1. Introduction
Head and neck squamous cell carcinoma (HNSCC) represents a substantial challenge to healthcare systems globally, impacting over 16,000 individuals annually in Hong Kong alone [
1]. The five-year survival rate for late-stage patients is below 50%, and radiotherapy is the predominant treatment modality [
2]. Due to tumor heterogeneity, achieving effective locoregional control is challenging, often necessitating personalized treatment plans. Accurate prognosis prediction is essential for optimizing treatment strategies.
Traditionally, the tumor, node, and metastasis (TNM) staging system has been utilized for cancer prognosis, evaluating tumor size, lymph node involvement, and metastatic progression based on biopsy results. However, this process is often time-consuming and invasive. Recent advancements in medical imaging, particularly Computed Tomography (CT) and Magnetic Resonance Imaging (MRI), have produced high-quality images that facilitate more cost-effective tumor analysis. Moreover, breakthroughs in AI technology have accelerated the ability to obtain accurate cancer prognoses using radiomics.
Radiomics involves the quantitative extraction of image features, such as signal intensity and texture [
3]. These features are analyzed with AI to provide objective prognostic insights. Various studies have demonstrated the efficacy of radiomics, with some achieving 100% accuracy in distinguishing oral squamous cell carcinoma using support vector machines (SVM) [
4]. Machine learning algorithms, including generalized linear models, SVM, random forest (RF), decision trees (DT), and extreme boosting (EB), have shown promising results in cancer prognosis. For instance, Xiong et al. achieved an accuracy of 93.3% in esophageal cancer prognosis using an RF model [
5].
Recent research emphasizes that combining multiple algorithms can enhance prediction accuracy, which can be achieved through ensemble learning algorithms. A notable approach is the voted ensemble machine learning algorithm (VEML), which integrates predictions from various classifiers, yielding an accuracy of 88.3% in HNSCC prognosis prediction [
6].
While significant advancements in AI and ensemble learning have been made, the application of stacking ensemble learning methods in cancer prognosis, particularly in radiomics, remains underexplored. This study aims to fill this gap by employing a stacking ensemble approach to improve HNSCC prognosis prediction.
2. Methodology
2.1. Research Workflow
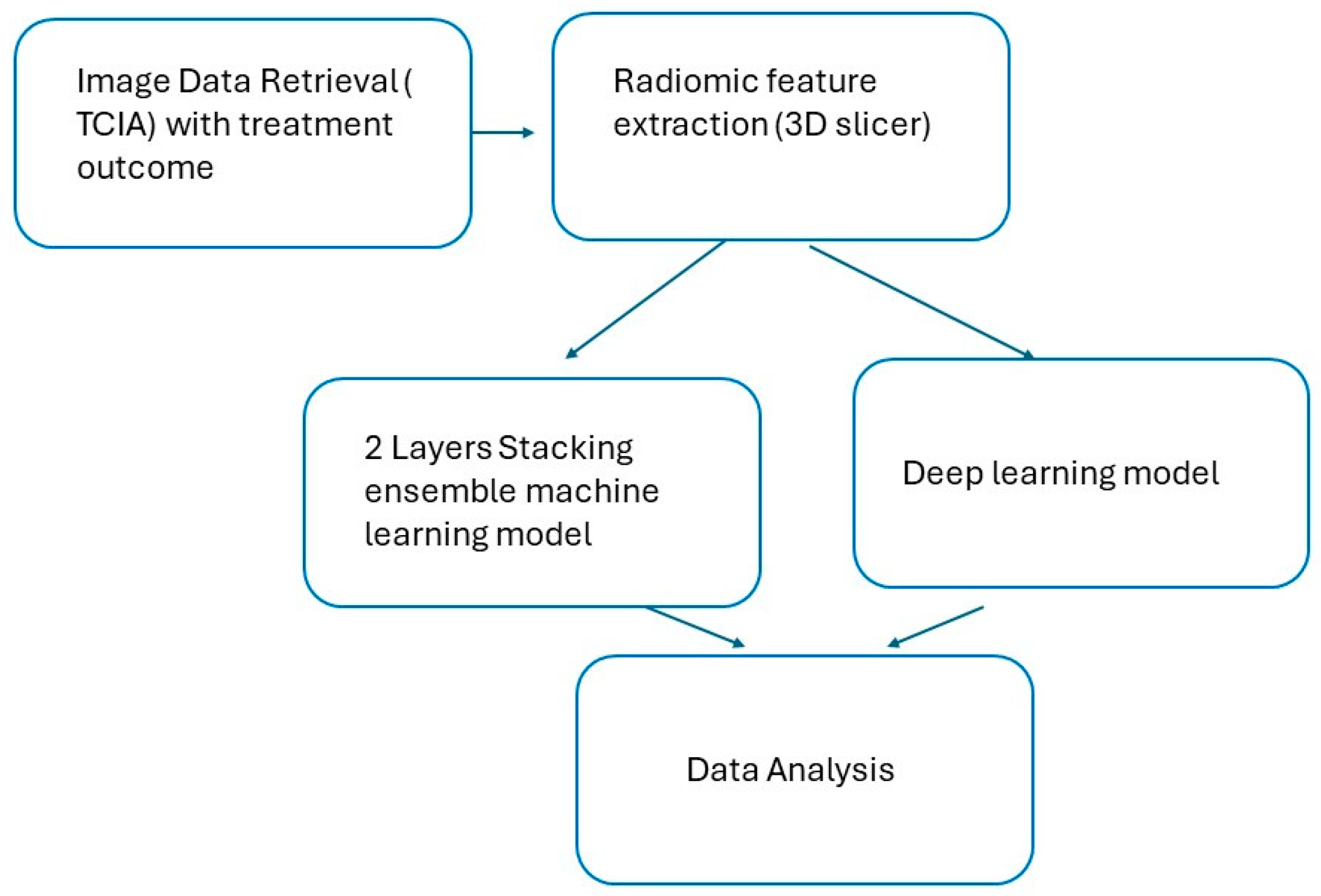
The research workflow included data retrieval, raw data filtering, image importation, and feature extraction. Two models were utilized: a two-layer stacking ensemble model and deep learning models. The performance of both models was subsequently compared (
Figure 1).
2.2. Patient Data
Data for this study were sourced from The Cancer Imaging Archive (TCIA), a publicly accessible repository for cancer medical images maintained by the National Cancer Institute (NCI). The dataset, labeled "HNSCC," comprised planning CT images of patients who underwent radical radiotherapy for HNSCC between 2003 and 2013. It included radiotherapy structures such as GTV and PTV, alongside clinical data including age, sex, diagnosis, smoking status, staging, and three- and five-year survival outcomes.
A thorough quality check resulted in a final sample of 164 cases from the original 215 collected.
Radiomics features were extracted from the GTV and PTV structures using 3D Slicer (version 4.10.2) with the PyRadiomics extension. The features were categorized into various groups, including shape, first-order statistics, gray-level zone matrix (GLSZM), gray-level dependence matrix (GLDM), gray-level run-length matrix (GLRLM), gray-level co-occurrence matrix (GLCM), and neighborhood gray-tone difference matrix (GLTDM). A total of 107 radiomics features were extracted for analysis.
2.3. Machine Learning Models
Cancer prognosis is typically assessed based on specific time points, such as the five-year survival rate [
13]. This allows for objective comparisons across different cancer studies, as patients who survive five years post-treatment are generally classified as "cancer survivors."
In this study, the five-year survival rate was selected as the treatment outcome to ensure comparability with other research. Of the 164 cases, 118 patients survived, while 46 did not. An overfitting test was performed by randomly selecting 46 cases from the 117 patients who survived to balance the sample outcomes.
2.4. Machine Learning Process
Two machine learning models were employed: The Two-layer Stacking Ensemble Machine Learning Model and Deep Learning Machine Learning Model
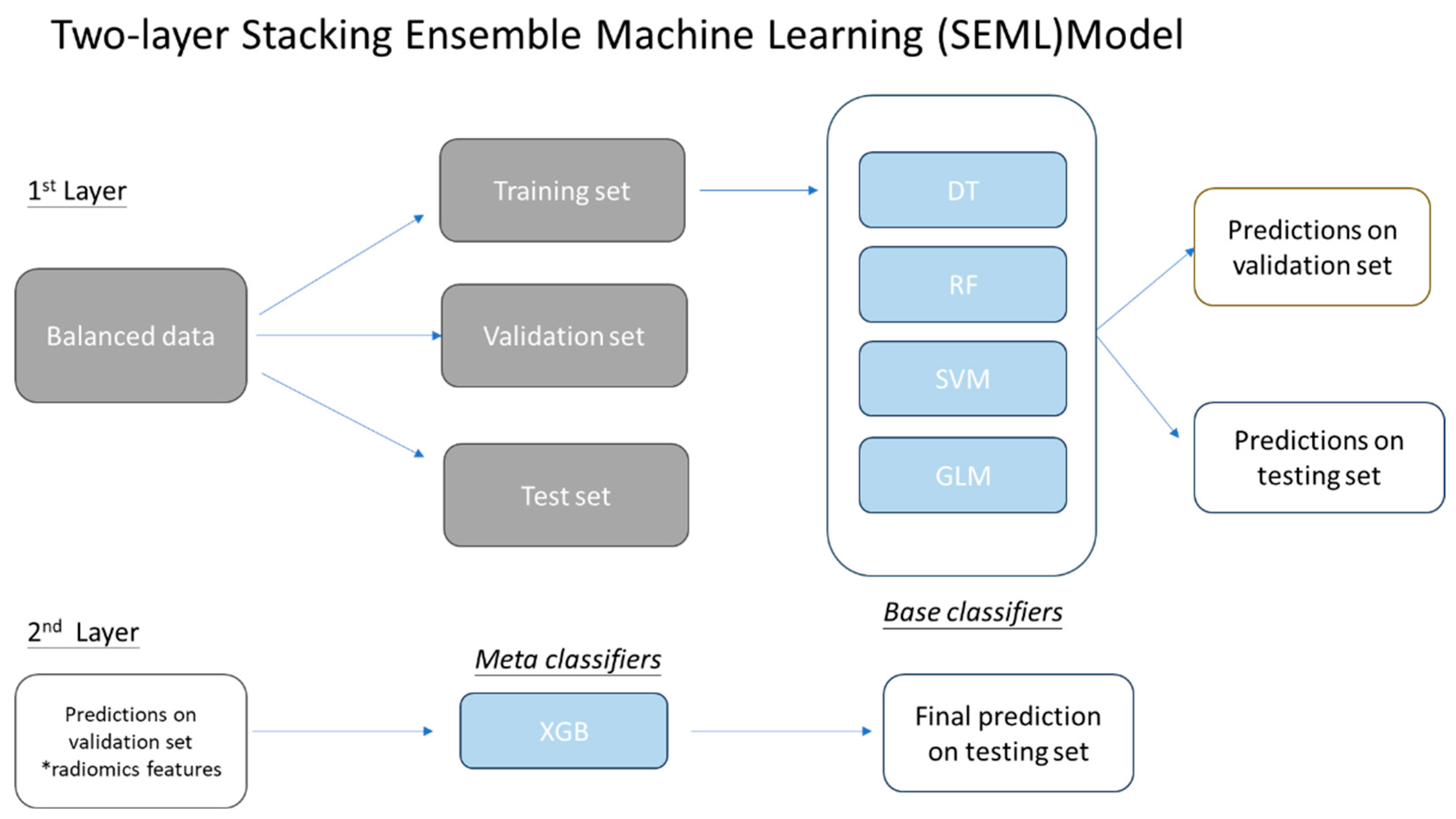
2.4.1. Two-layer Stacking Ensemble Machine Learning (SEML) Model
In this model, the radiomics data were divided into three sets: training (70%), validation (15%), and testing (15%). Four classifiers were utilized: decision trees (DT), random forests (RF), support vector machine (SVM), and generalized linear model (GLM). The training set was used to train the models, which were then validated with the validation set to generate predictions. Predicted outcomes were quantified numerically, with values of 0 representing survival beyond five years or death from other causes, and 1 indicating death within five years of diagnosis.
The stacking ensemble model consists of heterogeneous classifiers organized in a two-layer structure. The base classifiers were initially trained with the radiomics data. The meta-learner was trained using the prediction results from the base classifiers, with XGBoost selected as the meta-classifier. The selection of an appropriate meta-classifier is crucial for model performance. Previous studies have indicated that XGBoost is optimal for recurrent HNSCC prognosis [
14,
15,
16]. The final prediction outcome was the training result of the XGboost. Details of protocol were illustrated in
Figure 2.
2.4.2. Overfitting Test
As the dataset exhibited an unbalanced distribution of outcomes, an overfitting test was conducted using a balanced sample with an equal number of cases for each treatment outcome.
2.4.3. Deep Learning Machine Learning Models (DLML)
To compare the SEML model with deep learning methods, a deep learning machine learning model comprising three fully connected layers was trained. Each layer, except the last, utilized a Rectified Linear Unit (ReLU) activation function, while the final layer employed a sigmoid activation function to produce outputs in the range [0, 1]. A 70/30 split for training and testing was utilized.
2.5. Data Analysis
The predicted outcomes from the two-layer stacking model were compared with those from the base classifiers. Model performance was evaluated using the Receiver Operating Characteristic (ROC) curve, with metrics including the area under the ROC curve (AUC), accuracy, specificity, and sensitivity calculated using ROCkit from the University of Chicago (1995).
3. Results
3.1. Demographic Cohort
The dataset comprised 215 patients, of which 51 were excluded due to missing data, resulting in 164 cases for analysis. Both PTV and GTV CT datasets were collected. Demographic details are summarized in
Table 1.
3.2. Prediction Performance of SEML and DLML Models
The SEML model demonstrated exceptional performance, with AUC ranging from 0.82 to 0.982 across all target volumes. For PTV radiomic features, the AUC reached 0.982. The model exhibited sensitivity of 100%, specificity of 83%, and accuracy of 93%. In contrast, the AUC for GTV features was slightly lower at 0.820, with sensitivity, specificity, and accuracy at 62.5%, 85.7%, and 73.3%, respectively. In comparison, the DLML model showed an AUC ranging from 0.605 to 0.774, with accuracy between 0.655 and 0.724, indicating superior performance of the SEML model (Table 2).
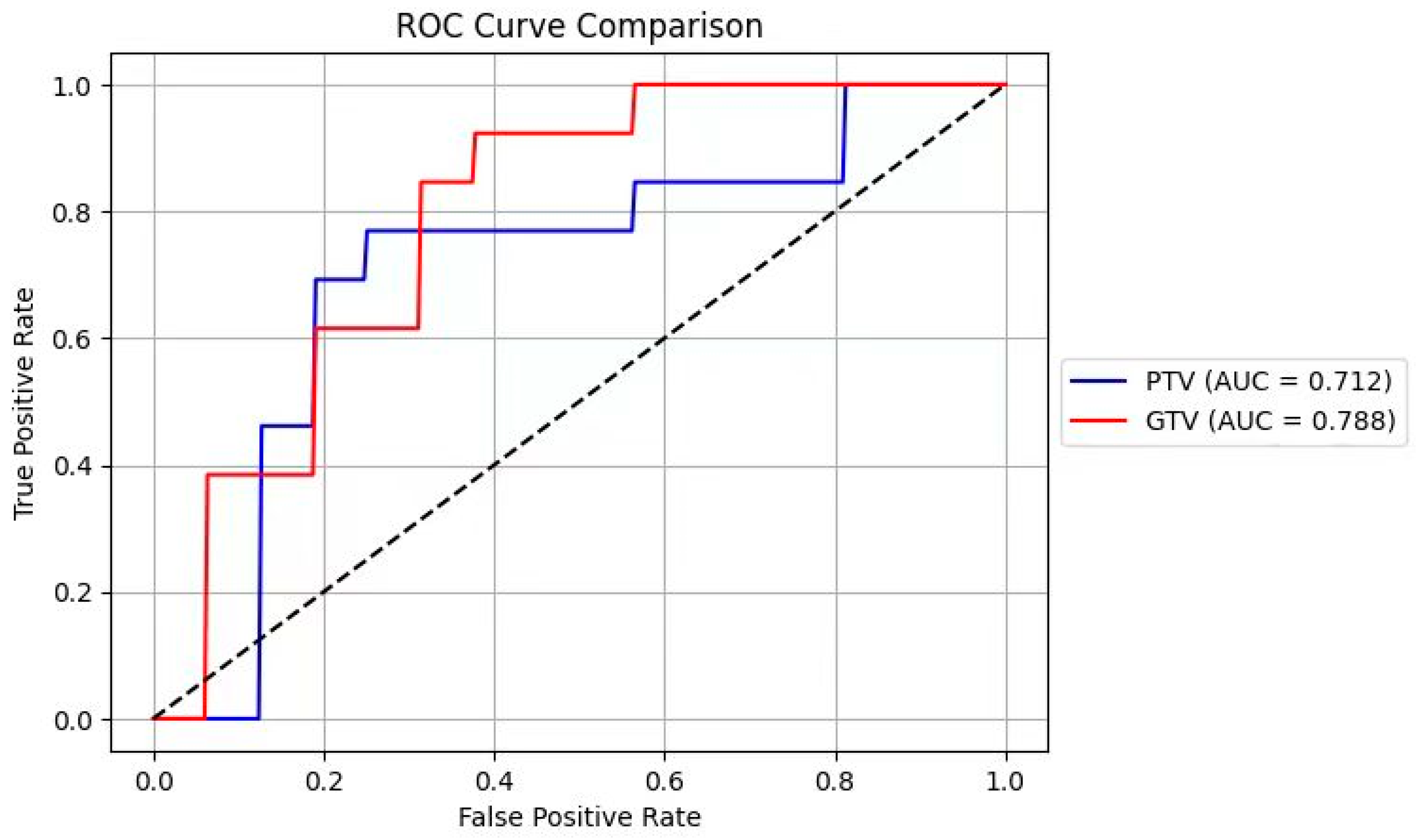
3.3. ROC Analysis
Despite the PTV radiomics features yielding better predictions, ROC analysis indicated no significant difference between PTV and GTV features (chi-square test, p > 0.05).(
Figure 3)
3.4. Comparison of SEML and DLML Models
The SEML model consistently outperformed the DLML model in HNSCC prognosis prediction regarding sensitivity, specificity, accuracy, and AUC. ROC analysis further confirmed this finding, showing no significant difference between the SEML and SVM models (chi-square test, p > 0.05)(
Figure 4).
4. Discussion
This study evaluates the performance of the SEML and deep learning models in predicting five-year survival in HNSCC patients using radiomics features. While no significant difference was observed between the models, the SEML demonstrated a notable improvement in prognostic prediction.
The SEML model employed in this research is the first to quantitatively explore a stacking ensemble approach for enhancing cancer prognosis predictions based on CT radiomics. Prior studies have highlighted the utility of machine learning in cancer prognosis, with notable successes such as an AUC of 0.61 for head and neck cancer using RF models [
18] and a C-index of 0.782 for laryngeal squamous cell carcinoma prognosis [
19].
The SEML model's superior performance compared to the DLML model underscores its potential in integrating various algorithms for enhanced prognostic accuracy. This approach leverages the strengths of individual classifiers, leading to significant improvements in predictive performance.
The findings align with previous research on stacking ensemble learning, which has demonstrated improved accuracy and higher AUC compared to single machine learning models [
9,
10,
11,
12]. The limitations of current applications in cancer prognosis highlight the need for further exploration of the stacking ensemble approach.
Incorporating clinical and genomic data alongside radiomics features could enhance predictive capabilities. Recent studies have indicated that radiomics-clinical (RC) models yield higher accuracy compared to radiomics-only models [
21,
22,
23]. Similarly, radiomics-genomics (RG) models have shown promise in improving survival predictions [24,25]. This opens avenues for developing integrated models that utilize multi-modal data sources, enhancing the overall predictive power.
5. Conclusions
This study is the first to employ a stacking ensemble learning approach in a predictive model for forecasting cancer prognosis in HNSCC patients. The SEML model demonstrated high accuracy (98%), sensitivity (100%), and specificity (83%) in predicting five-year survival. The results affirm the effectiveness of the stacking ensemble approach in enhancing prognostic accuracy, laying a foundation for its clinical application and potential to facilitate personalized treatment for cancer patients.
Author Contributions
Conceptualization, FH Tang. And HYT Wong.; methodology, FH Tang and C Xue.; software, FH Tang, C Xue.; validation, CCY Chan., VTY Li; SWY Lee.; formal analysis, F.H.Tang, C Xue.; investigation, CCY Chan VTY Li SWY Lee.; resources, HYT Wong.; writing—original draft preparation, CCY Chan, V TY Li , SWY Lee.; writing—review and editing, HYT Wong, FH Tang.; ; supervision, HYT Wong FH Tang.; project administration, FH Tang.; funding acquisition, FH Tang. All authors have read and agreed to the published version of the manuscript.
Funding
This project is supported by UGC Research Matching Grant 2021-02-75 RMGS210201.
Data Availability Statement
References
- Tsui, T., Cheung, K. M., Chow, J. C. H., & Wong, K. H. (2022). Risk Factors for Early Mortality in Head and Neck Cancer Patients Undergoing Definitive Chemoradiation. Hong Kong Journal of Radiology, 25(2), 127.
- Gormley M, Creaney G, Schache A, Ingarfield K, Conway DI. Reviewing the epidemiology of head and neck cancer: Definitions, trends and risk factors. Br Dent J 2022;233:780-786. [CrossRef]
- Huang S, Franc BL, Harnish RJ et al. Exploration of PET and MRI radiomic features for decoding breast cancer phenotypes and prognosis. NPJ Breast Cancer 2018;4:1-13. [CrossRef]
- Alabi, R. O., et al. (2021). Machine learning in oral squamous cell carcinoma: Current status, clinical concerns and prospects for future—A systematic review. Artificial Intelligence in Medicine, 115, 102060.
- Xiong, J., et al. (2018). The role of PET-based radiomic features in predicting local control of esophageal cancer treated with concurrent chemoradiotherapy. Scientific Reports, 8(1), 9902.
- Tang, F. H., et al. (2022). Radiomics from various tumour volume sizes for prognosis prediction of head and neck squamous cell carcinoma: a voted ensemble machine learning approach. Life, 12(9), 1380. [CrossRef]
- Feng, Y., et al. (2021). A heterogeneous ensemble learning method for neuroblastoma survival prediction. IEEE Journal of Biomedical and Health Informatics, 26(4), 1472-1483.
- Lui, V. W., et al. (2013). Frequent mutation of the PI3K pathway in head and neck cancer defines predictive biomarkers. Cancer Discovery, 3(7), 761-769.
- Yan, F., & Feng, Y. (2022). A two-stage stacked-based heterogeneous ensemble learning for cancer survival prediction. Complex Intelligent Systems, 8, 4619–4639. [CrossRef]
- Kumar, M., et al. (2022). Optimized stacking ensemble learning model for breast cancer detection and classification using machine learning. Sustainability, 14(21), 13998.
- Lee J, et al. (2023). Machine learning-based radiomics models for prediction of locoregional recurrence in patients with breast cancer. Oncology Letters, 26:1-10. [CrossRef]
- Zhao S, et al. (2023). Stacking ensemble learning-based [18F] FDG PET radiomics for outcome prediction in diffuse large B-cell lymphoma. J Nucl Med, 64:160-1609. [CrossRef]
- National Research Council. (2006). From Cancer Patient to Cancer Survivor: Lost in Transition. Washington, DC: The National Academies Press.
- Kwon, H., et al. (2019). Stacking ensemble technique for classifying breast cancer. Healthcare Informatics Research, 25(4), 283-288.
- Agarwal, A. (2020). Breast Cancer Prognosis Using Stacking Ensemble (Doctoral dissertation, State University of New York at Binghamton).
- Owusu DK, Nyarko PK. (2023). Stacked ensemble model for recurrent head and neck squamous cell carcinoma prognosis based on clinicopathologic and genomic markers. J Math Prob Equations Stat, 4:121-134.
- Marzorati, C., et al. (2017). Who is a cancer survivor? A systematic review of published definitions. Journal of Cancer Education, 32, 228-237.
- Parmar, C., et al. (2015). Radiomic machine-learning classifiers for prognostic biomarkers of head and neck cancer. Frontiers in Oncology, 5, 272.
- Chen, L., et al. (2020). Evaluation of CT-based radiomics signature and nomogram as prognostic markers in patients with laryngeal squamous cell carcinoma. Cancer Imaging, 20, 1-9. [CrossRef]
- ang FH, et al. (2021). Radiomics AI prediction for head and neck squamous cell carcinoma (HNSCC) prognosis and recurrence with target volume approach. BJR| Open, 3(1), 20200073.
- Bao D, et al. (2021). Prognostic and predictive value of radiomics features at MRI in nasopharyngeal carcinoma. Discov Oncol, 12:1-13. [CrossRef]
- Ching JCF, et al. (2023). Integrating CT-based radiomic model with clinical features improves long-term prognostication in high-risk prostate cancer. Front Oncol, 13:1-12. [CrossRef]
- Tang FH, et al. (2023). Radiomics-clinical AI model with probability weighted strategy for prognosis prediction in non-small cell lung cancer. Biomedicines, 11:1-12.
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).