Submitted:
24 January 2025
Posted:
27 January 2025
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Computing Power
2.3.3. D Structure Modeling and MM2 Energy Minimization
2.4. Network Pharmacology and Target Receptor Identification
2.5. Molecular Docking Simulation
2.6. Molecular Dynamics (MD) Simulation
2.7. Molecular Mechanics/Poisson–Boltzmann Surface Area (MM/PBSA) Calculations
- ΔG_binding: the binding free energy associated with forming the protein-protein complex.
- ΔG_complex: the free energy of the fully solvated ligand-receptor protein-receptor complex.
- ΔG_ligand/protein: the free energy of ligand/protein in its solvated state when unbound.
- ΔG_receptor: the free energy of the receptor in its solvated state when unbound.
2.8. Pharmacophore Modeling and In-Silico Toxicity Assessment
3. Results
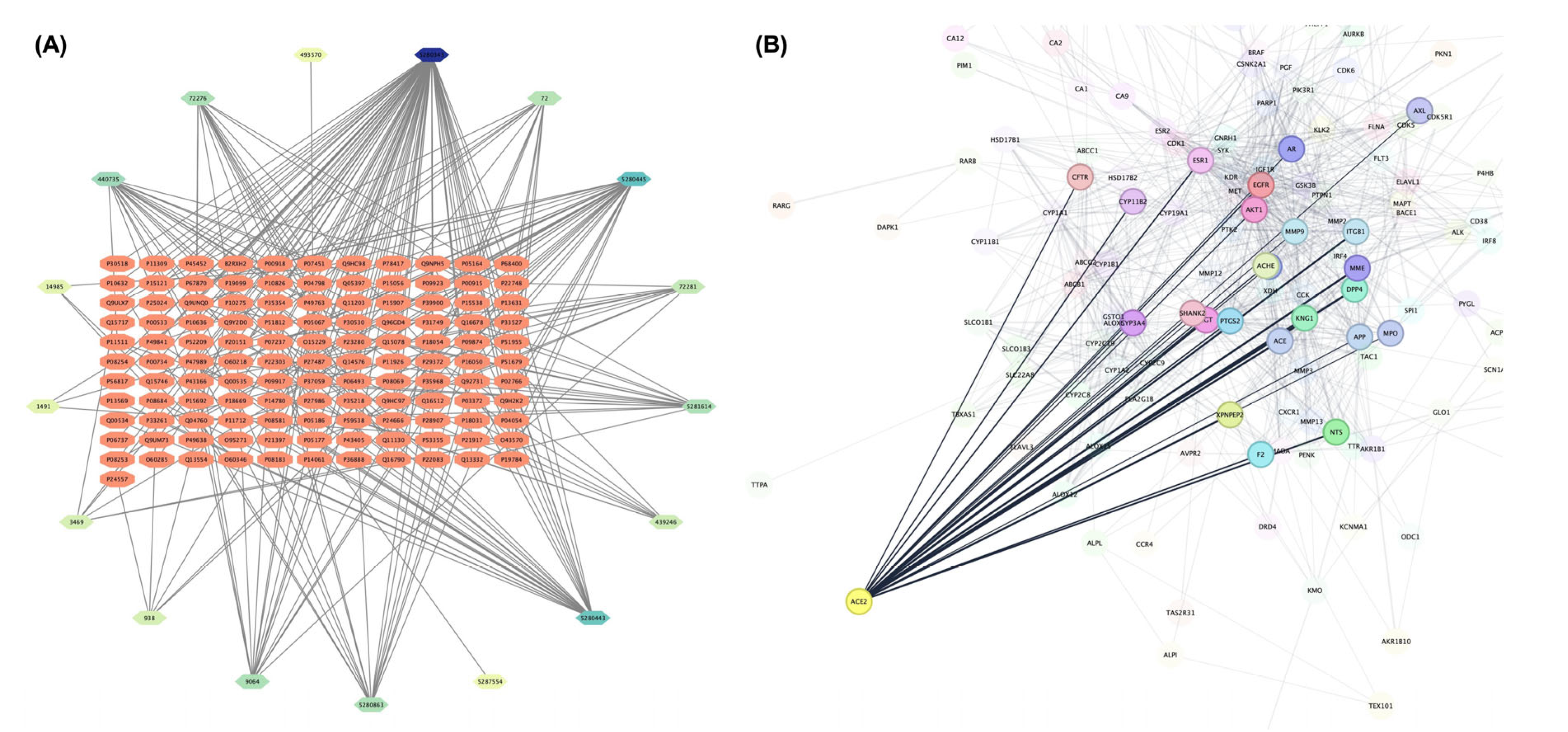
3.1. Identification of Drug-Target Interactions and Determination of Target Receptor
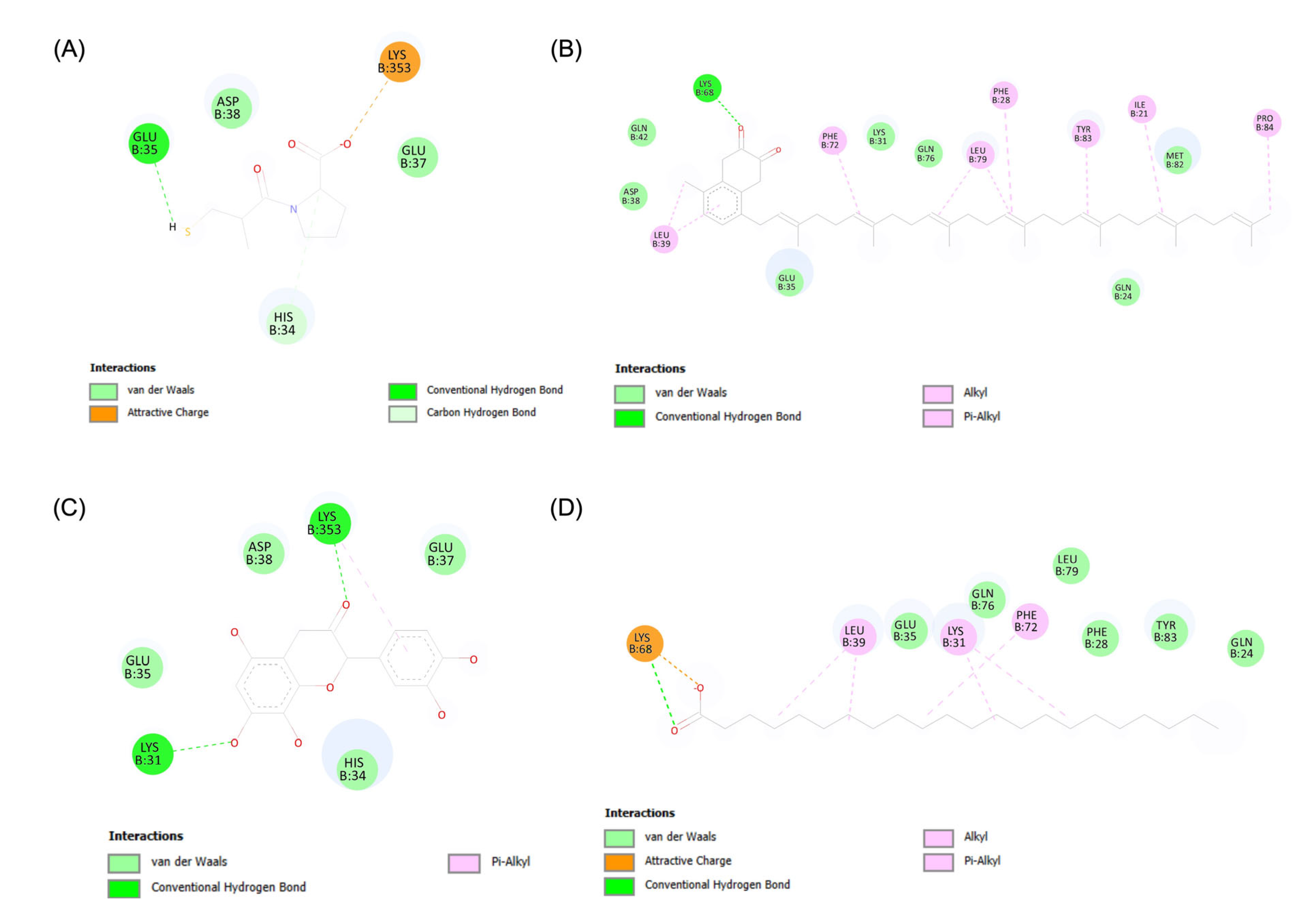
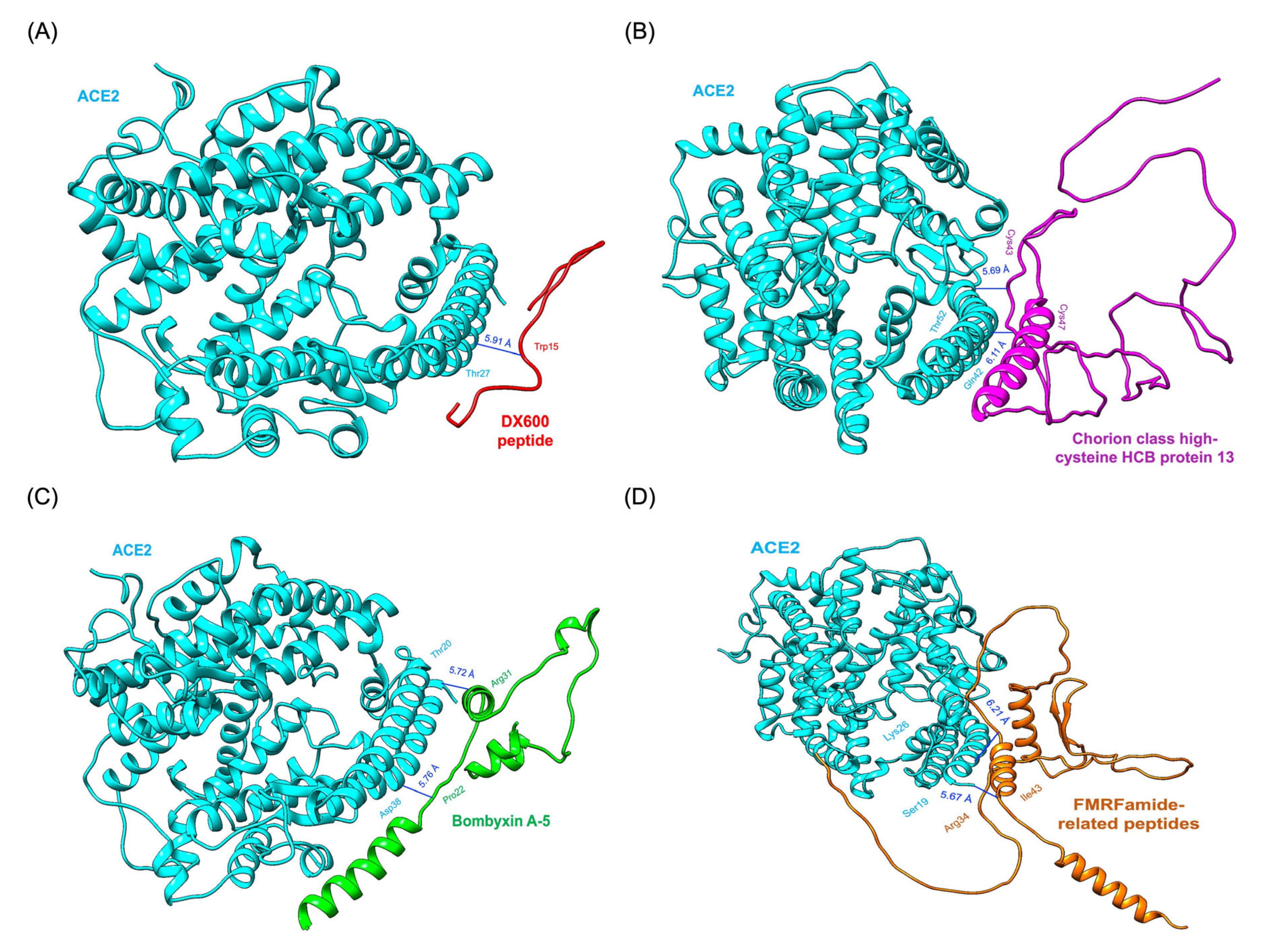
3.2. Analysis of Molecular Interactions through Molecular Docking Simulations
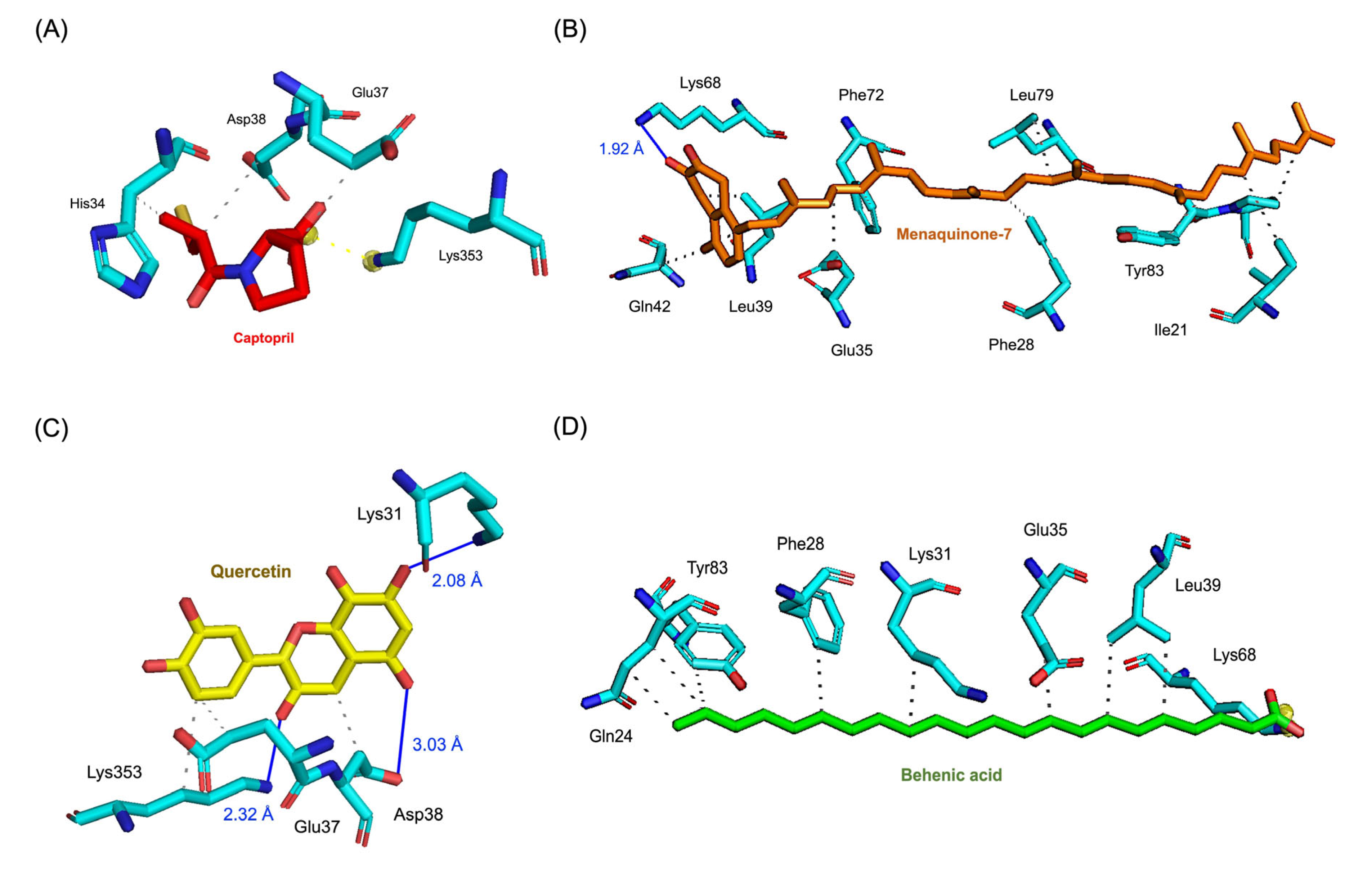
3.3. Evaluation of Molecular Dynamics: Stability, Interactions, and Binding Affinity
3.4. Pharmacophore Modeling and Toxicity Profile Assessment
4. Discussion
5. Limitations, Clinical Implications, and Future Works
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| ADORA2A | Adenosine A2A receptor |
| ACE2 | Angiotensin-converting enzyme 2 |
| BC | Betweenness centrality |
| CASTp | Computed atlas of surface topography of proteins |
| CC | Closeness centrality |
| COX2 | Cyclooxygenase 2 |
| CVDs | Cardiovascular diseases |
| DC | Degree centrality |
| DL | Drug-likeness |
| EC | Eigenvector centrality |
| GAFF2 | General amber force field |
| HADDOCK | High ambiguity driven protein-protein docking |
| HBA | Hydrogen bond acceptor |
| HBD | Hydrogen bond donor |
| ICs | Intermolecular contacts |
| IHD | Ischemic heart disease |
| MD | Molecular dynamics |
| MM/PBSA | Molecular mechanics/poisson-boltzmann surface area |
| MMP9 | Matrix metalloproteinase 9 |
| NIS | Non-interacting surface areas |
| NO | Nitric oxide |
| NOS | Nitric oxide synthase |
| NPT | Number of particles, pressure, and temperature |
| NVT | Number of particles, volume, and temperature |
| OB | Oral bioavailability |
| OPLS-AA/L | Optimized potentials for liquid simulations |
| PME | Particle mesh Ewald |
| PPIs | Protein-protein interactions |
| RMSD | Root mean square deviation |
| RMSF | Root mean square fluctuation |
| RoG | Radius of gyration |
| SEA | Similarity ensemble approach |
| SMILES | Simplified molecular input line entry system |
| SPCE | Single point charge extended |
| TC | Tanimoto coefficient |
References
- Roth, G.A. , et al., Global Burden of Cardiovascular Diseases and Risk Factors, 1990-2019: Update From the GBD 2019 Study. J Am Coll Cardiol, 2020. 76(25): p. 2982-3021.
- Di Cesare, M. , et al., The Heart of the World. Glob Heart, 2024. 19(1): p. 11.
- Balakumar, P., K. Maung-U, and G. Jagadeesh, Prevalence and prevention of cardiovascular disease and diabetes mellitus. Pharmacological Research, 2016. 113: p. 600-609.
- Pandey, R. , et al., A review study on blood in human coronary artery: Numerical approach. Computer Methods and Programs in Biomedicine, 2020. 187: p. 105243.
- Mortada, E.M. , Evidence-Based Complementary and Alternative Medicine in Current Medical Practice. Cureus, 2024. 16(1): p. e52041.
- Yuan, H. , et al., The Traditional Medicine and Modern Medicine from Natural Products. Molecules, 2016. 21(5).
- Rizvi, S.A.A. , et al., Introduction to Traditional Medicine and Their Role in Prevention and Treatment of Emerging and Re-Emerging Diseases. Biomolecules, 2022. 12(10).
- Ahmed, R. , et al., Bombyx mori Cocoon (Abresham): A potent Unani drug. International Journal of Unani and Integrative Medicine, 2022. 6: p. 33-36.
- Ali, M.M. and S.B. Arumugam, Effect of crude extract of Bombyx mori coccoons in hyperlipidemia and atherosclerosis. J Ayurveda Integr Med, 2011. 2(2): p. 72-8.
- Goyal, S. , et al., Cardioprotective effect of 'Khamira Abresham Hakim Arshad Wala' a unani formulation in isoproterenol-induced myocardial necrosis in rats. Exp Toxicol Pathol, 2010. 62(1): p. 61-74.
- Nazmi, A. , et al., Protective effects of 'Khamira Abresham Hakim Arshad Wala', a unani formulation against doxorubicin-induced cardiotoxicity and nephrotoxicity. Toxicology mechanisms and methods, 2010. 21: p. 41-7.
- Biganeh, H. , et al., Bombyx mori cocoon as a promising pharmacological agent: A review of ethnopharmacology, chemistry, and biological activities. Heliyon, 2022. 8(9): p. e10496.
- Kunz, R.I. , et al., Silkworm Sericin: Properties and Biomedical Applications. Biomed Res Int, 2016. 2016: p. 8175701.
- Yeruva, T. , et al., Profiling of nutrients and bioactive compounds in the pupae of silkworm, Bombyx mori. Food Chemistry Advances, 2023. 3: p. 100382.
- Zafar, M. and K. Al Samadani, Potential use of natural silk for bio-dental applications. Journal of Taibah University Medical Sciences, 2014. 9.
- Hu, L. , et al., Untargeted screening and differential analysis of bioactive compounds in male and female silkworm (Bombyx mori) pupae through Orbitrap Exploris mass spectrometry. Food Chemistry, 2025. 469: p. 142584.
- Saini, R.S. , et al., Dental biomaterials redefined: molecular docking and dynamics-driven dental resin composite optimization. BMC Oral Health, 2024. 24(1): p. 557.
- Saini, R.S. , et al., In silico assessment of biocompatibility and toxicity: molecular docking and dynamics simulation of PMMA-based dental materials for interim prosthetic restorations. Journal of Materials Science: Materials in Medicine, 2024. 35(1): p. 28.
- Chen, S., G. Fan, and J. Li, Improving completeness and accuracy of 3D point clouds by using deep learning for applications of digital twins to civil structures. Adv Eng Inform, 2023. 58: p. 1-13.
- Davis, E.M. , et al., SequencErr: measuring and suppressing sequencer errors in next-generation sequencing data. Genome Biol, 2021. 22(1): p. 1-18.
- Alotaiq, N. and D. Dermawan Evaluation of Structure Prediction and Molecular Docking Tools for Therapeutic Peptides in Clinical Use and Trials Targeting Coronary Artery Disease. International Journal of Molecular Sciences, 2025. 26. [CrossRef]
- Jumper, J. , et al., Highly accurate protein structure prediction with AlphaFold. Nature, 2021. 596(7873): p. 583-589.
- Ye, B. , et al., CASTpFold: Computed Atlas of Surface Topography of the universe of protein Folds. Nucleic Acids Research, 2024. 52(W1): p. W194-W199.
- Lan, J. , et al., Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature, 2020. 581(7807): p. 215-220.
- Sun, T. , et al., Network Pharmacology-Based Strategy Combined with Molecular Docking and in vitro Validation Study to Explore the Underlying Mechanism of Huo Luo Xiao Ling Dan in Treating Atherosclerosis. Drug Des Devel Ther, 2022. 16: p. 1621-1645.
- Zhang, M. , et al., Network Pharmacology Analysis of Bioactive Components and Mechanisms of Action of Qi Wei Wan Formula for Treating Non-Small Cell Lung Carcinoma. Natural Product Communications, 2022. 17: p. 1934578X2211202.
- Daina, A., O. Michielin, and V. Zoete, SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Scientific Reports, 2017. 7(1): p. 42717.
- Molsoft, ICM: Integrated Computational Microscope (ICM). 2024, Molsoft LLC: La Jolla, USA.
- Keiser, M.J. , et al., Relating protein pharmacology by ligand chemistry. Nat Biotechnol, 2007. 25(2): p. 197-206.
- Vilar, S. , et al., Computational Drug Target Screening through Protein Interaction Profiles. Sci Rep, 2016. 6: p. 36969.
- Rebhan, M. , et al., GeneCards: a novel functional genomics compendium with automated data mining and query reformulation support. Bioinformatics, 1998. 14(8): p. 656-664.
- Shannon, P. , et al., Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res, 2003. 13(11): p. 2498-504.
- Doncheva, N.T. , et al., Cytoscape StringApp: Network Analysis and Visualization of Proteomics Data. J Proteome Res, 2019. 18(2): p. 623-632.
- Szklarczyk, D. , et al., The STRING database in 2021: customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res, 2021. 49(D1): p. D605-d612.
- Laskowski, R.A. , et al., PDBsum: Structural summaries of PDB entries. Protein Sci, 2018. 27(1): p. 129-134.
- Guex, N. and M.C. Peitsch, SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis, 1997. 18(15): p. 2714-23.
- Darwish, M.M. , et al., In-depth in silico and in vitro screening of selected edible plants for identification of selective C-domain ACE-1 inhibitor and its synergistic effect with captopril. Food Bioscience, 2024. 59: p. 104115.
- Huang, L. , et al., Novel peptide inhibitors of angiotensin-converting enzyme 2. J Biol Chem, 2003. 278(18): p. 15532-40.
- Dominguez, C., R. Boelens, and A.M.J.J. Bonvin, HADDOCK: A Protein−Protein Docking Approach Based on Biochemical or Biophysical Information. Journal of the American Chemical Society, 2003. 125(7): p. 1731-1737.
- van Zundert, G.C.P. , et al., The HADDOCK2.2 Web Server: User-Friendly Integrative Modeling of Biomolecular Complexes. J Mol Biol, 2016. 428(4): p. 720-725.
- Pronk, S. , et al., GROMACS 4.5: a high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics, 2013. 29(7): p. 845-854.
- Robertson, M.J., J. Tirado-Rives, and W.L. Jorgensen, Improved Peptide and Protein Torsional Energetics with the OPLSAA Force Field. J Chem Theory Comput, 2015. 11(7): p. 3499-509.
- Yuet, P. and D. Blankschtein, Molecular Dynamics Simulation Study of Water Surfaces: Comparison of Flexible Water Models. The journal of physical chemistry. B, 2010. 114: p. 13786-13795.
- Schrödinger, The PyMOL Molecular Graphics System. 2020.
- BIOVIA, D.S. , BIOVIA Discovery Studio. 2024, Dassault Systèmes: San Diego, USA.
- Pettersen, E.F. , et al., UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem, 2004. 25(13): p. 1605-1612.
- Alotaiq, N., D. Dermawan, and N.E. Elwali, Leveraging Therapeutic Proteins and Peptides from Lumbricus Earthworms: Targeting SOCS2 E3 Ligase for Cardiovascular Therapy through Molecular Dynamics Simulations. International Journal of Molecular Sciences, 2024. 25(19): p. 10818.
- Valdés-Tresanco, M. , et al., gmx_MMPBSA: A New Tool to Perform End-State Free Energy Calculations with GROMACS. Journal of Chemical Theory and Computation, 2021. 17.
- Wolber, G. and T. Langer, LigandScout: 3-D pharmacophores derived from protein-bound ligands and their use as virtual screening filters. J Chem Inf Model, 2005. 45(1): p. 160-9.
- Sander, T. , et al., DataWarrior: an open-source program for chemistry aware data visualization and analysis. J Chem Inf Model, 2015. 55(2): p. 460-73.
- Sandoo, A. , et al., The endothelium and its role in regulating vascular tone. Open Cardiovasc Med J, 2010. 4: p. 302-12.
- Virdis, A. , et al., Cyclooxygenase-2 Inhibition Improves Vascular Endothelial Dysfunction in a Rat Model of Endotoxic Shock: Role of Inducible Nitric-Oxide Synthase and Oxidative Stress. The Journal of Pharmacology and Experimental Therapeutics, 2005. 312(3): p. 945-953.
- Mustafa, S.J. , et al., Adenosine receptors and the heart: role in regulation of coronary blood flow and cardiac electrophysiology. Handb Exp Pharmacol, 2009(193): p. 161-88.
- Wang, X. and R.A. Khalil, Matrix Metalloproteinases, Vascular Remodeling, and Vascular Disease. Adv Pharmacol, 2018. 81: p. 241-330.
- García-Llorca, A. , et al., Carbonic anhydrase, its inhibitors and vascular function. Front Mol Biosci, 2024. 11: p. 1338528.
- Kuriakose, J., A. C. Montezano, and R.M. Touyz, ACE2/Ang-(1-7)/Mas1 axis and the vascular system: vasoprotection to COVID-19-associated vascular disease. Clin Sci (Lond), 2021. 135(2): p. 387-407.
- Li, T. , et al., The Role of Matrix Metalloproteinase-9 in Atherosclerotic Plaque Instability. Mediators Inflamm, 2020. 2020: p. 3872367.
- Khayat, M.T. and M.A. Nayeem, The Role of Adenosine A(2A) Receptor, CYP450s, and PPARs in the Regulation of Vascular Tone. Biomed Res Int, 2017. 2017: p. 1720920.
- Yacouba Moukeila, M.B. , et al., Adenosine 2 receptor regulates autophagy and apoptosis to alleviate ischemia reperfusion injury in type 2 diabetes via IRE-1 signaling. BMC Cardiovascular Disorders, 2023. 23(1): p. 154.
- de Chaves, E.P. and V. Narayanaswami, Apolipoprotein E and cholesterol in aging and disease in the brain. Future Lipidol, 2008. 3(5): p. 505-530.
- Kalmijn, S. , et al., Cerebrovascular disease, the apolipoprotein e4 allele, and cognitive decline in a community-based study of elderly men. Stroke, 1996. 27(12): p. 2230-5.
- Van Langendonckt, A., F. Casanas-Roux, and J. Donnez, Oxidative stress and peritoneal endometriosis. Fertility and Sterility, 2002. 77(5): p. 861-870.
- Tran, N. , et al., Endothelial Nitric Oxide Synthase (eNOS) and the Cardiovascular System: in Physiology and in Disease States. Am J Biomed Sci Res, 2022. 15(2): p. 153-177.
- Kumar, G., S. K. Dey, and S. Kundu, Functional implications of vascular endothelium in regulation of endothelial nitric oxide synthesis to control blood pressure and cardiac functions. Life Sciences, 2020. 259: p. 118377.
- Doni Dermawan, F.A. , Nasr Eldin Elwali, Nasser Alotaiq, Therapeutic potential of earthworm-derived proteins: Targeting NEDD4 for cardiovascular disease intervention. Vol. Volume: 15. 2024: Issue: 1. 216-232.
- Lazniewski, M. , et al., Drug repurposing for identification of potential spike inhibitors for SARS-CoV-2 using molecular docking and molecular dynamics simulations. Methods, 2022. 203: p. 498-510.
- Zhang, W. , et al., Research progress of quercetin in cardiovascular disease. Front Cardiovasc Med, 2023. 10: p. 1203713.
- Papakyriakopoulou, P. , et al., Potential Pharmaceutical Applications of Quercetin in Cardiovascular Diseases. Pharmaceuticals (Basel), 2022. 15(8).
- Aghababaei, F. and M. Hadidi, Recent Advances in Potential Health Benefits of Quercetin. Pharmaceuticals, 2023. 16(7): p. 1020.
- Ho, W.Y. , et al., Therapeutic implications of quercetin and its derived-products in COVID-19 protection and prophylactic. Heliyon, 2024. 10(9): p. e30080.
- Häckl, L. , et al., Inhibition of Angiotensin-Converting Enzyme by Quercetin Alters the Vascular Response to Bradykinin and Angiotensin I. Pharmacology, 2002. 65: p. 182-6.
- Kim, D.W. , et al., Effect of silk fibroin peptide derived from silkworm Bombyx mori on the anti-inflammatory effect of Tat-SOD in a mice edema model. BMB Rep, 2011. 44(12): p. 787-92.
- Kawabe, Y., H. Waterson, and A. Mizoguchi, Bombyxin (Bombyx Insulin-Like Peptide) Increases the Respiration Rate Through Facilitation of Carbohydrate Catabolism in Bombyx mori. Front Endocrinol (Lausanne), 2019. 10: p. 150.
- Cermeño, M. , et al., Identification of peptides from edible silkworm pupae (Bombyx mori) protein hydrolysates with antioxidant activity. Journal of Functional Foods, 2022. 92: p. 105052.
- Thiemermann, C. , et al., FMRF-amide and L-Arg-L-Phe increase blood pressure and heart rate in the anaesthetised rat by central stimulation of the sympathetic nervous system. Biochemical and Biophysical Research Communications, 1991. 175(1): p. 318-324.
- Nichols, R. , et al., Human RFamide-related peptide-1 diminishes cellular and integrated cardiac contractile performance. Peptides, 2010. 31(11): p. 2067-74.
- Dagher, O. , et al., Therapeutic Potential of Quercetin to Alleviate Endothelial Dysfunction in Age-Related Cardiovascular Diseases. Front Cardiovasc Med, 2021. 8: p. 658400.

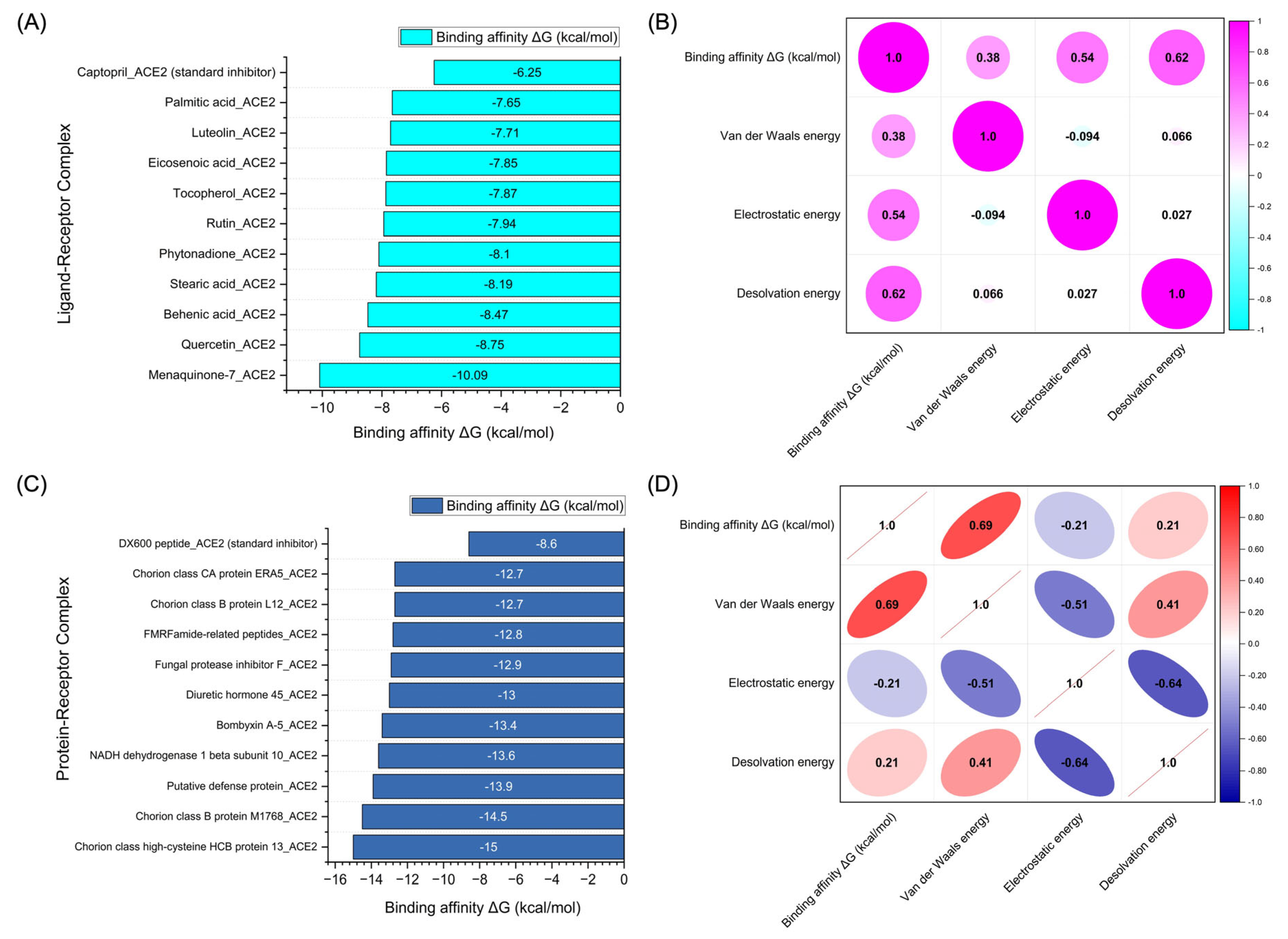
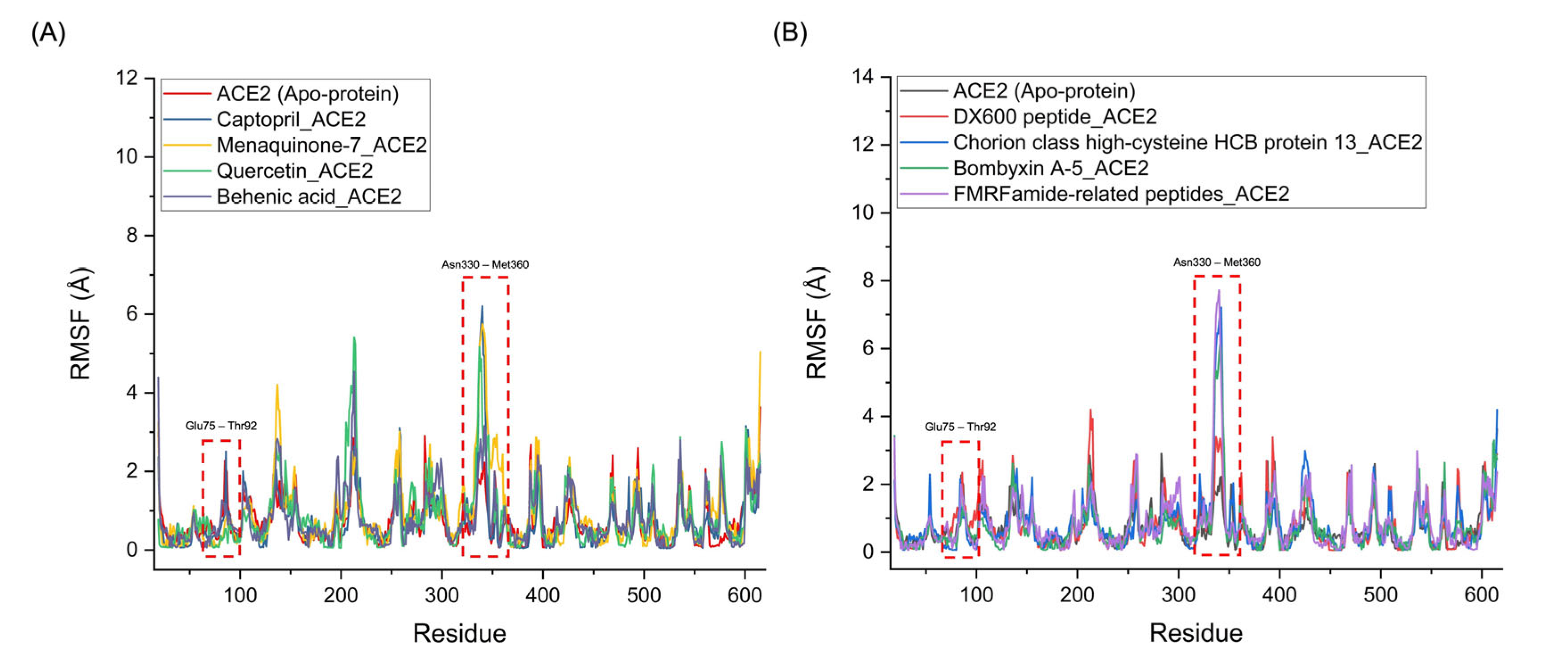
| Complex | HADDOCK score (a.u.) | Binding energy (kcal/mol) | Van der Waals energy | Electrostatic energy | Desolvation energy | RMSD |
|---|---|---|---|---|---|---|
| Captopril_ACE2 (standard inhibitor) | -11.2 +/- 1.8 | -6.25 | -12.6 +/- 0.4 | -40.5 +/- 7.3 | -1.5 +/- 0.5 | 0.5 +/- 0.0 |
| Menaquinone-7_ACE2 | -42.0 +/- 3.0 | -10.09 | -24.1 +/- 2.3 | -88.3 +/- 4.0 | -9.4 +/- 1.2 | 0.4 +/- 0.0 |
| Quercetin_ACE2 | -20.9 +/- 0.2 | -8.75 | -18.0 +/- 0.2 | -37.2 +/- 2.2 | 0.7 +/- 0.3 | 0.6 +/- 0.0 |
| Behenic acid_ACE2 | -25.6 +/- 1.7 | -8.47 | -14.2 +/- 2.5 | -94.2 +/- 10.7 | -6.4 +/- 0.8 | 0.2 +/- 0.0 |
| Stearic acid_ACE2 | -24.5 +/- 3.3 | -8.19 | -13.7 +/- 0.7 | -88.2 +/- 13.8 | -5.0 +/- 0.4 | 0.2 +/- 0.0 |
| Phytonadione_ACE2 | -31.6 +/- 1.2 | -8.10 | -24.6 +/- 0.1 | -43.3 +/- 4.4 | -3.6 +/- 0.8 | 0.5 +/- 0.0 |
| Rutin_ACE2 | -31.8 +/- 0.1 | -7.94 | -26.1 +/- 0.4 | -22.6 +/- 10.9 | -3.4 +/- 0.9 | 0.6 +/- 0.0 |
| Tocopherol_ACE2 | -33.7 +/- 1.4 | -7.87 | -26.1 +/- 0.3 | 5.6 +/- 0.2 | -9.1 +/- 0.3 | 0.1 +/- 0.0 |
| Eicosenoic acid_ACE2 | -26.0 +/- 3.5 | -7.85 | -17.1 +/- 1.9 | -99.1 +/- 17.7 | -2.4 +/- 1.0 | 0.8 +/- 0.0 |
| Luteolin_ACE2 | -19.5 +/- 0.5 | -7.71 | -16.8 +/- 0.4 | -26.2 +/- 2.3 | -0.1 +/- 0.3 | 0.9 +/- 0.0 |
| Palmitic acid_ACE2 | -25.7 +/- 0.7 | -7.65 | -18.4 +/- 0.3 | -61.6 +/- 5.5 | -5.8 +/- 0.1 | 0.6 +/- 0.0 |
| Complex | HADDOCK score (a.u.) | Binding energy (kcal/mol) | Van der Waals energy | Electrostatic energy | Desolvation energy | RMSD |
|---|---|---|---|---|---|---|
| DX600 peptide_ACE2 (standard inhibitor) | −76.8 +/- 0.9 | −8.6 | −38.7 +/- 2.9 | −79.0 +/- 28.3 | −26.1 +/- 4.2 | 1.4 +/- 0.2 |
| Chorion class high-cysteine HCB protein 13_ACE2 | −71.4 +/- 18.1 | −15.0 | −58.4 +/- 12.0 | −178.2 +/- 42.5 | 0.2 +/- 3.5 | 2.0 +/- 0.5 |
| Chorion class B protein M1768_ACE2 | −89.6 +/- 9.1 | −14.5 | −68.4 +/- 6.3 | −111.0 +/- 21.3 | −14.5 +/- 2.2 | 2.1 +/- 0.1 |
| Putative defense protein_ACE2 | −110.2 +/- 6.6 | −13.9 | −61.7 +/- 8.4 | −344.6 +/- 15.9 | 10.9 +/- 1.4 | 0.8 +/- 0.6 |
| NADH dehydrogenase 1 beta subunit 10_ACE2 | −81.3 +/- 12.3 | −13.6 | −37.5 +/- 12.5 | −290.1 +/- 20.3 | −5.0 +/- 2.8 | 2.3 +/- 0.2 |
| Bombyxin A-5_ACE2 | −113.2 +/- 10.0 | −13.4 | −63.7 +/- 4.4 | −185.9 +/- 32.4 | −26.3 +/- 3.6 | 0.9 +/- 0.9 |
| Diuretic hormone 45_ACE2 | −98.9 +/- 14.7 | −13.0 | −61.0 +/- 8.8 | −130.7 +/- 54.8 | −27.8 +/- 4.3 | 1.1 +/- 0.7 |
| Fungal protease inhibitor F_ACE2 | −82.0 +/- 12.0 | −12.9 | −44.4 +/- 12.0 | −169.1 +/- 14.3 | −7.4 +/- 2.8 | 2.4 +/- 0.3 |
| FMRFamide-related peptides_ACE2 | −65.3 +/- 12.9 | −12.8 | −60.0 +/- 7.5 | −120.4 +/- 50.1 | −9.1 +/- 2.5 | 1.3 +/- 0.8 |
| Chorion class B protein L12_ACE2 | −95.1 +/- 4.4 | −12.7 | −67.6 +/- 6.2 | −194.7 +/- 16.4 | −12.2 +/- 1.5 | 1.2 +/- 0.1 |
| Chorion class CA protein ERA.5_ACE2 | −70.2 +/- 5.6 | −12.7 | −56.9 +/- 3.6 | −134.4 +/- 41.3 | −6.7 +/- 1.8 | 1.9 +/- 0.3 |
| Complex | ICs charged-charged | ICs charged-polar | ICs charged-apolar | ICs polar-polar | ICs polar-apolar | ICs apolar-apolar | NIS charged | NIS apolar |
|---|---|---|---|---|---|---|---|---|
| DX600 peptide_ACE2 (standard inhibitor) | 3 | 3 | 14 | 0 | 5 | 6 | 27.88 | 33.63 |
| Chorion class high-cysteine HCB protein 13_ACE2 | 6 | 18 | 26 | 5 | 32 | 13 | 22.20 | 39.23 |
| Chorion class B protein M1768_ACE2 | 2 | 6 | 36 | 1 | 26 | 19 | 23.96 | 40.83 |
| Putative defense protein_ACE2 | 17 | 13 | 16 | 8 | 30 | 9 | 26.80 | 36.51 |
| NADH dehydrogenase 1 beta subunit 10_ACE2 | 13 | 13 | 25 | 2 | 21 | 12 | 28.84 | 34.81 |
| Bombyxin A-5_ACE2 | 3 | 6 | 25 | 0 | 22 | 18 | 26.73 | 35.25 |
| Diuretic hormone 45_ACE2 | 3 | 2 | 25 | 3 | 25 | 17 | 26.23 | 38.20 |
| Fungal protease inhibitor F_ACE2 | 8 | 10 | 24 | 1 | 19 | 5 | 26.31 | 35.54 |
| FMRFamide-related peptides_ACE2 | 9 | 10 | 27 | 2 | 19 | 14 | 28.45 | 35.61 |
| Chorion class B protein L12_ACE2 | 3 | 8 | 31 | 1 | 21 | 10 | 22.47 | 43.22 |
| Chorion class CA protein ERA.5_ACE2 | 2 | 11 | 27 | 4 | 24 | 13 | 22.63 | 41.19 |
| Complex | Average RMSD (Å) | Average RMSF (Å) | Average RoG (Å) | Number of Hydrogen Bonds Between the Ligand-Receptor |
|---|---|---|---|---|
| Chemical compounds | ||||
| Captopril_ACE2 (standard inhibitor) | 1.250 | 0.746 | 2.185 | 4 |
| Menaquinone-7_ACE2 | 1.381 | 0.936 | 2.212 | 5 |
| Quercetin_ACE2 | 1.317 | 0.836 | 2.205 | 4 |
| Behenic acid_ACE2 | 1.482 | 0.912 | 2.198 | 3 |
| Stearic acid_ACE2 | 1.412 | 0.884 | 2.203 | 3 |
| Phytonadione_ACE2 | 1.358 | 0.815 | 2.214 | 5 |
| Rutin_ACE2 | 1.452 | 0.922 | 2.197 | 6 |
| Tocopherol_ACE2 | 1.393 | 0.858 | 2.209 | 4 |
| Eicosenoic acid_ACE2 | 1.424 | 0.891 | 2.201 | 3 |
| Luteolin_ACE2 | 1.336 | 0.829 | 2.210 | 4 |
| Palmitic acid_ACE2 | 1.372 | 0.847 | 2.202 | 3 |
| Protein-based compounds | ||||
| DX600 peptide_ACE2 (standard inhibitor) | 3.152 | 1.802 | 2.792 | 8 |
| Chorion class high-cysteine HCB protein 13_ACE2 | 3.284 | 1.833 | 2.805 | 13 |
| Chorion class B protein M1768_ACE2 | 3.344 | 1.889 | 2.822 | 10 |
| Putative defense protein_ACE2 | 3.421 | 1.902 | 2.818 | 11 |
| NADH dehydrogenase 1 beta subunit 10_ACE2 | 3.381 | 1.854 | 2.809 | 10 |
| Bombyxin A-5_ACE2 | 3.224 | 1.776 | 2.798 | 10 |
| Diuretic hormone 45_ACE2 | 3.463 | 1.915 | 2.814 | 11 |
| Fungal protease inhibitor F_ACE2 | 3.322 | 1.845 | 2.810 | 10 |
| FMRFamide-related peptides_ACE2 | 3.381 | 1.875 | 2.806 | 12 |
| Chorion class B protein L12_ACE2 | 3.294 | 1.812 | 2.794 | 9 |
| Chorion class CA protein ERA.5_ACE2 | 3.302 | 1.821 | 2.799 | 9 |
| Complex | MM/PBSA Calculation ResultsΔG_binding (kcal/mol) |
|---|---|
| Chemical compounds | |
| Captopril_ACE2 (standard inhibitor) | −21.08 |
| Menaquinone-7_ACE2 | −35.12 |
| Quercetin_ACE2 | −29.98 |
| Behenic acid_ACE2 | −27.76 |
| Stearic acid_ACE2 | −27.01 |
| Phytonadione_ACE2 | −27.31 |
| Rutin_ACE2 | −26.88 |
| Tocopherol_ACE2 | −26.36 |
| Eicosenoic acid_ACE2 | −26.49 |
| Luteolin_ACE2 | −25.89 |
| Palmitic acid_ACE2 | −25.66 |
| Protein-based compounds | |
| DX600 peptide_ACE2 (standard inhibitor) | −81.93 |
| Chorion class high-cysteine HCB protein 13_ACE2 | −212.43 |
| Chorion class B protein M1768_ACE2 | −195.04 |
| Putative defense protein_ACE2 | −162.63 |
| NADH dehydrogenase 1 beta subunit 10_ACE2 | −198.03 |
| Bombyxin A-5_ACE2 | −209.36 |
| Diuretic hormone 45_ACE2 | −176.48 |
| Fungal protease inhibitor F_ACE2 | −171.07 |
| FMRFamide-related peptides_ACE2 | −198.93 |
| Chorion class B protein L12_ACE2 | −193.50 |
| Chorion class CA protein ERA.5_ACE2 | −140.36 |
| Molecule | Lipinski violation | Drug-likeness | Mutagenic | Tumorigenic | Reproductiveeffective | Irritant |
|---|---|---|---|---|---|---|
| Menaquinone-7_ACE2 | 2 violations: MW > 500 g/mol LogP > 5 |
0.62 | None | None | None | None |
| Quercetin_ACE2 | 0 | 0.52 | High | High | None | None |
| Behenic acid_ACE2 | 1 violation: LogP > 5 |
0.54 | None | None | None | None |
| Stearic acid_ACE2 | 0 | 0.54 | High | High | None | High |
| Phytonadione_ACE2 | 1 violation: LogP > 5 |
0.93 | None | None | None | None |
| Rutin_ACE2 | 3 violations: MW > 500 g/mol HBA > 10 HBD > 5 |
0.91 | None | None | None | None |
| Tocopherol_ACE2 | 1 violation: LogP > 5 |
0.48 | None | None | None | None |
| Eicosenoic acid_ACE2 | 1 violation: LogP > 5 |
-0.30 | None | None | None | None |
| Luteolin_ACE2 | 0 | 0.38 | None | None | None | None |
| Palmitic acid_ACE2 | 1 violation: LogP > 5 |
-0.54 | None | High | None | High |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





