Submitted:
28 March 2025
Posted:
31 March 2025
You are already at the latest version
Abstract
Keywords:
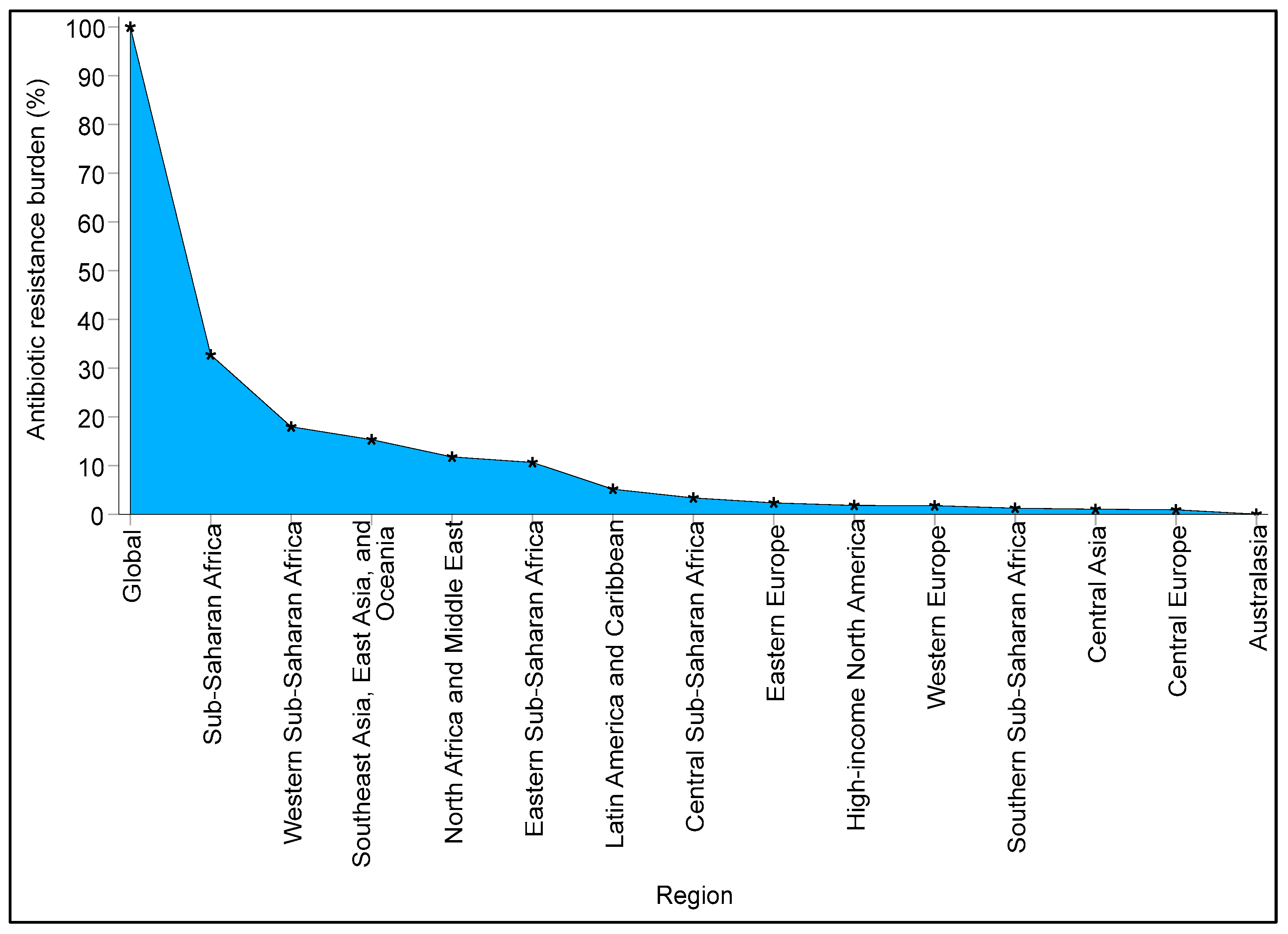
1. Introduction
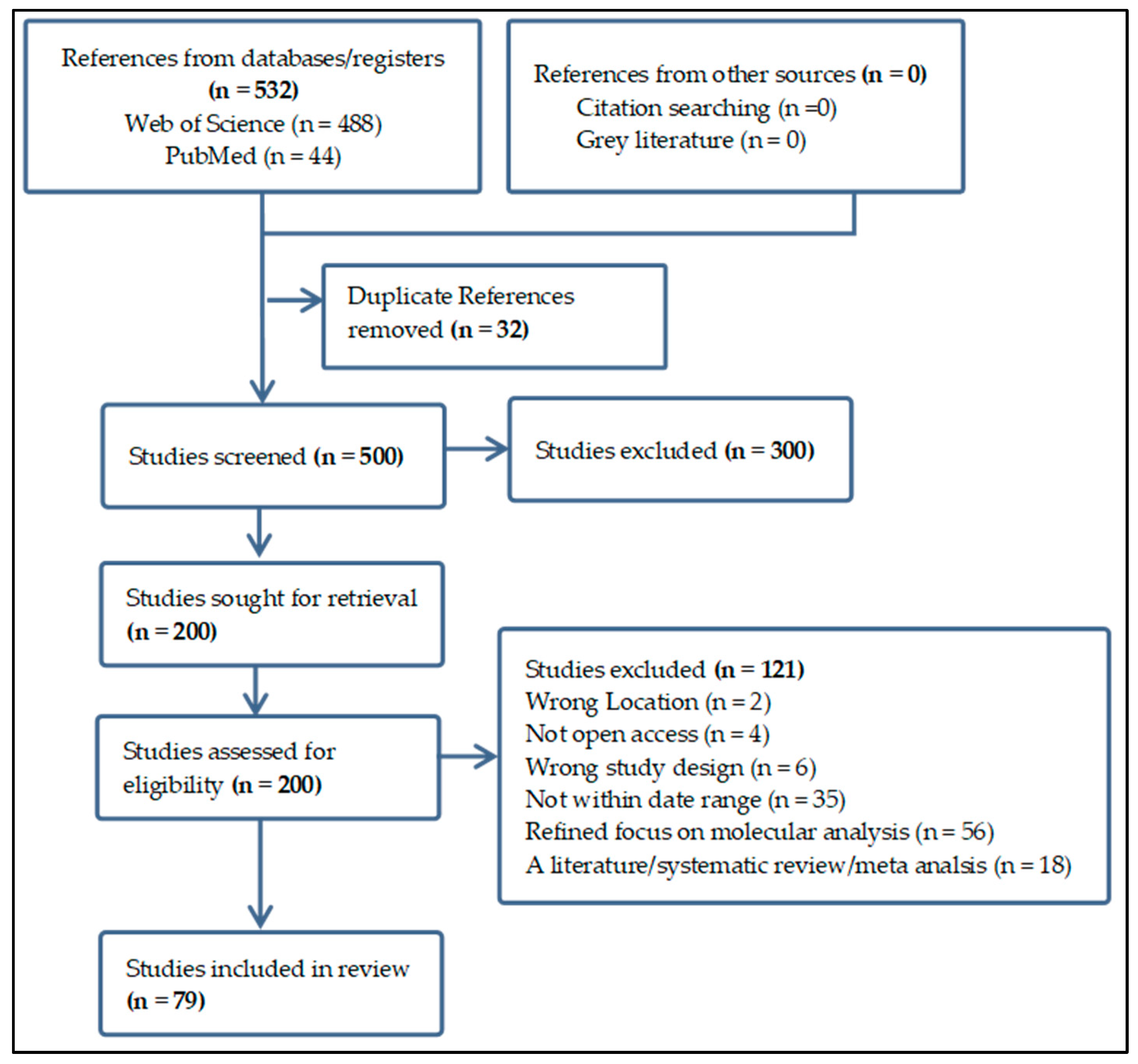
2. Materials and Methods
3. Results and Discussion
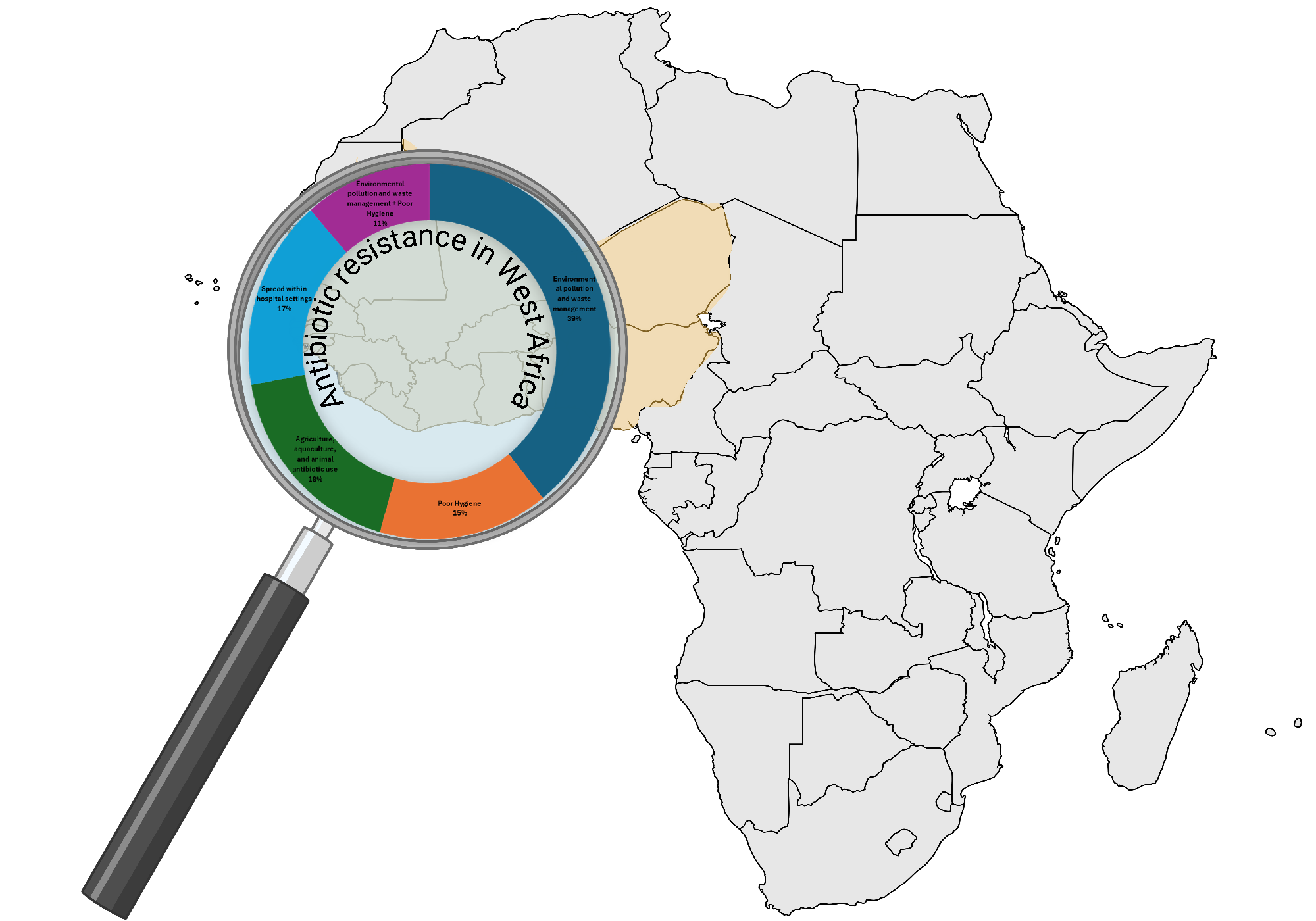
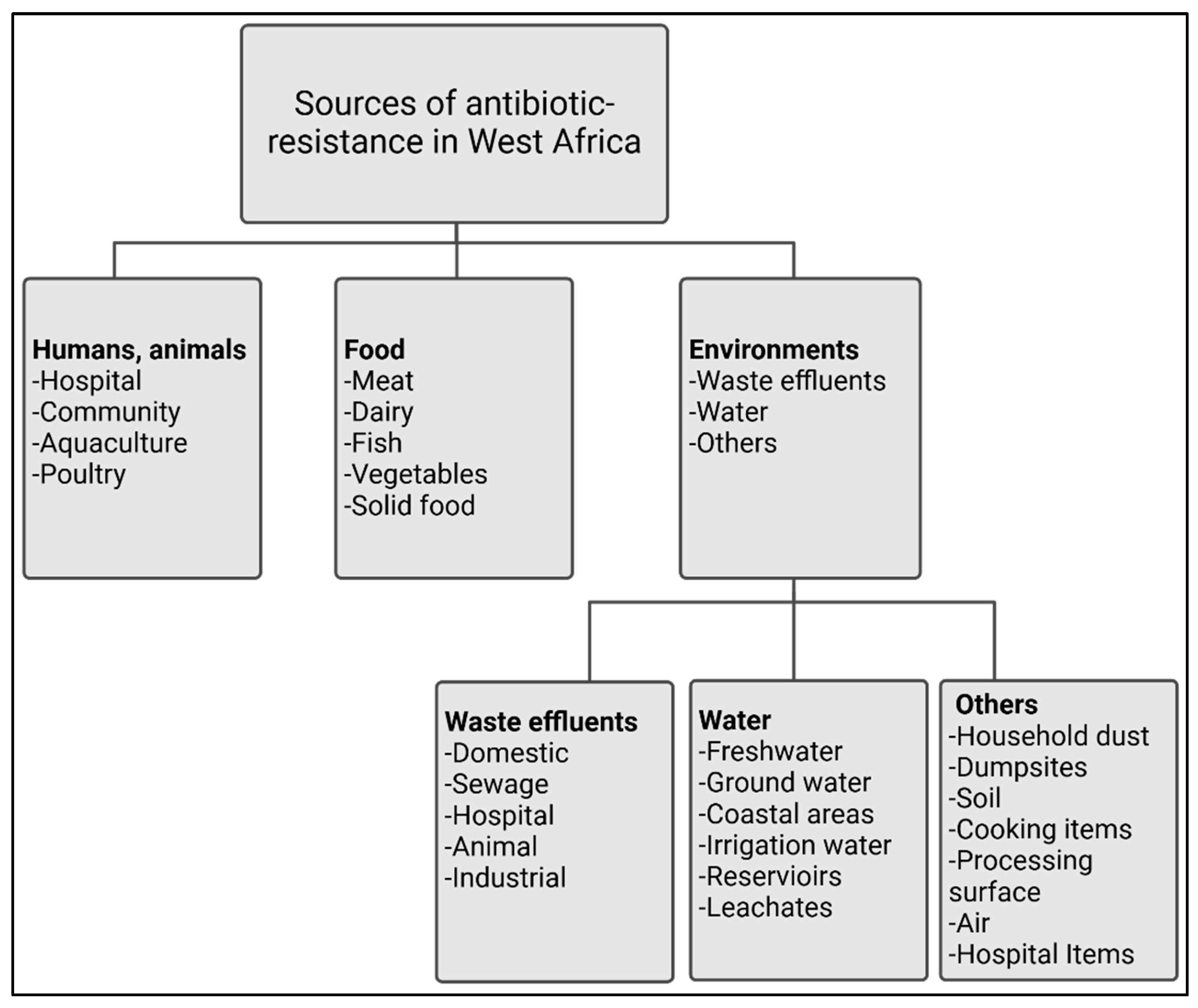
3.1. Sources of Antibiotic Resistance in West Africa
3.2. Antibiotics Under Study in West Africa
3.3. Antibiotic-Resistant Bacterial Strains, Along with Their Associated Resistance Genes
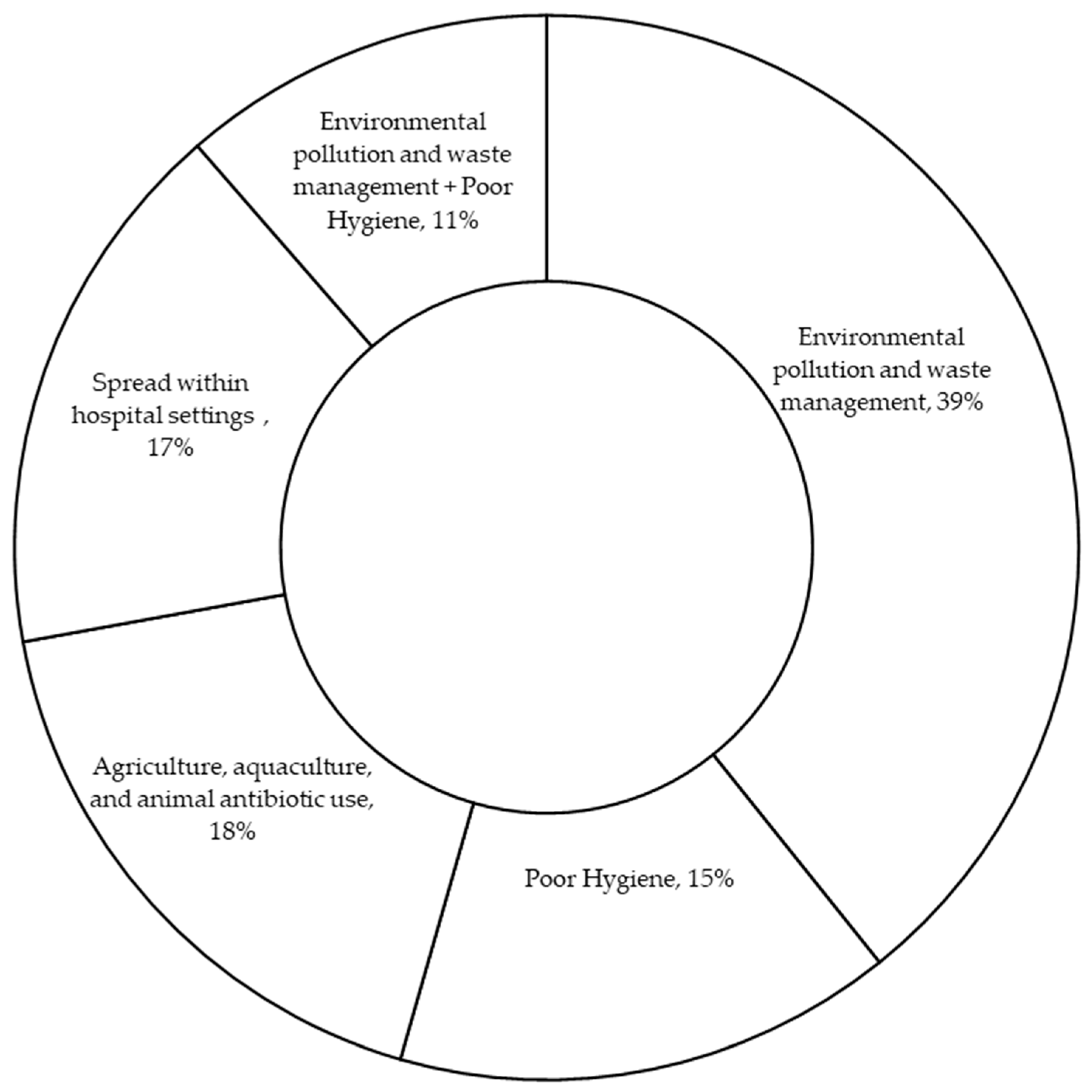
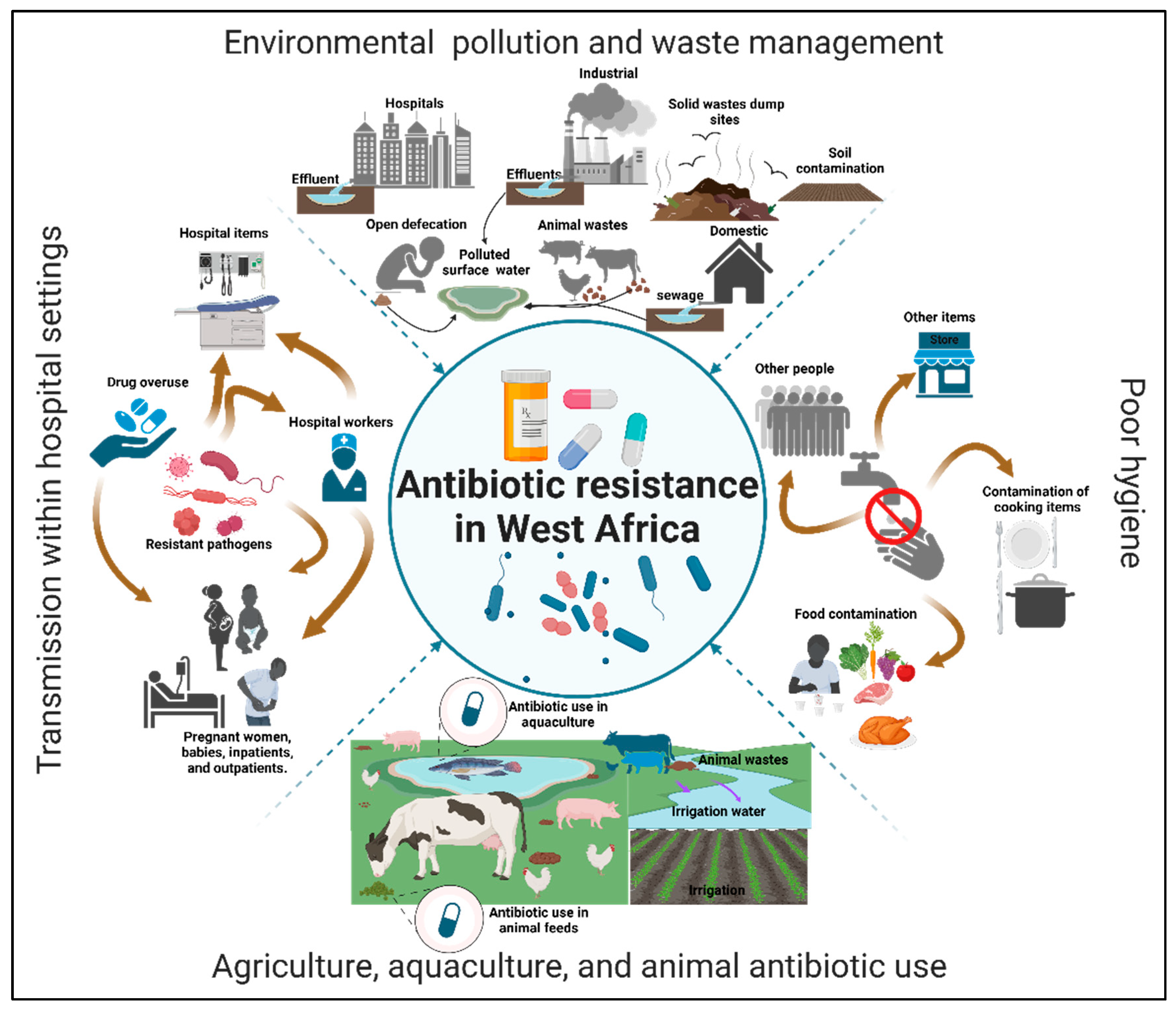
3.3. Environmental Risk Factors Contributing to The Spread of Antibiotic Resistance in West Africa
3.3.1. Environmental Pollution and Waste Management
3.3.2. Poor Hygiene
3.3.3. Agriculture, Aquaculture, and Animal Antibiotic Use
3.3.4. Transmission Within Hospital Settings
4. Challenges and Limitations Encountered by Researchers in Studying Antibiotic Resistance in West Africa
5. Conclusions
Supplementary Materials
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Murray, C.J.L.; et al. Global burden of bacterial antimicrobial resistance in 2019: A systematic analysis. The lancet 2022, 399, 629–655. [Google Scholar] [CrossRef] [PubMed]
- Roca, I.; et al. The global threat of antimicrobial resistance: Science for intervention. New Microbes New Infect 2015, 6, 22–29. [Google Scholar] [CrossRef] [PubMed]
- Hutchings, M.I.; Truman, A.W.; Wilkinson, B. Antibiotics: Past, present and future. Curr Opin Microbiol 2019, 51, 72–80. [Google Scholar] [CrossRef] [PubMed]
- Spellberg, B.; Gilbert, D.N. The future of antibiotics and resistance: A tribute to a career of leadership by John Bartlett. Clinical infectious diseases 2014, 59, S71–S75. [Google Scholar] [CrossRef]
- Tang, K.W.K.; Millar, B.C.; Moore, J.E. Antimicrobial resistance (AMR). Br J Biomed Sci 2023, 80, 11387. [Google Scholar] [CrossRef]
- Doan, T.; et al. Macrolide and Nonmacrolide Resistance with Mass Azithromycin Distribution. N Engl J Med 2020, 383, 1941–1950. [Google Scholar] [CrossRef]
- Tiseo, K.; Huber, L.; Gilbert, M.; Robinson, T.P.; Van Boeckel, T.P. Global trends in antimicrobial use in food animals from 2017 to 2030. Antibiotics 2020, 9, 918. [Google Scholar] [CrossRef]
- Klein, E.Y.; et al. Global increase and geographic convergence in antibiotic consumption between 2000 and 2015. Proceedings of the National Academy of Sciences 2018, 115, E3463–E3470. [Google Scholar] [CrossRef]
- Gautam, A. Antimicrobial resistance: The next probable pandemic. JNMA J Nepal Med Assoc 2022, 60, 225. [Google Scholar] [CrossRef]
- Silbergeld, E.K.; Patrick, T.E. Environmental exposures, toxicologic mechanisms, and adverse pregnancy outcomes. Am J Obstet Gynecol 2022, 192, S11–S21. [Google Scholar] [CrossRef]
- Nwafia, I.N.; Ike, A.C.; Orabueze, I.N.; Nwafia, W.C. Carbapenemase producing Enterobacteriaceae: Environmental reservoirs as primary targets for control and prevention strategies. Nigerian Postgraduate Medical Journal 29, 183–191. [CrossRef] [PubMed]
- Ngbede, E.O.; et al. Concurrent Resistance to Carbapenem and Colistin Among Enterobacteriaceae Recovered From Human and Animal Sources in Nigeria Is Associated With Multiple Genetic Mechanisms. Front Microbiol 2021, 12. [Google Scholar] [CrossRef] [PubMed]
- Ayibieke, A.; et al. Prevalence and Characterization of Carbapenem-Hydrolyzing Class D β-Lactamase-Producing Acinetobacter Isolates From Ghana. Front Microbiol 2020, 11. [Google Scholar] [CrossRef] [PubMed]
- Founou, L.L.; Amoako, D.G.; Founou, R.C.; Essack, S.Y. Antibiotic resistance in food animals in Africa: A systematic review and meta-analysis. Microbial Drug Resistance 2018, 24, 648–665. [Google Scholar] [CrossRef]
- Kimera, Z.I.; Mshana, S.E.; Rweyemamu, M.M.; Mboera, L.E.G.; Matee, M.I.N. Antimicrobial use and resistance in food-producing animals and the environment: An African perspective. Antimicrob Resist Infect Control 2020, 9, 1–12. [Google Scholar] [CrossRef]
- Ogunlaja, A.; et al. Antibiotic resistomes and their chemical residues in aquatic environments in Africa. Environmental Pollution 2022, 312, 119783. [Google Scholar] [CrossRef]
- Founou, L.L.; Amoako, D.G.; Founou, R.C.; Essack, S.Y. Antibiotic resistance in food animals in Africa: A systematic review and meta-analysis. Microbial Drug Resistance 2018, 24, 648–665. [Google Scholar] [CrossRef]
- Bah, S.Y.; et al. Acquisition and carriage of genetically diverse multi-drug resistant gram-negative bacilli in hospitalised newborns in The Gambia. Communications Medicine 2023, 3. [Google Scholar] [CrossRef]
- Sintondji, K.; et al. Prevalence and characterization of ESBL-producing Escherichia coli in healthy pregnant women and hospital environments in Benin: An approach based on Tricycle. Front Public Health 2023, 11. [Google Scholar] [CrossRef] [PubMed]
- Bekoe, S.O.; et al. Reservoir of Antibiotic Residues and Resistant Coagulase Negative Staphylococci in a Healthy Population in the Greater Accra Region, Ghana. Antibiotics 2022, 11. [Google Scholar] [CrossRef]
- Socohou, A.; et al. Pathogenicity and Molecular Characterization of Staphylococcus aureus Strains Isolated from the Hospital Environment of CHU-Z Abomey-Calavi/So-Ava (Benin). Biomed Res Int 2021, 2021. [Google Scholar] [CrossRef] [PubMed]
- Acolatse, J.E.E.; et al. Environmental surveillance of ESBL and carbapenemase-producing gram-negative bacteria in a Ghanaian Tertiary Hospital. Antimicrob Resist Infect Control 2022, 11. [Google Scholar] [CrossRef] [PubMed]
- Acolatse, J.E.E.; et al. Environmental surveillance of ESBL and carbapenemase-producing gram-negative bacteria in a Ghanaian Tertiary Hospital. Antimicrob Resist Infect Control 2022, 11. [Google Scholar] [CrossRef] [PubMed]
- Olowe, O.A.; et al. Phylogenetic grouping and biofilm formation of multidrug resistant Escherichia coli isolates from humans, animals and food products in South-West Nigeria. Sci Afr 2019, 6. [Google Scholar] [CrossRef]
- Adzitey, F.; Huda, N.; Shariff, A.H.M. Phenotypic Antimicrobial Susceptibility of Escherichia coli from Raw Meats, Ready-to-Eat Meats, and Their Related Samples in One Health Context. Microorganisms 2021, 9. [Google Scholar] [CrossRef]
- Dougnon, V.; et al. Assessment of the Presence of Resistance Genes Detected from the Environment and Selected Food Products in Benin. J Environ Public Health 2021, 2021. [Google Scholar] [CrossRef]
- Abegewi, U.A.; Esemu, S.N.; Ndip, R.N.; Ndip, L.M. Prevalence and risk factors of coliform-associated mastitis and antibiotic resistance of coliforms from lactating dairy cows in North West Cameroon. PLoS ONE 2022, 17. [Google Scholar] [CrossRef]
- Beshiru, A.; Igbinosa, E.O. Surveillance of Vibrio parahaemolyticus pathogens recovered from ready-to-eat foods. Sci Rep 2023, 13. [Google Scholar] [CrossRef]
- Quarcoo, G.; et al. What Is in the Salad? Escherichia coli and Antibiotic Resistance in Lettuce Irrigated with Various Water Sources in Ghana. Int J Environ Res Public Health 2022, 19. [Google Scholar] [CrossRef]
- Deguenon, E.; et al. Hospital effluents as sources of antibiotics residues, resistant bacteria and heavy metals in Benin. SN Appl Sci 2022, 4. [Google Scholar] [CrossRef]
- Kagambèga, A.B.; et al. Detection and Characterization of Carbapenemase-Producing Escherichia coli and Klebsiella pneumoniae from Hospital Effluents of Ouagadougou, Burkina Faso. Antibiotics 2023, 12. [Google Scholar] [CrossRef] [PubMed]
- Adelowo, O.O.; Helbig, T.; Knecht, C.; Reincke, F.; Mäusezahl, I.; Müller, J.A. High abundances of class 1 integrase and sulfonamide resistance genes, and characterisation of class 1 integron gene cassettes in four urban wetlands in Nigeria. PLoS ONE 2018, 13. [Google Scholar] [CrossRef] [PubMed]
- OO, A.; et al. A survey of extended-spectrum beta-lactamase-producing Enterobacteriaceae in urban wetlands in southwestern Nigeria as a step towards generating prevalence maps of antimicrobial resistance. PLoS ONE 2020, 15, e0229451. [Google Scholar] [CrossRef]
- Adekanmbi, A.O.; Adeleke, O.J.; Aremu, O.O.; Victoria, A. Molecular characterization, antibiogram and distribution of zntA gene in zinc-resistant Escherichia coli population recovered from anthropogenically-influenced surface water sources in Nigeria. Meta Gene 2020, 26. [Google Scholar] [CrossRef]
- AO, A.; et al. Solid waste dumpsite leachate and contiguous surface water contain multidrug-resistant ESBL-producing Escherichia coli carrying Extended Spectrum β-Lactamase (ESBL) genes. BMC Microbiol 2024, 24, 308. [Google Scholar] [CrossRef]
- Babalola, T.F.; Olowomofe, T.O.; Omodara, T.R.; Ogunyemi, T.Y. Antibiotic Resistance Pattern and Plasmid Profile of Bacteria Isolates from Household Water Distribution Tanks in Ado-Ekiti. J Pure Appl Microbiol 2021, 15, 1697–1704. [Google Scholar] [CrossRef]
- Tsekleves, E.; de Souza, D.; Pickup, R.; Ahorlu, C.; Darby, A. Developing home cleaning intervention through community engagement to reduce infections and antimicrobial resistance in Ghanaian homes. Sci Rep 2023, 13. [Google Scholar] [CrossRef]
- Adeyemi, F.M.; et al. Integrated poultry-fish farming system encourages multidrug-resistant gram-negative bacteria dissemination in pond environment and fishes. Aquaculture 2022, 548. [Google Scholar] [CrossRef]
- Ekwanzala, M.D.; Dewar, J.B.; Kamika, I.; Momba, M.N.B. Systematic review in South Africa reveals antibiotic resistance genes shared between clinical and environmental settings. Infect Drug Resist 2018, 1907–1920. [Google Scholar] [CrossRef]
- Chokshi, A.; Sifri, Z.; Cennimo, D.; Horng, H. Global contributors to antibiotic resistance. J Glob Infect Dis 2019, 11, 36–42. [Google Scholar] [CrossRef]
- Haenni, M.; et al. Environmental contamination in a high-income country (France) by antibiotics, antibiotic-resistant bacteria, and antibiotic resistance genes: Status and possible causes. Environ Int 2022, 159, 107047. [Google Scholar] [CrossRef] [PubMed]
- Ehsan, H. Antibiotic Resistance in Developing Countries: Emerging Threats and Policy Responses. Public Health Challenges 2025, 4, e70034. [Google Scholar] [CrossRef]
- Adomako, L.A.B.; et al. Reduced Bacterial Counts from a Sewage Treatment Plant but Increased Counts and Antibiotic Resistance in the Recipient Stream in Accra, Ghana-A Cross-Sectional Study. Trop Med Infect Dis 2021, 6. [Google Scholar] [CrossRef]
- Appau, A.A.A.; Ofori, L.A. Antibiotic Resistance Profile of E. coli Isolates from Lettuce, Poultry Manure, Irrigation Water, and Soil in Kumasi, Ghana. Int J Microbiol 2024, 2024. [Google Scholar] [CrossRef]
- Al-Mustapha, A.I.; et al. extensively drug-resistant Escherichia coli isolated from broilers in Ilorin, North Central Nigeria. J Glob Antimicrob Resist 2022, 31, 337–344. [Google Scholar] [CrossRef]
- Igbinosa, E.O.; Beshiru, A.; Igbinosa, I.H.; Okoh, A.I. Antimicrobial resistance and genetic characterisation of Salmonella enterica from retail poultry meats in Benin City, Nigeria. LWT-Food Science and Technology 2022, 169. [Google Scholar] [CrossRef]
- Somda, N.S.; et al. Safety of ready-to-eat chicken in Burkina Faso: Microbiological quality, antibiotic resistance, and virulence genes in Escherichia coli isolated from chicken samples of Ouagadougou. Food Sci Nutr 2018, 6, 1077–1084. [Google Scholar] [CrossRef]
- Chigor, V.; Ibangha, I.A.; Chigor, C.; Titilawo, Y. Treated wastewater used in fresh produce irrigation in Nsukka, Southeast Nigeria is a reservoir of enterotoxigenic and multidrug-resistant Escherichia coli. Heliyon 2020, 6. [Google Scholar] [CrossRef]
- Ukwuoma, C.I.; Asikong, B.E.E.; Ebob, T. Prevalence of Vibrio species in Sea Foods and Water Sources in Cross River State. Annu Res Rev Biol 2022, 63–78. [Google Scholar] [CrossRef]
- Bah, S.Y.; et al. Acquisition and carriage of genetically diverse multi-drug resistant gram-negative bacilli in hospitalised newborns in The Gambia. Communications Medicine 2023, 3. [Google Scholar] [CrossRef]
- Djim-Adjim-Ngana, K.; et al. Prevalence of extended-spectrum beta-lactamase-producing enterobacterial urinary infections and associated risk factors in small children of Garoua, Northern Cameroon. Pan African Medical Journal 2020, 36. [Google Scholar] [CrossRef] [PubMed]
- Paterson, D.L.; Bonomo, R.A. Extended-spectrum β-lactamases: A clinical update. Clin Microbiol Rev 2005, 18, 657–686. [Google Scholar] [CrossRef] [PubMed]
- Quarcoo, G.; et al. What Is in the Salad? Escherichia coli and Antibiotic Resistance in Lettuce Irrigated with Various Water Sources in Ghana. Int J Environ Res Public Health 2022, 19. [Google Scholar] [CrossRef] [PubMed]
- Alhaji, N.B.; Odetokun, I.A.; Jibrin, M.S.; Lawan, M.K.; Kwaga, J. Antibiotic resistance and mitigation using One Health lens in aquaculture of Northern Nigeria. Onderstepoort Journal of Veterinary Research 2024, 91, 2165. [Google Scholar] [CrossRef]
- Oche, C.; Aladetoun, N.F.; Barde, I.J. Use and abuse of antibiotics in Eriwe farms in Ijebu Ode, Ogun State, Nigeria: A case report. Global Journal of Fisheries Science 2024, 6, 97–106. [Google Scholar] [CrossRef]
- Oloso, N.O.; Adeyemo, I.A.; van Heerden, H.; Fasanmi, O.G.; Fasina, F.O. Antimicrobial Drug Administration and Antimicrobial Resistance of Salmonella Isolates Originating from the Broiler Production Value Chain in Nigeria. Antibiotics 2019, 8. [Google Scholar] [CrossRef]
- Obeng-Nkrumah, N.; et al. High level of colonization with third-generation cephalosporin-resistant Enterobacterales in African community settings, Ghana. Diagn Microbiol Infect Dis 2023, 106. [Google Scholar] [CrossRef]
- JK, C.; et al. Population structure and antimicrobial resistance among Klebsiella isolates sampled from human, animal, and environmental sources in Ghana: A cross-sectional genomic One Health study. Lancet Microbe 2023, 4, e943–e952. [Google Scholar] [CrossRef]
- Ayibieke, A.; et al. Molecular characterisation of the NDM-1-encoding plasmid p2189-NDM in an Escherichia coli ST410 clinical isolate from Ghana. PLoS ONE 2018, 13. [Google Scholar] [CrossRef]
- Muhigwa, M.; et al. Characterization of extended-spectrum beta-lactamase and carbapenemase genes in bacteria from environment in Burkina Faso. J Infect Dev Ctries 2023, 17, 1714–1721. [Google Scholar] [CrossRef]
- Adelowo, O.O.; Vollmers, J.; Mäusezahl, I.; Kaster, A.K.; Müller, J.A. Detection of the carbapenemase gene blaVIM-5 in members of the Pseudomonas putida group isolated from polluted Nigerian wetlands. Sci Rep 2018, 8. [Google Scholar] [CrossRef] [PubMed]
- Akinola, O.T.; Onyeaghasiri, F.U.; Oluranti, O.O.; Elutade, O.O. Assessment of well water as a reservoir for extended-spectrum β-lactamases (ESBL) and carbapenem resistant Enterobacteriaceae from Iwo, Osun state, Nigeria. Iran J Microbiol 2022, 14, 351–361. [Google Scholar] [CrossRef] [PubMed]
- Banu, R.A.; et al. Extended Spectrum Beta-Lactamase Escherichia coli in River Waters Collected from Two Cities in Ghana, 2018-2020. Trop Med Infect Dis 2021, 6. [Google Scholar] [CrossRef]
- Mahazu, S.; et al. Insights and genetic features of extended-spectrum beta-lactamase producing Escherichia coli isolates from two hospitals in Ghana. Sci Rep 2022, 12, 1843. [Google Scholar] [CrossRef]
- Founou, L.L.; Founou, R.C.; Allam, M.; Ismail, A.; Djoko, C.F.; Essack, S.Y. Genome Sequencing of Extended-Spectrum β-Lactamase (ESBL)-Producing Klebsiella pneumoniae Isolated from Pigs and Abattoir Workers in Cameroon. Front Microbiol 2018, 9. [Google Scholar] [CrossRef]
- Adekanmbi, A.O.; Oluwaseyi, T.A.; Oyelade, A.A. Dumpsite leachate as a hotspot of multidrug resistant Enterobacteriaceae harbouring extended spectrum and AmpC β-lactamase genes; a case study of Awotan municipal solid waste dumpsite in Southwest Nigeria. Meta Gene 2021, 28. [Google Scholar] [CrossRef]
- Murray, C.J.L.; et al. Global burden of bacterial antimicrobial resistance in 2019: A systematic analysis. The lancet 2022, 399, 629–655. [Google Scholar] [CrossRef] [PubMed]
- Labi, A.K.; et al. High Carriage Rates of Multidrug-Resistant Gram-Negative Bacteria in Neonatal Intensive Care Units From Ghana. Open Forum Infect Dis 2020, 7. [Google Scholar] [CrossRef]
- John-Onwe, B.N.; et al. Prevalence and multidrug-resistant ESBL-producing E. coli in urinary tract infection cases of HIV patients attending Federal Teaching Hospital, Abakaliki, Nigeria. Afr J Microbiol Res 2022, 16, 196–201. [Google Scholar] [CrossRef]
- Adegoke, A.A.; Ikott, W.E.; Okoh, A.I. Carbapenem resistance associated with coliuria among outpatient and hospitalised urology patients. New Microbes New Infect 2022, 48. [Google Scholar] [CrossRef]
- Adegoke, A.A.; Ikott, W.E.; Okoh, A.I. Carbapenem resistance associated with coliuria among outpatient and hospitalised urology patients. New Microbes New Infect 2022, 48. [Google Scholar] [CrossRef] [PubMed]
- Egbule, O.S.; Iweriebor, B.C.; Odum, E.I. Beta-Lactamase-Producing Escherichia coli Isolates Recovered from Pig Handlers in Retail Shops and Abattoirs in Selected Localities in Southern Nigeria: Implications for Public Health. Antibiotics 2021, 10. [Google Scholar] [CrossRef] [PubMed]
- Obeng-Nkrumah, N.; et al. High level of colonization with third-generation cephalosporin-resistant Enterobacterales in African community settings, Ghana. Diagn Microbiol Infect Dis 2023, 106. [Google Scholar] [CrossRef]
- Ayibieke, A.; et al. Molecular characterisation of the NDM-1-encoding plasmid p2189-NDM in an Escherichia coli ST410 clinical isolate from Ghana. PLoS ONE 2018, 13. [Google Scholar] [CrossRef]
- Bisi-Johnson, M.A.; Adedeji, A.A.; Sulaiman, A.A.; Adefisoye, M.A.; Okoh, A.I. Isolation and genotypic characterization of extended-spectrum beta-lactamase-producing Escherichia coli O157:H7 and Aeromonas hydrophila from selected freshwater sources in Southwest Nigeria. Sci Rep 2023, 13. [Google Scholar] [CrossRef]
- Aworh, M.K.; et al. Extended-spectrum β-lactamase-producing Escherichia coli among humans, chickens and poultry environments in Abuja, Nigeria. One Health Outlook 2020, 2. [Google Scholar] [CrossRef]
- Atobatele, B.O.; Akinola, O.T.; Olutona, G.O. Molecular characterization and detection of multidrug-resistant gene in bacterial strains in a health care centre located in Iwo, Osun State, Nigeria. Sci Afr 2023, 21. [Google Scholar] [CrossRef]
- Nsofor, C.A.; Moses, A.; Onyeakazi, C.M.; Okeke, C.J.; Ikegbunam, M.N. Detection of blaCTX-M, blaTEM, and blaSHV genes in clinical isolates of Escherichia coli and Klebsiella pneumoniae from Nigeria. Reviews and Research in Medical Microbiology 2023, 34, 66–72. [Google Scholar] [CrossRef]
- Hansen, K.H.; Andreasen, M.R.; Pedersen, M.S.; Westh, H.; Jelsbak, L.; Schønning, K. Resistance to piperacillin/tazobactam in Escherichia coli resulting from extensive IS 26-associated gene amplification of bla TEM-1. Journal of Antimicrobial Chemotherapy 2019, 74, 3179–3183. [Google Scholar] [CrossRef]
- Portal, E.A.R.; et al. Characterisation of colistin resistance in Gram-negative microbiota of pregnant women and neonates in Nigeria. Nat Commun 2024, 15, 2302. [Google Scholar] [CrossRef]
- Mohamadou, M.; et al. High prevalence of Panton-Valentine leukocidin positive, multidrug resistant, Methicillin-resistant Staphylococcus aureus strains circulating among clinical setups in Adamawa and Far North regions of Cameroon. PLoS ONE 2022, 17. [Google Scholar] [CrossRef] [PubMed]
- Atobatele, B.O.; Akinola, O.T.; Olutona, G.O. Molecular characterization and detection of multidrug-resistant gene in bacterial strains in a health care centre located in Iwo, Osun State, Nigeria. Sci Afr 2023, 21. [Google Scholar] [CrossRef]
- Oladipo, A.O.; Oladipo, O.G.; Bezuidenhout, C.C. Multi-drug resistance traits of methicillin-resistant Staphylococcus aureus and other Staphylococcal species from clinical and environmental sources. J Water Health 2019, 17, 930–943. [Google Scholar] [CrossRef] [PubMed]
- Adekanmbi, A.O.; Adejoba, A.T.; Banjo, O.A.; Saki, M. Detection of sul1 and sul2 genes in sulfonamide-resistant bacteria (SRB) from sewage, aquaculture sources, animal wastes and hospital wastewater in South-West Nigeria. Gene Rep 2020, 20. [Google Scholar] [CrossRef]
- Oladipo, A.O.; Oladipo, O.G.; Bezuidenhout, C.C. Multi-drug resistance traits of methicillin-resistant Staphylococcus aureus and other Staphylococcal species from clinical and environmental sources. J Water Health 2019, 17, 930–943. [Google Scholar] [CrossRef]
- Tettey, R.; et al. Genomic analysis of multidrug-resistant Escherichia coli from Urban Environmental water sources in Accra, Ghana, Provides Insights into public health implications. PLoS ONE 2024, 19, e0301531. [Google Scholar] [CrossRef]
- Abana, D.; et al. Investigating the virulence genes and antibiotic susceptibility patterns of Vibrio cholerae O1 in environmental and clinical isolates in Accra, Ghana. BMC Infect Dis 2019, 19, 76. [Google Scholar] [CrossRef]
- Adekanmbi, A.O.; Oluwaseyi, T.A.; Oyelade, A.A. Dumpsite leachate as a hotspot of multidrug resistant Enterobacteriaceae harbouring extended spectrum and AmpC β-lactamase genes; a case study of Awotan municipal solid waste dumpsite in Southwest Nigeria. Meta Gene 2021, 28. [Google Scholar] [CrossRef]
- Azaglo, G.S.K.; et al. Bacteria and Their Antibiotic Resistance Profiles in Ambient Air in Accra, Ghana, February 2020: A Cross-Sectional Study. Trop Med Infect Dis 2021, 6. [Google Scholar] [CrossRef]
- Banu, R.A.; et al. Extended Spectrum Beta-Lactamase Escherichia coli in River Waters Collected from Two Cities in Ghana, 2018-2020. Trop Med Infect Dis 2021, 6. [Google Scholar] [CrossRef]
- Greenland, K.; Huberts, J.D.-W.; Wright, R.; Hawkes, L.; Ekor, C.; Biran, A. A cross-sectional survey to assess household sanitation practices associated with uptake of ‘Clean Team’ serviced home toilets in Kumasi, Ghana. Environ Urban 2016, 28, 583–598. [Google Scholar] [CrossRef]
- Abubakar, I.R. Exploring the determinants of open defecation in Nigeria using demographic and health survey data. Science of The Total Environment 2018, 637–638, 1455–1465. [Google Scholar] [CrossRef] [PubMed]
- Gayawan, E.; Somo-Aina, O.; Kuti, O. Analysis of the space-time trends in open defecation in Nigeria. Environmental Science and Pollution Research 2023, 30, 68524–68535. [Google Scholar] [CrossRef]
- Johnson, N.G.; Agada, D.U.; John-Nwagwu, H.O.; Aremu, R.; Guma, E.P. Assessing the Spatial Distribution of Public Toilets in Lokoja Metropolis, Kogi State, Nigeria, Mitigating Open Defecation. Journal of Spatial Information Sciences 2024, 1, 111–126. [Google Scholar]
- Nwafia, I.N.; Ike, A.C.; Orabueze, I.N.; Nwafia, W.C. Carbapenemase producing Enterobacteriaceae: Environmental reservoirs as primary targets for control and prevention strategies. Nigerian Postgraduate Medical Journal 2022, 29, 183–191. [Google Scholar] [CrossRef]
- Mo, A.I.-J. Incidence of antibiotic resistance genotypes of Vibrio species recovered from selected freshwaters in Southwest Nigeria. Sci Rep 2022, 12, 18912. [Google Scholar] [CrossRef]
- Agboola, T.D.; Nmema, E.E.; Samuel, B.T.; Odetoyin, B.W. Multiple Antibiotic Resistant Vibrio Pathotypes with the Incidence of V. Cholerae and V. parahaemolyticus in Fish and Fish Storage Water in Okitipupa and Igbokoda Areas, Nigeria. South Asian Journal of Research in Microbiology 2022, 13, 11–23. [Google Scholar] [CrossRef]
- Azike, C.A.; Agi, V.N.; Nwokah, E.G.; Ollor, A.O.; Nyenke, C.U.; Wachukwu, C.K. Antibiogram, Genomic and Phylogeny of Stool and Seafood Isolates from Some Cholera-Prone Coastal Communities in Rivers State, Nigeria. J Biosci Med 2023, 11, 385–406. [Google Scholar] [CrossRef]
- Mphasa, M.; et al. Urban waste piles are reservoirs for human pathogenic bacteria with high levels of multidrug resistance against last resort antibiotics: A comprehensive temporal and geographic field analysis. J Hazard Mater 2022, 484, 136639. [Google Scholar] [CrossRef]
- Edet, U.O.; et al. Impact of ‘sachet water’ microplastic on agricultural soil physicochemistry, antibiotics resistance, bacteria diversity and function. SN Appl Sci 2022, 4, 1–16. [Google Scholar] [CrossRef]
- Borquaye, L.S.; et al. Occurrence of Antibiotics and Antibiotic-Resistant Bacteria in Landfill Sites in Kumasi, Ghana. J Chem 2019, 2019. [Google Scholar] [CrossRef]
- Akpan, S.N.; Odeniyi, O.A.; Adebowale, O.O.; Alarape, S.A.; Adeyemo, O.K. Antibiotic resistance profile of Gram-negative bacteria isolated from Lafenwa abattoir effluent and its receiving water (Ogun River) in Abeokuta, Ogun state, Nigeria. Onderstepoort Journal of Veterinary Research 2020, 87. [Google Scholar] [CrossRef] [PubMed]
- Bisi-Johnson, M.A.; Adedeji, A.A.; Sulaiman, A.A.; Adefisoye, M.A.; Okoh, A.I. Isolation and genotypic characterization of extended-spectrum beta-lactamase-producing Escherichia coli O157:H7 and Aeromonas hydrophila from selected freshwater sources in Southwest Nigeria. Sci Rep 2023, 13. [Google Scholar] [CrossRef] [PubMed]
- Baah, D.A.; Kotey, F.C.N.; Dayie, N.; Codjoe, F.S.; Tetteh-Quarcoo, P.B.; Donkor, E.S. Multidrug-Resistant Gram-Negative Bacteria Contaminating Raw Meat Sold in Accra, Ghana. Pathogens 2022, 11. [Google Scholar] [CrossRef]
- Abas, R.; Cobbina, S.J.; Abakari, G. Microbial quality and antibiotic sensitivity of bacterial isolates in ‘Tuo-Zaafi’ vended in the central business district of tamale. Food Sci Nutr 2019, 7, 3613–3621. [Google Scholar] [CrossRef]
- Egbule, O.S.; Iweriebor, B.C.; Odum, E.I. Beta-Lactamase-Producing Escherichia coli Isolates Recovered from Pig Handlers in Retail Shops and Abattoirs in Selected Localities in Southern Nigeria: Implications for Public Health. Antibiotics 2021, 10. [Google Scholar] [CrossRef]
- Igbinosa, E.O.; Beshiru, A.; Igbinosa, I.H.; Ogofure, A.G.; Uwhuba, K.E. Prevalence and Characterization of Food-Borne Vibrio parahaemolyticus From African Salad in Southern Nigeria. Front Microbiol 2021, 12. [Google Scholar] [CrossRef]
- Somda, N.S.; et al. Safety of ready-to-eat chicken in Burkina Faso: Microbiological quality, antibiotic resistance, and virulence genes in Escherichia coli isolated from chicken samples of Ouagadougou. Food Sci Nutr 2018, 6, 1077–1084. [Google Scholar] [CrossRef]
- Founou, L.L.; Founou, R.C.; Allam, M.; Ismail, A.; Djoko, C.F.; Essack, S.Y. Genome Sequencing of Extended-Spectrum β-Lactamase (ESBL)-Producing Klebsiella pneumoniae Isolated from Pigs and Abattoir Workers in Cameroon. Front Microbiol 2018, 9. [Google Scholar] [CrossRef]
- Beshiru, A.; et al. Biofilm and antimicrobial resistance profile of extended-spectrum β-lactamase (ESBL) and AmpC β-lactamase producing Enterobacteriaceae in vegetables and salads. LWT-Food Science and Technology 2023, 182. [Google Scholar] [CrossRef]
- Dougnon, V.; et al. Assessment of the Presence of Resistance Genes Detected from the Environment and Selected Food Products in Benin. J Environ Public Health 2021, 2021. [Google Scholar] [CrossRef]
- Darboe, S.; et al. Prevalence of Panton-Valentine Leukocidin (PVL) and Antimicrobial Resistance in Community-Acquired Clinical Staphylococcus aureus in an Urban Gambian Hospital: A 11-Year Period Retrospective Pilot Study. Front Cell Infect Microbiol 2019, 9, 170. [Google Scholar] [CrossRef] [PubMed]
- Bekoe, S.O.; et al. Reservoir of Antibiotic Residues and Resistant Coagulase Negative Staphylococci in a Healthy Population in the Greater Accra Region, Ghana. Antibiotics 2022, 11. [Google Scholar] [CrossRef] [PubMed]
- Appau, A.A.A.; Ofori, L.A. Antibiotic Resistance Profile of E. coli Isolates from Lettuce, Poultry Manure, Irrigation Water, and Soil in Kumasi, Ghana. Int J Microbiol 2024, 2024. [Google Scholar] [CrossRef]
- Manasfi, R.; Brienza, M.; Ait-Mouheb, N.; Montemurro, N.; Perez, S.; Chiron, S. Impact of long-term irrigation with municipal reclaimed wastewater on the uptake and degradation of organic contaminants in lettuce and leek. Science of the Total Environment 2021, 765. [Google Scholar] [CrossRef]
- Chigor, V.; Ibangha, I.A.; Chigor, C.; Titilawo, Y. Treated wastewater used in fresh produce irrigation in Nsukka, Southeast Nigeria is a reservoir of enterotoxigenic and multidrug-resistant Escherichia coli. Heliyon 2020, 6. [Google Scholar] [CrossRef]
- Manasfi, R.; Brienza, M.; Ait-Mouheb, N.; Montemurro, N.; Perez, S.; Chiron, S. Impact of long-term irrigation with municipal reclaimed wastewater on the uptake and degradation of organic contaminants in lettuce and leek. Science of the Total Environment 2021, 765. [Google Scholar] [CrossRef]
- Matchawe, C.; Bonny, P.; Yandang, G.; Mafo, H.C.Y.; Nsawir, B.J. Water shortages: Cause of water safety in sub-Saharan Africa. in Drought-impacts and management, IntechOpen, 2022.
- Inyinbor, A.A.; Tsopmo, A.; Udenigwe, C.C. Antibiotics threats on vegetables and the perils of low income nations practices. Sustain Chem Pharm 2021, 21, 100448. [Google Scholar] [CrossRef]
- Mapfumo, B. Regional review on status and trends in aquaculture development in sub-Saharan Africa–2020. 2022.
- Ogunji, J.; Wuertz, S. Aquaculture Development in Nigeria: The Second Biggest Aquaculture Producer in Africa. Water 2023, 15, 4224. [Google Scholar] [CrossRef]
- Oche, C.; Aladetoun, N.F.; Barde, I.J. Use and abuse of antibiotics in Eriwe farms in Ijebu Ode, Ogun State, Nigeria: A case report. Global Journal of Fisheries Science 2024, 6, 97–106. [Google Scholar] [CrossRef]
- Alhaji, N.B.; Maikai, B.V.; Kwaga, J.K.P. Antimicrobial use, residue and resistance dissemination in freshwater fish farms of north-central Nigeria: One health implications. Food Control 2021, 130. [Google Scholar] [CrossRef]
- Michael, A.; Polycarp, M.; Sanda, M.K.; David, S.A. An Analysis of Fish Farmers’ Management Practices and Information Needs in Adamawa State, Nigeria. Scientific Papers Series Management, Economic Engineering in Agriculture & Rural Development 2021, 21. [Google Scholar]
- Adah, D.A.; Saidu, L.; Oniye, S.J.; Adah, A.S.; Daoudu, O.B.; Ola-Fadunsin, S.D. Molecular characterization and antibiotics resistance of Aeromonas species isolated from farmed African catfish Clarias gariepinus Burchell, 1822. BMC Vet Res 2024, 20. [Google Scholar] [CrossRef] [PubMed]
- Ngbede, E.O.; et al. Concurrent Resistance to Carbapenem and Colistin Among Enterobacteriaceae Recovered From Human and Animal Sources in Nigeria Is Associated With Multiple Genetic Mechanisms. Front Microbiol 2021, 12. [Google Scholar] [CrossRef]
- Oloso, N.O.; Adeyemo, I.A.; van Heerden, H.; Fasanmi, O.G.; Fasina, F.O. Antimicrobial Drug Administration and Antimicrobial Resistance of Salmonella Isolates Originating from the Broiler Production Value Chain in Nigeria. Antibiotics 2019, 8. [Google Scholar] [CrossRef]
- Yakubu, Y.; et al. Understanding the awareness of antimicrobial resistance amongst commercial poultry farmers in northwestern Nigeria. Prev Vet Med 2024, 228, 106226. [Google Scholar] [CrossRef]
- WHO, “WHO Strategic and Technical Advisory Group for Antimicrobial Resistance (STAG- AMR): Report of the fourth meeting 11-13 June 2024. Geneva: World Health Organization; 2024. Licence: CC BY-NC-SA 3.0 IGO.. Jun. 2024.
- Ayibieke, A.; et al. Prevalence and Characterization of Carbapenem-Hydrolyzing Class D β-Lactamase-Producing Acinetobacter Isolates From Ghana. Front Microbiol 2020, 11. [Google Scholar] [CrossRef]
- Labi, A.K.; et al. High Carriage Rates of Multidrug-Resistant Gram-Negative Bacteria in Neonatal Intensive Care Units From Ghana. Open Forum Infect Dis 2020, 7. [Google Scholar] [CrossRef]
- Odewale, G.; Jibola-Shittu, M.Y.; Ojurongbe, O.; Olowe, R.A.; Olowe, O.A. Genotypic Determination of Extended Spectrum β-Lactamases and Carbapenemase Production in Clinical Isolates of Klebsiella pneumoniae in Southwest Nigeria. Infect Dis Rep 2023, 15, 339–353. [Google Scholar] [CrossRef]
- Samia, N.I.; Robicsek, A.; Heesterbeek, H.; Peterson, L.R. Methicillin-resistant staphylococcus aureus nosocomial infection has a distinct epidemiological position and acts as a marker for overall hospital-acquired infection trends. Sci Rep 2022, 12, 17007. [Google Scholar] [CrossRef]
- Socohou, A.; et al. Pathogenicity and Molecular Characterization of Staphylococcus aureus Strains Isolated from the Hospital Environment of CHU-Z Abomey-Calavi/So-Ava (Benin). Biomed Res Int 2021, 2021. [Google Scholar] [CrossRef] [PubMed]
- Igbinosa, E.O.; Beshiru, A.; Igbinosa, I.H.; Ogofure, A.G.; Ekundayo, T.C.; Okoh, A.I. Prevalence, multiple antibiotic resistance and virulence profile of methicillin-resistant Staphylococcus aureus (MRSA) in retail poultry meat from Edo, Nigeria. Front Cell Infect Microbiol 2023, 13. [Google Scholar] [CrossRef] [PubMed]
- Franco-Duarte, R.; et al. Advances in chemical and biological methods to identify microorganisms—From past to present. Microorganisms 2019, 7, 130. [Google Scholar] [CrossRef] [PubMed]
- Fluit, A.C.; Visser, M.R.; Schmitz, F.-J. Molecular detection of antimicrobial resistance. Clin Microbiol Rev 2001, 14, 836–871. [Google Scholar] [CrossRef]
- Zamudio, R.; et al. Global transmission of extended-spectrum cephalosporin resistance in Escherichia coli driven by epidemic plasmids. EBioMedicine 2024, 103. [Google Scholar] [CrossRef]
- Anjum, M.F.; Zankari, E.; Hasman, H. Molecular methods for detection of antimicrobial resistance. Antimicrobial resistance in bacteria from livestock and companion animals, pp. 33–50, 2018.
- Akinyemi, K.O.; et al. Whole genome sequencing of Salmonella enterica serovars isolated from humans, animals, and the environment in Lagos, Nigeria. BMC Microbiol 2023, 23. [Google Scholar] [CrossRef]
- Cerdeira, J.; Mesquita, J.; Vieira, E.S. International research collaboration: Is Africa different? A cross-country panel data analysis. Scientometrics 2023, 128, 2145–2174. [Google Scholar] [CrossRef]
- Caelers, D.; Okoth, D. Research funding in Africa: Navigating sustainability and shifting perspectives. Nat Africa 2023, 10. [Google Scholar] [CrossRef]
- Jones, E.R.; et al. Sub-Saharan Africa will increasingly become the dominant hotspot of surface water pollution. Nature Water 2023, 1, 602–613. [Google Scholar] [CrossRef]
- Omohwovo, E.J. Wastewater management in Africa: Challenges and recommendations. Environ Health Insights 2024, 18, 11786302241289680. [Google Scholar] [CrossRef]
- Adzitey, F.; Ekli, R.; Aduah, M. Incidence and antibiotic susceptibility of Staphylococcus aureus isolated from ready-to-eat meats in the environs of Bolgatanga Municipality of Ghana. Cogent Environ Sci 2020, 6. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




