-Loubaton, S.; Gatt, M.E.; Gur, C.; et al. Identification of resistance pathways and therapeutic targets in relapsed multiple myeloma patients through single-cell sequencing. Nature Medicine 2021, 27, 491–503. [Google Scholar] [CrossRef] [PubMed]
- Ledergor, G.; Weiner, A.; Zada, M.; Wang, S.Y.; Cohen, Y.C.; Gatt, M.E.; Snir, N.; Magen, H.; Koren-Michowitz, M.; Herzog-Tzarfati, K.; et al. Single cell dissection of plasma cell heterogeneity in symptomatic and asymptomatic myeloma. Nat Med 2018, 24, 1867–1876. [Google Scholar] [CrossRef]
- Boiarsky, R.; Haradhvala, N.J.; Alberge, J.B.; Sklavenitis-Pistofidis, R.; Mouhieddine, T.H.; Zavidij, O.; Shih, M.C.; Firer, D.; Miller, M.; El-Khoury, H.; et al. Single cell characterization of myeloma and its precursor conditions reveals transcriptional signatures of early tumorigenesis. Nat Commun 2022, 13, 7040. [Google Scholar] [CrossRef]
- Cenzano, I.; Cócera, M.; Bantan, A.; Larrayoz, M.; Vilas-Zornoza, A.; San-Martin, P.; Aguirre-Ruiz, P.; Alignani, D.; Lopez, A.; Barrios, M.M.; et al. Transcriptional Remodeling of the Stromal and Endothelial Microenvironment in MGUS to Multiple Myeloma Progression. bioRxiv 2024. [Google Scholar] [CrossRef]
- Chen, M.; Wan, Y.; Li, X.; Xiang, J.; Chen, X.; Jiang, J.; Han, X.; Zhong, L.; Xiao, F.; Liu, J.; et al. Dynamic single-cell RNA-seq analysis reveals distinct tumor program associated with microenvironmental remodeling and drug sensitivity in multiple myeloma. Cell Biosci 2023, 13, 19. [Google Scholar] [CrossRef]
- Croucher, D.C.; Richards, L.M.; Tsofack, S.P.; Waller, D.; Li, Z.; Wei, E.N.; Huang, X.F.; Chesi, M.; Bergsagel, P.L.; Sebag, M.; et al. Longitudinal single-cell analysis of a myeloma mouse model identifies subclonal molecular programs associated with progression. Nature Communications 2021, 12, 6322. [Google Scholar] [CrossRef]
- Cui, J.; Li, X.; Deng, S.; Du, C.; Fan, H.; Yan, W.; Xu, J.; Li, X.; Yu, T.; Zhang, S.; et al. Identification of Therapy-Induced Clonal Evolution and Resistance Pathways in Minimal Residual Clones in Multiple Myeloma through Single-Cell Sequencing. Clin Cancer Res 2024, 30, 3919–3936. [Google Scholar] [CrossRef] [PubMed]
- Dang, M.; Lee, H.C.; Bansal, H.; Acevedo-Calado, M.J.; Qin, L.; Tan, W.; Moreno Rueda, L.Y.; Berrios, D.; Pasvolsky, O.; Gaballa, M.R.; et al. Multi-Omic Single-Cell Characterization of Paired and Serial Samples in Responding and Non-Responding Patients Receiving BCMA Bispecific Antibody Therapy for Multiple Myeloma. Blood 2024, 144, 3272–3272. [Google Scholar] [CrossRef]
- Dang, M.; Wang, R.; Lee, H.C.; Patel, K.K.; Becnel, M.R.; Han, G.; Thomas, S.K.; Hao, D.; Chu, Y.; Weber, D.M.; et al. Single cell clonotypic and transcriptional evolution of multiple myeloma precursor disease. Cancer Cell 2023, 41, 1032–1047. [Google Scholar] [CrossRef]
- de Jong, M.M.E.; Kellermayer, Z.; Papazian, N.; Tahri, S.; Hofste Op Bruinink, D.; Hoogenboezem, R.; Sanders, M.A.; van de Woestijne, P.C.; Bos, P.K.; Khandanpour, C.; et al. The multiple myeloma microenvironment is defined by an inflammatory stromal cell landscape. Nat Immunol 2021, 22, 769–780. [Google Scholar] [CrossRef]
- He, H.; Li, Z.; Lu, J.; Qiang, W.; Jiang, S.; Xu, Y.; Fu, W.; Zhai, X.; Zhou, L.; Qian, M.; et al. Single-cell RNA-seq reveals clonal diversity and prognostic genes of relapsed multiple myeloma. Clin Transl Med 2022, 12, e757. [Google Scholar] [CrossRef] [PubMed]
- John, L.; Poos, A.M.; Brobeil, A.; Schinke, C.; Huhn, S.; Prokoph, N.; Lutz, R.; Wagner, B.; Zangari, M.; Tirier, S.M.; et al. Resolving the spatial architecture of myeloma and its microenvironment at the single-cell level. Nat Commun 2023, 14, 5011. [Google Scholar] [CrossRef] [PubMed]
- Johnson, T.S.; Sudha, P.; Liu, E.; Becker, N.; Robertson, S.; Blaney, P.; Morgan, G.; Chopra, V.S.; Dos Santos, C.; Nixon, M.; et al. 1q amplification and PHF19 expressing high-risk cells are associated with relapsed/refractory multiple myeloma. Nat Commun 2024, 15, 4144. [Google Scholar] [CrossRef] [PubMed]
- Kumar, H.; Mazumder, S.; Chakravarti, S.; Sharma, N.; Mukherjee, U.K.; Kumar, S.; Baughn, L.B.; Van Ness, B.G.; Mitra, A.K. secDrug: a pipeline to discover novel drug combinations to kill drug-resistant multiple myeloma cells using a greedy set cover algorithm and single-cell multi-omics. Blood Cancer Journal 2022, 12, 39. [Google Scholar] [CrossRef]
- Lannes, R.; Samur, M.; Perrot, A.; Mazzotti, C.; Divoux, M.; Cazaubiel, T.; Leleu, X.; Schavgoulidze, A.; Chretien, M.L.; Manier, S.; et al. In Multiple Myeloma, High-Risk Secondary Genetic Events Observed at Relapse Are Present From Diagnosis in Tiny, Undetectable Subclonal Populations. J Clin Oncol 2023, 41, 1695–1702. [Google Scholar] [CrossRef]
- Larrayoz, M.; Arriazu, E.; Zabaleta, A.; Roncal, C.; Tamayo, I.; Llopiz, D.; Jimenez, M.; Castro, C.; Celay, J.; Vicente, C. An IFNγ-Mediated Immune Inflamed Microenvironment State in Multiple Myeloma with TP53 Loss That Can be Therapeutically Exploited. Blood 2024, 144, 670. [Google Scholar] [CrossRef]
- Liang, Y.; He, H.; Wang, W.; Wang, H.; Mo, S.; Fu, R.; Liu, X.; Song, Q.; Xia, Z.; Wang, L. Malignant clonal evolution drives multiple myeloma cellular ecological diversity and microenvironment reprogramming. Mol Cancer 2022, 21, 182. [Google Scholar] [CrossRef]
- Liu, R.; Gao, Q.; Foltz, S.M.; Fowles, J.S.; Yao, L.; Wang, J.T.; Cao, S.; Sun, H.; Wendl, M.C.; Sethuraman, S.; et al. Co-evolution of tumor and immune cells during progression of multiple myeloma. Nature Communications 2021, 12, 2559. [Google Scholar] [CrossRef]
- Pilcher, W.; Thomas, B.E.; Bhasin, S.S.; Jayasinghe, R.G.; Yao, L.; Gonzalez-Kozlova, E.; Dasari, S.; Kim-Schulze, S.; Rahman, A.; Patton, J.; et al. Cross center single-cell RNA sequencing study of the immune microenvironment in rapid progressing multiple myeloma. NPJ Genom Med 2023, 8, 3. [Google Scholar] [CrossRef]
- Poos, A.M.; Prokoph, N.; Przybilla, M.J.; Mallm, J.P.; Steiger, S.; Seufert, I.; John, L.; Tirier, S.M.; Bauer, K.; Baumann, A.; et al. Resolving therapy resistance mechanisms in multiple myeloma by multiomics subclone analysis. Blood 2023, 142, 1633–1646. [Google Scholar] [CrossRef] [PubMed]
- Rade, M.; Grieb, N.; Weiss, R.; Sia, J.; Fischer, L.; Born, P.; Boldt, A.; Fricke, S.; Franz, P.; Scolnick, J.; et al. Single-cell multiomic dissection of response and resistance to chimeric antigen receptor T cells against BCMA in relapsed multiple myeloma. Nat Cancer 2024, 5, 1318–1333. [Google Scholar] [CrossRef]
- Tirier, S.M.; Mallm, J.P.; Steiger, S.; Poos, A.M.; Awwad, M.H.S.; Giesen, N.; Casiraghi, N.; Susak, H.; Bauer, K.; Baumann, A.; et al. Subclone-specific microenvironmental impact and drug response in refractory multiple myeloma revealed by single-cell transcriptomics. Nat Commun 2021, 12, 6960. [Google Scholar] [CrossRef] [PubMed]
- Verheye, E.; Kancheva, D.; Satilmis, H.; Vandewalle, N.; Fan, R.; Bardet, P.M.R.; Clappaert, E.J.; Verstaen, K.; De Becker, A.; Vanderkerken, K.; et al. A single-cell transcriptomic map of the murine and human multiple myeloma immune microenvironment across disease stages. J Hematol Oncol 2024, 17, 107. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Peng, Y.; Yang, C.; Xiong, D.; Wang, Z.; Peng, H.; Wu, X.; Xiao, X.; Liu, J. Single-cell sequencing analysis of multiple myeloma heterogeneity and identification of new theranostic targets. Cell Death Dis 2024, 15, 672. [Google Scholar] [CrossRef]
- Yao, L.; Wang, J.T.; Jayasinghe, R.G.; O'Neal, J.; Tsai, C.F.; Rettig, M.P.; Song, Y.; Liu, R.; Zhao, Y.; Ibrahim, O.M.; et al. Single-Cell Discovery and Multiomic Characterization of Therapeutic Targets in Multiple Myeloma. Cancer Res 2023, 83, 1214–1233. [Google Scholar] [CrossRef]
- Zavidij, O.; Haradhvala, N.J.; Mouhieddine, T.H.; Sklavenitis-Pistofidis, R.; Cai, S.; Reidy, M.; Rahmat, M.; Flaifel, A.; Ferland, B.; Su, N.K.; et al. Single-cell RNA sequencing reveals compromised immune microenvironment in precursor stages of multiple myeloma. Nature Cancer 2020, 1, 493–506. [Google Scholar] [CrossRef]
- Borsi, E.; Vigliotta, I.; Poletti, A.; Mazzocchetti, G.; Solli, V.; Zazzeroni, L.; Martello, M.; Armuzzi, S.; Taurisano, B.; Kanapari, A.; et al. Single-Cell DNA Sequencing Reveals an Evolutionary Pattern of CHIP in Transplant Eligible Multiple Myeloma Patients. Cells 2024, 13. [Google Scholar] [CrossRef]
- Koh, Y.; Park, C.; Cho, G.; Ryu, G.; Park, J.; Yoon, H.; Oh, Y.M.; Lee, C.; An, H.; Sun, C.-H. Impact of Clonal Hematopoiesis on the Carcinogenic Process of Multiple Myeloma. 2024.
- Avigan, J.I.; Gagler, D.C.; Ghamlouch, H.; Blaney, P.; Zhang, D.; Davies, F.E.; Morgan, G. Bone Marrow Fibroblasts at the Single-Cell Resolution Level in Multiple Myeloma. Blood 2024, 144, 4636–4636. [Google Scholar] [CrossRef]
- Botrugno, O.A.; Tonon, G. Genomic Instability and Replicative Stress in Multiple Myeloma: The Final Curtain? Cancers (Basel) 2021, 14. [Google Scholar] [CrossRef]
- Beksac, M.; Balli, S.; Akcora Yildiz, D. Drug Targeting of Genomic Instability in Multiple Myeloma. Front Genet 2020, 11, 228. [Google Scholar] [CrossRef] [PubMed]
- Rasche, L.; Schinke, C.; Maura, F.; Bauer, M.A.; Ashby, C.; Deshpande, S.; Poos, A.M.; Zangari, M.; Thanendrarajan, S.; Davies, F.E.; et al. The spatio-temporal evolution of multiple myeloma from baseline to relapse-refractory states. Nat Commun 2022, 13, 4517. [Google Scholar] [CrossRef] [PubMed]
- Popek-Marciniec, S.; Styk, W.; Wojcierowska-Litwin, M.; Chocholska, S.; Szudy-Szczyrek, A.; Samardakiewicz, M.; Swiderska-Kolacz, G.; Czerwik-Marcinkowska, J.; Zmorzynski, S. Association of Chromosome 17 Aneuploidy, TP53 Deletion, Expression and Its rs1042522 Variant with Multiple Myeloma Risk and Response to Thalidomide/Bortezomib Treatment. Cancers (Basel) 2023, 15. [Google Scholar] [CrossRef] [PubMed]
- Misund, K.; Hofste op Bruinink, D.; Coward, E.; Hoogenboezem, R.M.; Rustad, E.H.; Sanders, M.A.; Rye, M.; Sponaas, A.-M.; van der Holt, B.; Zweegman, S.; et al. Clonal evolution after treatment pressure in multiple myeloma: heterogenous genomic aberrations and transcriptomic convergence. Leukemia 2022, 36, 1887–1897. [Google Scholar] [CrossRef] [PubMed]
- Flynt, E.; Bisht, K.; Sridharan, V.; Ortiz, M.; Towfic, F.; Thakurta, A. Prognosis, Biology, and Targeting of TP53 Dysregulation in Multiple Myeloma. Cells 2020, 9. [Google Scholar] [CrossRef]
- Punke, A.P.; Waddell, J.A.; Solimando, D.A., Jr. Lenalidomide, Bortezomib, and Dexamethasone (RVD) Regimen for Multiple Myeloma. Hosp Pharm 2017, 52, 27–32. [Google Scholar] [CrossRef]
- Facon, T.; Lee, J.H.; Moreau, P.; Niesvizky, R.; Dimopoulos, M.; Hajek, R.; Pour, L.; Jurczyszyn, A.; Qiu, L.; Klippel, Z.; et al. Carfilzomib or bortezomib with melphalan-prednisone for transplant-ineligible patients with newly diagnosed multiple myeloma. Blood 2019, 133, 1953–1963. [Google Scholar] [CrossRef]
- Pawlyn, C.; Davies, F.; Cairns, D.; Striha, A.; Hockaday, A.; Kishore, B.; Garg, M.; Williams, C.; Karunanithi, K.; Lindsay, J.; et al. Quadruplet KCRD (Carfilzomib, Cyclophosphamide, Lenalidomide and Dexamethasone) Induction for Newly Diagnosed Myeloma Patients. Clinical Lymphoma, Myeloma and Leukemia 2019, 19, e2. [Google Scholar] [CrossRef]
- Venkatesh, P.; Bellman, P.; Shatnawi, S.; Shatnawi, Y.; Shaikh, H.; Strouse, C.S.; Atrash, S.; Hashmi, H.; Mushtaq, M.U.; Ahmed, N.; et al. VD-PACE As Salvage and Bridging Therapy to Transplant and Cellular Therapy in Triple Class Relapsed/Refractory Multiple Myeloma. Transplantation and Cellular Therapy, Official Publication of the American Society for Transplantation and Cellular Therapy 2024, 30, S389–S390. [Google Scholar] [CrossRef]
- Voorhees, P.M.; Sborov, D.W.; Laubach, J.; Kaufman, J.L.; Reeves, B.; Rodriguez, C.; Chari, A.; Silbermann, R.; Costa, L.J.; Anderson, L.D., Jr.; et al. Addition of daratumumab to lenalidomide, bortezomib, and dexamethasone for transplantation-eligible patients with newly diagnosed multiple myeloma (GRIFFIN): final analysis of an open-label, randomised, phase 2 trial. Lancet Haematol 2023, 10, e825–e837. [Google Scholar] [CrossRef]
- Jakubowiak, A.; Usmani, S.Z.; Krishnan, A.; Lonial, S.; Comenzo, R.L.; Wang, J.; de Boer, C.; Deraedt, W.; Weiss, B.M.; Schecter, J.M.; et al. Daratumumab Plus Carfilzomib, Lenalidomide, and Dexamethasone in Patients With Newly Diagnosed Multiple Myeloma. Clin Lymphoma Myeloma Leuk 2021, 21, 701–710. [Google Scholar] [CrossRef] [PubMed]
- Dingli, D.; Ailawadhi, S.; Bergsagel, P.L.; Buadi, F.K.; Dispenzieri, A.; Fonseca, R.; Gertz, M.A.; Gonsalves, W.I.; Hayman, S.R.; Kapoor, P.; et al. Therapy for Relapsed Multiple Myeloma: Guidelines From the Mayo Stratification for Myeloma and Risk-Adapted Therapy. Mayo Clinic Proceedings 2017, 92, 578–598. [Google Scholar] [CrossRef] [PubMed]
- Ciriello, G.; Magnani, L.; Aitken, S.J.; Akkari, L.; Behjati, S.; Hanahan, D.; Landau, D.A.; Lopez-Bigas, N.; Lupianez, D.G.; Marine, J.C.; et al. Cancer Evolution: A Multifaceted Affair. Cancer Discov 2024, 14, 36–48. [Google Scholar] [CrossRef]
- Yaccoby, S. The phenotypic plasticity of myeloma plasma cells as expressed by dedifferentiation into an immature, resilient, and apoptosis-resistant phenotype. Clin Cancer Res 2005, 11, 7599–7606. [Google Scholar] [CrossRef] [PubMed]
- Fonseca, R.; Blood, E.A.; Oken, M.M.; Kyle, R.A.; Dewald, G.W.; Bailey, R.J.; Van Wier, S.A.; Henderson, K.J.; Hoyer, J.D.; Harrington, D.; et al. Myeloma and the t(11;14)(q13;q32); evidence for a biologically defined unique subset of patients. Blood 2002, 99, 3735–3741. [Google Scholar] [CrossRef]
- Walker, B.A.; Wardell, C.P.; Johnson, D.C.; Kaiser, M.F.; Begum, D.B.; Dahir, N.B.; Ross, F.M.; Davies, F.E.; Gonzalez, D.; Morgan, G.J. Characterization of IGH locus breakpoints in multiple myeloma indicates a subset of translocations appear to occur in pregerminal center B cells. Blood 2013, 121, 3413–3419. [Google Scholar] [CrossRef]
- Sobh, A.; Encinas, E.; Patel, A.; Surapaneni, G.; Bonilla, E.; Kaestner, C.; Poullard, J.; Clerio, M.; Vasan, K.; Freeman, T.; et al. NSD2 drives t(4;14) myeloma cell dependence on adenylate kinase 2 by diverting one-carbon metabolism to the epigenome. Blood 2024, 144, 283–295. [Google Scholar] [CrossRef]
- Jiang, Y.; Zhang, C.; Lu, L.; Wang, X.; Liu, H.; Jiang, Y.; Hong, L.; Chen, Y.; Huang, H.; Guo, D. The Prognostic Role of Cyclin D1 in Multiple Myeloma: A Systematic Review and Meta-Analysis. Technol Cancer Res Treat 2022, 21, 15330338211065252. [Google Scholar] [CrossRef]
- Qiang, Y.W.; Ye, S.; Chen, Y.; Buros, A.F.; Edmonson, R.; van Rhee, F.; Barlogie, B.; Epstein, J.; Morgan, G.J.; Davies, F.E. MAF protein mediates innate resistance to proteasome inhibition therapy in multiple myeloma. Blood 2016, 128, 2919–2930. [Google Scholar] [CrossRef]
- Schavgoulidze, A.; Perrot, A.; Cazaubiel, T.; Leleu, X.; Montes, L.; Jacquet, C.; Belhadj, K.; Brechignac, S.; Frenzel, L.; Chalopin, T.; et al. Prognostic impact of translocation t(14;16) in multiple myeloma according to the presence of additional genetic lesions. Blood Cancer J 2023, 13, 160. [Google Scholar] [CrossRef]
- Cardona-Benavides, I.J.; de Ramon, C.; Gutierrez, N.C. Genetic Abnormalities in Multiple Myeloma: Prognostic and Therapeutic Implications. Cells 2021, 10. [Google Scholar] [CrossRef] [PubMed]
- Ross, F.M.; Chiecchio, L.; Dagrada, G.; Protheroe, R.K.; Stockley, D.M.; Harrison, C.J.; Cross, N.C.; Szubert, A.J.; Drayson, M.T.; Morgan, G.J.; et al. The t(14;20) is a poor prognostic factor in myeloma but is associated with long-term stable disease in monoclonal gammopathies of undetermined significance. Haematologica 2010, 95, 1221–1225. [Google Scholar] [CrossRef] [PubMed]
- Bal, S.; Kumar, S.K.; Fonseca, R.; Gay, F.; Hungria, V.T.; Dogan, A.; Costa, L.J. Multiple myeloma with t(11;14): unique biology and evolving landscape. Am J Cancer Res 2022, 12, 2950–2965. [Google Scholar] [PubMed]
- Caprio, C.; Sacco, A.; Giustini, V.; Roccaro, A.M. Epigenetic Aberrations in Multiple Myeloma. Cancers (Basel) 2020, 12. [Google Scholar] [CrossRef] [PubMed]
- Bolli, N.; Avet-Loiseau, H.; Wedge, D.C.; Van Loo, P.; Alexandrov, L.B.; Martincorena, I.; Dawson, K.J.; Iorio, F.; Nik-Zainal, S.; Bignell, G.R.; et al. Heterogeneity of genomic evolution and mutational profiles in multiple myeloma. Nat Commun 2014, 5, 2997. [Google Scholar] [CrossRef]
- Lohr, J.G.; Stojanov, P.; Carter, S.L.; Cruz-Gordillo, P.; Lawrence, M.S.; Auclair, D.; Sougnez, C.; Knoechel, B.; Gould, J.; Saksena, G.; et al. Widespread genetic heterogeneity in multiple myeloma: implications for targeted therapy. Cancer Cell 2014, 25, 91–101. [Google Scholar] [CrossRef]
- Forster, S.; Radpour, R.; Ochsenbein, A.F. Molecular and immunological mechanisms of clonal evolution in multiple myeloma. Front Immunol 2023, 14, 1243997. [Google Scholar] [CrossRef]
- Rasche, L.; Chavan, S.S.; Stephens, O.W.; Patel, P.H.; Tytarenko, R.; Ashby, C.; Bauer, M.; Stein, C.; Deshpande, S.; Wardell, C.; et al. Spatial genomic heterogeneity in multiple myeloma revealed by multi-region sequencing. Nat Commun 2017, 8, 268. [Google Scholar] [CrossRef]
- Walker, B.A.; Wardell, C.P.; Melchor, L.; Hulkki, S.; Potter, N.E.; Johnson, D.C.; Fenwick, K.; Kozarewa, I.; Gonzalez, D.; Lord, C.J.; et al. Intraclonal heterogeneity and distinct molecular mechanisms characterize the development of t(4;14) and t(11;14) myeloma. Blood 2012, 120, 1077–1086. [Google Scholar] [CrossRef]
- Schavgoulidze, A.; Cazaubiel, T.; Perrot, A.; Avet-Loiseau, H.; Corre, J. Multiple Myeloma: Heterogeneous in Every Way. Cancers (Basel) 2021, 13. [Google Scholar] [CrossRef]
- Medina, A.; Puig, N.; Flores-Montero, J.; Jimenez, C.; Sarasquete, M.E.; Garcia-Alvarez, M.; Prieto-Conde, I.; Chillon, C.; Alcoceba, M.; Gutierrez, N.C.; et al. Comparison of next-generation sequencing (NGS) and next-generation flow (NGF) for minimal residual disease (MRD) assessment in multiple myeloma. Blood Cancer J 2020, 10, 108. [Google Scholar] [CrossRef] [PubMed]
- Ghermezi, M.; Li, M.; Vardanyan, S.; Harutyunyan, N.M.; Gottlieb, J.; Berenson, A.; Spektor, T.M.; Andreu-Vieyra, C.; Petraki, S.; Sanchez, E.; et al. Serum B-cell maturation antigen: a novel biomarker to predict outcomes for multiple myeloma patients. Haematologica 2017, 102, 785–795. [Google Scholar] [CrossRef]
- Kadam Amare, P.; Nikalje Khasnis, S.; Hande, P.; Lele, H.; Wable, N.; Kaskar, S.; Nikam Gujar, N.; Gardi, N.; Prabhudesai, A.; Todi, K.; et al. Cytogenetic Abnormalities in Multiple Myeloma: Incidence, Prognostic Significance, and Geographic Heterogeneity in Indian and Western Populations. Cytogenet Genome Res 2022, 162, 529–540. [Google Scholar] [CrossRef] [PubMed]
- Sonneveld, P.; Avet-Loiseau, H.; Lonial, S.; Usmani, S.; Siegel, D.; Anderson, K.C.; Chng, W.J.; Moreau, P.; Attal, M.; Kyle, R.A.; et al. Treatment of multiple myeloma with high-risk cytogenetics: a consensus of the International Myeloma Working Group. Blood 2016, 127, 2955–2962. [Google Scholar] [CrossRef]
- Sanchez, E.; Li, M.; Kitto, A.; Li, J.; Wang, C.S.; Kirk, D.T.; Yellin, O.; Nichols, C.M.; Dreyer, M.P.; Ahles, C.P.; et al. Serum B-cell maturation antigen is elevated in multiple myeloma and correlates with disease status and survival. Br J Haematol 2012, 158, 727–738. [Google Scholar] [CrossRef]
- Avigan, Z.M.; Mitsiades, C.S.; Lagana, A. The role of 1q abnormalities in multiple myeloma: Genomic insights, clinical implications, and therapeutic challenges. Semin Hematol 2024. [Google Scholar] [CrossRef]
- Trasanidis, N.; Katsarou, A.; Ponnusamy, K.; Shen, Y.A.; Kostopoulos, I.V.; Bergonia, B.; Keren, K.; Reema, P.; Xiao, X.; Szydlo, R.M.; et al. Systems medicine dissection of chr1q-amp reveals a novel PBX1-FOXM1 axis for targeted therapy in multiple myeloma. Blood 2022, 139, 1939–1953. [Google Scholar] [CrossRef]
- Sudha, P.; Ahsan, A.; Ashby, C.; Kausar, T.; Khera, A.; Kazeroun, M.H.; Hsu, C.C.; Wang, L.; Fitzsimons, E.; Salminen, O.; et al. Myeloma Genome Project Panel is a Comprehensive Targeted Genomics Panel for Molecular Profiling of Patients with Multiple Myeloma. Clin Cancer Res 2022, 28, 2854–2864. [Google Scholar] [CrossRef]
- Merz, M.; Wei, L.; Hu, Q.; Merz, A.M.A.; Wang, J.; Belal, A.; Alberico, R.; Rondeau, C.; Celotto, K.; Block, A.W.; et al. Clinical Significance of Spatial Heterogeneity in Newly Diagnosed and Relapsed Multiple Myeloma. Blood 2021, 138, 1607–1607. [Google Scholar] [CrossRef]
- van de Donk, N. Sequencing multiple myeloma therapies with and after antibody therapies. Hematology Am Soc Hematol Educ Program 2020, 2020, 248–258. [Google Scholar] [CrossRef]
- Rasche, L.; Kortum, K.M.; Raab, M.S.; Weinhold, N. The Impact of Tumor Heterogeneity on Diagnostics and Novel Therapeutic Strategies in Multiple Myeloma. Int J Mol Sci 2019, 20. [Google Scholar] [CrossRef] [PubMed]
- Teo, W.Z.Y.; Ong, I.Y.E.; Tong, J.W.Y.; Ong, W.L.; Lin, A.; Song, F.; Tai, B.C.; Ooi, M.; Seokojo, C.Y.; Chen, Y.; et al. Response-Adapted Therapy for Newly Diagnosed Multiple Myeloma. Curr Hematol Malig Rep 2023, 18, 190–200. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Kaufman, J.L.; Gasparetto, C.; Mikhael, J.; Vij, R.; Pegourie, B.; Benboubker, L.; Facon, T.; Amiot, M.; Moreau, P.; et al. Efficacy of venetoclax as targeted therapy for relapsed/refractory t(11;14) multiple myeloma. Blood 2017, 130, 2401–2409. [Google Scholar] [CrossRef] [PubMed]
- Biancon, G.; Gimondi, S.; Vendramin, A.; Carniti, C.; Corradini, P. Noninvasive Molecular Monitoring in Multiple Myeloma Patients Using Cell-Free Tumor DNA: A Pilot Study. J Mol Diagn 2018, 20, 859–870. [Google Scholar] [CrossRef]
- Garces, J.J.; Cedena, M.T.; Puig, N.; Burgos, L.; Perez, J.J.; Cordon, L.; Flores-Montero, J.; Sanoja-Flores, L.; Calasanz, M.J.; Ortiol, A.; et al. Circulating Tumor Cells for the Staging of Patients With Newly Diagnosed Transplant-Eligible Multiple Myeloma. J Clin Oncol 2022, 40, 3151–3161. [Google Scholar] [CrossRef]
- Vrabel, D.; Sedlarikova, L.; Besse, L.; Rihova, L.; Bezdekova, R.; Almasi, M.; Kubaczkova, V.; Brozova, L.; Jarkovsky, J.; Plonkova, H.; et al. Dynamics of tumor-specific cfDNA in response to therapy in multiple myeloma patients. Eur J Haematol 2020, 104, 190–197. [Google Scholar] [CrossRef]
- Ferreira, B.; Caetano, J.; Barahona, F.; Lopes, R.; Carneiro, E.; Costa-Silva, B.; Joao, C. Liquid biopsies for multiple myeloma in a time of precision medicine. J Mol Med (Berl) 2020, 98, 513–525. [Google Scholar] [CrossRef]
- Li, S.; Zhang, E.; Cai, Z. Liquid biopsy by analysis of circulating myeloma cells and cell-free nucleic acids: a novel noninvasive approach of disease evaluation in multiple myeloma. Biomark Res 2023, 11, 27. [Google Scholar] [CrossRef]
- Auclair, D. , et al., A Next Generation Liquid Biopsy Approach for Multiple Myeloma. Blood, 2020. 136(Supplement 1): p. 33-33.
- Yee, A.J. and N. Raje, Minimal residual disease in multiple myeloma: why, when, where. Hematology, 2021. 2021(1): p. 37-45.
- Kaur, G.; Jena, L.; Gupta, R.; Farswan, A.; Gupta, A.; Sriram, K. Correlation of changes in subclonal architecture with progression in the MMRF CoMMpass study. Transl Oncol 2022, 23, 101472. [Google Scholar] [CrossRef]
- Ansari-Pour, N.; Samur, M.; Flynt, E.; Gooding, S.; Towfic, F.; Stong, N.; Estevez, M.O.; Mavrommatis, K.; Walker, B.; Morgan, G.; et al. Whole-genome analysis identifies novel drivers and high-risk double-hit events in relapsed/refractory myeloma. Blood 2023, 141, 620–633. [Google Scholar] [CrossRef]
- Jones, J.R.; Weinhold, N.; Ashby, C.; Walker, B.A.; Wardell, C.; Pawlyn, C.; Rasche, L.; Melchor, L.; Cairns, D.A.; Gregory, W.M.; et al. Clonal evolution in myeloma: the impact of maintenance lenalidomide and depth of response on the genetics and sub-clonal structure of relapsed disease in uniformly treated newly diagnosed patients. Haematologica 2019, 104, 1440–1450. [Google Scholar] [CrossRef] [PubMed]
- Lee, L.X.; Li, S.C. Hunting down the dominating subclone of cancer stem cells as a potential new therapeutic target in multiple myeloma: An artificial intelligence perspective. World J Stem Cells 2020, 12, 706–720. [Google Scholar] [CrossRef] [PubMed]
- Sandmann, S.; Karsch, K.; Bartel, P.; Exeler, R.; Brix, T.J.; Mai, E.K.; Varghese, J.; Lenz, G.; Khandanpour, C. The Role of Clonal Evolution on Progression, Blood Parameters, and Response to Therapy in Multiple Myeloma. Front Oncol 2022, 12, 919278. [Google Scholar] [CrossRef] [PubMed]
- Guo, G.; Raje, N.S.; Seifer, C.; Kloeber, J.; Isenhart, R.; Ha, G.; Yee, A.J.; O'Donnell, E.K.; Tai, Y.T.; Richardson, P.G.; et al. Genomic discovery and clonal tracking in multiple myeloma by cell-free DNA sequencing. Leukemia 2018, 32, 1838–1841. [Google Scholar] [CrossRef]
- Salomon-Perzynski, A.; Jamroziak, K.; Glodkowska-Mrowka, E. Clonal Evolution of Multiple Myeloma-Clinical and Diagnostic Implications. Diagnostics (Basel) 2021, 11. [Google Scholar] [CrossRef]
- Keats, J.J.; Chesi, M.; Egan, J.B.; Garbitt, V.M.; Palmer, S.E.; Braggio, E.; Van Wier, S.; Blackburn, P.R.; Baker, A.S.; Dispenzieri, A.; et al. Clonal competition with alternating dominance in multiple myeloma. Blood 2012, 120, 1067–1076. [Google Scholar] [CrossRef]
- Dutta, A.K.; Fink, J.L.; Grady, J.P.; Morgan, G.J.; Mullighan, C.G.; To, L.B.; Hewett, D.R.; Zannettino, A.C.W. Subclonal evolution in disease progression from MGUS/SMM to multiple myeloma is characterised by clonal stability. Leukemia 2019, 33, 457–468. [Google Scholar] [CrossRef]
- Hitzler, J.K.; Martinez-Valdez, H.; Bergsage, D.B.; Minden, M.D.; Messner, H.A. Role of Interleukin-6 in the Proliferation of Human Multiple Myeloma Cell Lines OCI-My 1 to 7 Established From Patients With Advanced Stage of the Disease. Blood 1991, 78, 1996–2004. [Google Scholar] [CrossRef]
- Kawano, M.; Hirano, T.; Matsuda, T.; Taga, T.; Horii, Y.; Iwato, K.; Asaoku, H.; Tang, B.; Tanabe, O.; Tanaka, H.; et al. Autocrine generation and requirement of BSF-2/IL-6 for human multiple myelomas. Nature 1988, 332, 83–85. [Google Scholar] [CrossRef]
- Bjorklund, C.C.; Baladandayuthapani, V.; Lin, H.Y.; Jones, R.J.; Kuiatse, I.; Wang, H.; Yang, J.; Shah, J.J.; Thomas, S.K.; Wang, M.; et al. Evidence of a role for CD44 and cell adhesion in mediating resistance to lenalidomide in multiple myeloma: therapeutic implications. Leukemia 2014, 28, 373–383. [Google Scholar] [CrossRef]
- Swamydas, M.; Murphy, E.V.; Ignatz-Hoover, J.J.; Malek, E.; Driscoll, J.J. Deciphering mechanisms of immune escape to inform immunotherapeutic strategies in multiple myeloma. Journal of Hematology & Oncology 2022, 15, 17. [Google Scholar] [CrossRef]
- Lopes, R.; Caetano, J.; Ferreira, B.; Barahona, F.; Carneiro, E.A.; João, C. The Immune Microenvironment in Multiple Myeloma: Friend or Foe? Cancers 2021, 13, 625. [Google Scholar] [CrossRef] [PubMed]
- Lu, K.; Wang, W.; Liu, Y.; Xie, C.; Liu, J.; Xing, L. Advancements in microenvironment-based therapies: transforming the landscape of multiple myeloma treatment. Front Oncol 2024, 14, 1413494. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Wang, W.; Wang, M.; Deng, J.; Sun, C.; Hu, Y.; Luo, S. Different evasion strategies in multiple myeloma. Front Immunol 2024, 15, 1346211. [Google Scholar] [CrossRef]
- Bhowmick, K.; von Suskil, M.; Al-Odat, O.S.; Elbezanti, W.O.; Jonnalagadda, S.C.; Budak-Alpdogan, T.; Pandey, M.K. Pathways to therapy resistance: The sheltering effect of the bone marrow microenvironment to multiple myeloma cells. Heliyon 2024, 10, e33091. [Google Scholar] [CrossRef]
- Edwards, C.M.; Zhuang, J.; Mundy, G.R. The pathogenesis of the bone disease of multiple myeloma. Bone 2008, 42, 1007–1013. [Google Scholar] [CrossRef]
- Kumar, S.; Witzig, T.E.; Timm, M.; Haug, J.; Wellik, L.; Fonseca, R.; Greipp, P.R.; Rajkumar, S.V. Expression of VEGF and its receptors by myeloma cells. Leukemia 2003, 17, 2025–2031. [Google Scholar] [CrossRef]
- Musolino, C.; Allegra, A.; Innao, V.; Allegra, A.G.; Pioggia, G.; Gangemi, S. Inflammatory and Anti-Inflammatory Equilibrium, Proliferative and Antiproliferative Balance: The Role of Cytokines in Multiple Myeloma. Mediators of Inflammation 2017, 2017, 1852517. [Google Scholar] [CrossRef]
- Brown, C.O.; Salem, K.; Wagner, B.A.; Bera, S.; Singh, N.; Tiwari, A.; Choudhury, A.; Buettner, G.R.; Goel, A. Interleukin-6 counteracts therapy-induced cellular oxidative stress in multiple myeloma by up-regulating manganese superoxide dismutase. Biochem J 2012, 444, 515–527. [Google Scholar] [CrossRef]
- Liu, C.-D.; Chang, C.-C.; Huang, W.-H. The perspectives of interleukin-10 in the pathogenesis and therapeutics of multiple myeloma. Tzu Chi Medical Journal 2021, 33, 257–262. [Google Scholar] [CrossRef]
- Zhang, X.; Zhang, H.; Lan, H.; Wu, J.; Xiao, Y. CAR-T cell therapy in multiple myeloma: Current limitations and potential strategies. Front Immunol 2023, 14, 1101495. [Google Scholar] [CrossRef] [PubMed]
- van de Donk, N.W.C.J.; Themeli, M.; Usmani, S.Z. Determinants of Response and Mechanisms of Resistance of CAR T-cell Therapy in Multiple Myeloma. Blood Cancer Discovery 2021, 2, 302–318. [Google Scholar] [CrossRef] [PubMed]
- Sheykhhasan, M.; Ahmadieh-Yazdi, A.; Vicidomini, R.; Poondla, N.; Tanzadehpanah, H.; Dirbaziyan, A.; Mahaki, H.; Manoochehri, H.; Kalhor, N.; Dama, P. CAR T therapies in multiple myeloma: unleashing the future. Cancer Gene Ther 2024, 31, 667–686. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Guo, H.; Han, L.; Song, Y.; Zhou, K. Dual-targeted CAR T-cell immunotherapies for hematological malignancies: latest updates from the 2023 ASH annual meeting. Exp Hematol Oncol 2024, 13, 25. [Google Scholar] [CrossRef]
- Alabanza, L.M.; Xiong, Y.; Vu, B.; Webster, B.; Wu, D.; Hu, P.; Zhu, Z.; Dropulic, B.; Dash, P.; Schneider, D. Armored BCMA CAR T Cells Eliminate Multiple Myeloma and Are Resistant to the Suppressive Effects of TGF-β. Front Immunol 2022, 13, 832645. [Google Scholar] [CrossRef]
- Devasia, A.J.; Chari, A.; Lancman, G. Bispecific antibodies in the treatment of multiple myeloma. Blood Cancer Journal 2024, 14, 158. [Google Scholar] [CrossRef]
- Cipkar, C.; Chen, C.; Trudel, S. Antibodies and bispecifics for multiple myeloma: effective effector therapy. Hematology 2022, 2022, 163–172. [Google Scholar] [CrossRef]
- Letouzé, E.; Moreau, P.; Munshi, N.; Samur, M.; Minvielle, S.; Touzeau, C. Mechanisms of resistance to bispecific T-cell engagers in multiple myeloma and their clinical implications. Blood Advances 2024, 8, 2952–2959. [Google Scholar] [CrossRef]
- Ledergor, G.; Weiner, A.; Zada, M.; Wang, S.-Y.; Cohen, Y.C.; Gatt, M.E.; Snir, N.; Magen, H.; Koren-Michowitz, M.; Herzog-Tzarfati, K.; et al. Single cell dissection of plasma cell heterogeneity in symptomatic and asymptomatic myeloma. Nature Medicine 2018, 24, 1867–1876. [Google Scholar] [CrossRef]
- Flanders, A.; Stetler-Stevenson, M.; Landgren, O. Minimal residual disease testing in multiple myeloma by flow cytometry: major heterogeneity. Blood 2013, 122, 1088–1089. [Google Scholar] [CrossRef]
- Corre, J.; Cleynen, A.; Robiou du Pont, S.; Buisson, L.; Bolli, N.; Attal, M.; Munshi, N.; Avet-Loiseau, H. Multiple myeloma clonal evolution in homogeneously treated patients. Leukemia 2018, 32, 2636–2647. [Google Scholar] [CrossRef] [PubMed]
- Baysoy, A.; Bai, Z.; Satija, R.; Fan, R. The technological landscape and applications of single-cell multi-omics. Nature Reviews Molecular Cell Biology 2023, 24, 695–713. [Google Scholar] [CrossRef] [PubMed]
- Heumos, L.; Schaar, A.C.; Lance, C.; Litinetskaya, A.; Drost, F.; Zappia, L.; Lücken, M.D.; Strobl, D.C.; Henao, J.; Curion, F.; et al. Best practices for single-cell analysis across modalities. Nature Reviews Genetics 2023, 24, 550–572. [Google Scholar] [CrossRef] [PubMed]
- Kourelis, T.; Bansal, R.; Berdeja, J.; Siegel, D.; Patel, K.; Mailankody, S.; Htut, M.; Shah, N.; Wong, S.W.; Sidana, S.; et al. Ethical Challenges with Multiple Myeloma BCMA Chimeric Antigen Receptor T Cell Slot Allocation: A Multi-Institution Experience. Transplant Cell Ther 2023, 29, 255–258. [Google Scholar] [CrossRef]
| Author | Date | Platform | DOI | REF |
|---|---|---|---|---|
| Melchor et al. | 8/28/2014 | Fluidigm multiplex qPCR | 10.1038/leu.2014.13 | [13] |
| Ledergor et al. | 12/24/2018 | MARS-seq | 10.1038/s41591-018-0269-2 | [15] |
| Jang et al. | 1/3/2019 | Fluidigm C1 + MAP-RSeq | 10.1038/s41408-018-0160-x | [12] |
| Zavidij et al. | 4/27/2020 | 10X Genomics RNA (3’) | 10.1038/s43018-020-0053-3 | [39] |
| Cohen et al. | 2/22/2021 | MARS-seq | 10.1038/s41591-021-01232-w | [14] |
| Liu et al. | 5/7/2021 | 10X Genomics RNA (3’ and 5’) | 10.1038/s41467-021-22804-x | [31] |
| de Jong et al. | 5/20/2021 | 10X Genomics RNA (3’) | 10.1038/s41590-021-00931-3 | [23] |
| Croucher et al. | 11/3/2021 | 10X Genomics RNA (NA) | 10.1038/s41467-021-26598-w | [19] |
| Tirier et al. | 11/19/2021 | 10X Genomics RNA (3’) | 10.1038/s41467-021-26951-z | [35] |
| Kumar et al. | 3/9/2022 | 10X Genomics RNA (NA) | 10.1038/s41408-022-00636-2 | [27] |
| He et al. | 3/12/2022 | 10X Genomics RNA (5’) + V(D)J | 10.1002/ctm2.757 | [24] |
| Liang et al. | 9/22/2022 | 10X Genomics RNA (3’) | 10.1186/s12943-022-01648-z | [30] |
| Boiarsky et al. | 11/17/2022 | 10X Genomics RNA (3’) | 10.1038/s41467-022-33944-z | [16] |
| Pilcher et al. | 1/26/2023 | 10X Genomics RNA (3’) and CITE-seq | 10.1038/s41525-022-00340-x | [32] |
| Chen et al. | 1/30/2023 | 10X Genomics RNA (3’) | 10.1186/s13578-023-00971-2 | [18] |
| Lannes et al. | 3/20/2023 | 10X Genomics DNA (NA) | 10.1200/JCO.21.01987 | [28] |
| Yao et al. | 4/14/2023 | 10X Genomics RNA (3’) | 10.1158/0008-5472.CAN-22-1769 | [38] |
| John et al. | 8/17/2023 | 10X Genomics RNA (5’) + V(D)J and 10X Genomics ATAC | 10.1038/s41467-023-40584-4 | [25] |
| Dang et al. | 6/12/202311/5/2024 | 10X Genomics RNA (5’) + V(D)J (TCR or BCR) | 10.1182/blood-2024-21008010.1016/j.ccell.2023.05.007 | [21][22] |
| Poos et al. | 11/9/2023 | 10X Genomics RNA (3’) and 10X Genomics ATAC | 10.1182/blood.2023019758 | [33] |
| Borsi et al. | 4/9/2024 | Mission Bio Tapestry DNA | 10.3390/cells13080657 | [40] |
| Rade et al. | 4/19/2024 | 10X Genomics RNA (5’) + V(D)J (TCR) + V(D)J (BCR) + Protein (Ab) | 10.1038/s43018-024-00763-8 | [34] |
| Cenzano et al. | 4/24/2024 | 10X Genomics RNA (3’) | 10.1101/2024.04.24.589777 | [17] |
| Johnson et al. | 5/16/2024 | 10X Genomics RNA (NA) + ATAC | 10.1038/s41467-024-48327-9 | [26] |
| Koh et al. | 7/18/2024 | Mission Bio Tapestry DNA + Protein | 10.21203/rs.3.rs-4672454/v1 | [41] |
| Cui et al. | 9/3/2024 | 10X Genomics RNA (3’) | 10.1158/1078-0432.CCR-24-0545 | [20] |
| Wang et al. | 9/14/2024 | 10X Genomics RNA (3’) | 10.1038/s41419-024-07027-4 | [37] |
| Larrayoz et al. | 11/5/2024 | 10X Genomics RNA (NA) and V(D)J (TCR) | 10.1182/blood-2024-194041 | [29] |
| Avigan et al. | 11/5/2024 | scRNA-seq | 10.1182/blood-2024-194277 | [42] |
| Verheye et al. | 11/7/2024 | 10X Genomics RNA (3’) | 10.1186/s13045-024-01629-3 | [36] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





