1. Introduction
Microbial treatment especially simultaneous nitrification and denitrification (SND) processes for saline wastewater that is being ceaselessly created by industries, is of significant interest for mitigating water eutrophication and maintaining the natural nitrogen balance [
1,
2,
3,
4,
5]. For instance, wastewater generated from processes such as the steaming of ammonia in the coking industry, crude oil electro-desalting in the refining sector, pharmaceutical manufacturing, and the production of pickling and tartaric acid, typically exhibits elevated salinity levels ranging from 1% to over 10% [
6,
7,
8,
9,
10]. The elevated osmotic pressure of saline wastewater leads to the inhibition of microbial activity, induces plasmolysis, gives rise to bacterial demise. Up to now, most present halotolerant SND strains can only effectively remove nitrogen from wastewater with ≤ 5% NaCl [
11,
12,
13]. Recently, a robust bacterium
Exiguobacterium mexicanum demonstrated the ability to perform SND beyond 5% NaCl salinity but its growth was greatly inhibited [
14]. In addition, our previous work identified that
Halomonas salifodinae exhibited substantial SND capabilities at 15% salinity. This was achieved through the accumulation of intracellular compatible solutes, which facilitate water retention and enable the bacterium to withstand high osmotic pressure of the hypersaline wastewater [
15,
16].
Besides chloride salts, industrial wastewater usually contains various oxysalts such as sulfates (e.g., SO
42−), phosphates (e.g., HPO
42−), carbonates (e.g., HCO
3−), and nitrates (e.g., NO
3−) [
17,
18,
19,
20]. For example, wastewater from the hydrolysis of imidazole aldehyde in the pharmaceutical industry and from metal processing contain more than 10% phosphates and sulfates, respectively. In addition, swine wastewater contains ca. 1% carbonates, while wastewater from the nuclear industry contains over 5% nitrates due to the use of nitric acid in metal cleaning processes [
21,
22,
23]. Thus, it is desirable to remove nitrogen of industrial wastewater containing these oxysalts via SND processes. At present, research is mainly concentrated on determining the influences of sulfate salinity on the SND activities of mixed bacterial communities [
6,
11,
23,
24,
25]. When the concentration of Na
2SO
4 reached ca. 1.5% in the parallel bench reactors, the removal efficiency of ammonium nitrogen (NH
4+-N) exceeded 90%. In contrast, ca. 60% of total nitrogen (TN) was removed, attributed to the inhibition of nitrite oxidizing bacteria activity and the accumulation of nitrite nitrogen (NO
2–-N) under the stress induced by Na
2SO
4 [
25]. In the sequenced batch reactors at 3% salinity, the exposure of halophilic autotrophic nitrification sludge system to SO
42− instead of Cl
− resulted in an increased relative abundance of an ammonia oxidizing bacterium
Nitrosomonas, accompanied by a rise in extracellular polymeric substances to tolerate elevated osmotic pressure [
11]. In addition, half inhibitory concentrations of NaCl and Na
2SO
4 in freshwater Anammox system were determined to be 0.106 and 0.063 M, respectively, indicating that SO
42− has higher microbial activity inhibition than Cl
− [
6]. To date, efficient nitrogen removal has been reported in mixed bacterial systems containing ≤ 3% SO
42− [
11,
25,
26]. However, there is a paucity of robust single strains capable of the SND in saline wastewater with > 3% SO
42−, as well as even other oxysalts. A SND bacterium
Acinetobacter sp. TAC-1 isolated from pig farm wastewater exhibited half inhibition concentrations of 0.205 M for NaCl, 0.238 M for KCl, and 0.110 M for Na
2SO
4 [
27].
In this study, a robust bacterium Marinobacter sp. was separated from aerobic sludge in a saline wastewater treatment plant (WWTP). The SND activity of the separated bacterium at 11% Na2SO4 salinity was optimized via response surface methodology. The strain's exceptional tolerance to oxysalts was confirmed through comparative analysis with Pseudomonas sp. and Halomonas sp., which were isolated from aerobic sludges in petrochemical and pharmaceutical WWTP, respectively. A systematic investigation was conducted to assess the SND efficiencies in saline wastewater containing NaCl or oxysalts such as Na2SO4, Na2HPO4, NaHCO3, and NaNO3 at salinities ranging from 1% to 11%. Moreover, the SND efficiency of the separated bacterium at 11% Na2SO4 salinity, along with the associated functional genes and key enzymatic activities were studied to elucidate the nitrogen removal pathway and mechanism.
2. Materials and Methods
2.1. Isolation and Identification
Saline aerobic sludge supplied by Hubei Huisheng pharmaceutical WWTP (China) was used to isolate the SND bacteria. The control SND bacterium was separated from saline aerobic sludge sample in petrochemical refinery WWTP (China). The SND capabilities of the separated bacteria were assessed in heterotrophic nitrification (HN) media and aerobic denitrification (AD) media, the components of which are detailed in the Supplementary Material. Strain Y2 with high SND efficiencies and distinctive oxysalt-tolerant properties was selected for subsequent assays. The 16S rRNA of strain Y2 was amplified by polymerase chain reaction (PCR) and sequenced by TSINGKE Biotechnology Co., Ltd (Wuhan, China). The results were submitted to GenBank database in comparison with other microorganisms by BLAST. MEGA 7.0 software was applied to create a phylogenetic tree through NJ algorithm.
2.2. Response Surface Methodology Optimization
The influences of C/N ratio (8, 12 and 16), initial pH value (6, 8 and 10), temperature (20, 30 and 40 °C), and shaking speed (50, 150 and 250 rpm) on the SND activities of the isolated strain Y2 at 11% Na
2SO
4 salinity were investigated through response surface methodology with the Box-Behnken design. In these 29 experiments, bacterial suspensions were transferred into the HN media containing 11% Na
2SO
4, which were cultured for 72 h. The nitrogen removal efficiencies were measured by the equation:
where
RN is the nitrogen removal efficiencies,
C0 and
Ct are the NH
4+-N concentrations before and after the SND processes, respectively. Response surface plots along with the associated contour plots were generated according to the experimental models.
2.3. Assessment of Salt Tolerance
To evaluate the tolerance of microorganisms to different inorganic salts, including NaCl, Na
2SO
4, Na
2HPO
4, NaHCO
3, and NaNO
3, the isolated strain
Marinobacter sp. as well as
Pseudomonas sp. and
Halomonas sp. were cultivated at different salinities to determine bacterial growth and nitrogen removal efficiencies. In addition,
Marinobacter sp.,
Pseudomonas sp., and
Halomonas sp. were cultivated at 11%, 3%, and 11% NaCl for 96 h, 72 h, and 60 h, respectively. Subsequently, all the strains were collected and were individually inoculated into HN media containing 11%, 3%, and 11% NaNO
3 for further cultivation. The bacterial growth was measured at intervals and the bacterial morphology was observed by SEM (Regulus 8100, Hitachi). The salinity activity inhibition ratios were calculated according to the equation:
where
IRS is the salinity activity inhibition ratios,
νopt and
νs are the nitrogen removal rates at the optimal and other salinities, respectively.
2.4. Effects of Different Oxysalts on Bacterial Growth and Nitrogen Removal
Batch experiments were conducted to estimate the effects of Cl− and various oxysalts including SO42−, HPO42−, HCO3−, and NO3− on bacterial growth and nitrogen removal efficiencies of strain Y2. Suspensions of strain Y2 were transferred into either HN or AD media including NaCl, Na2SO4, Na2HPO4, NaHCO3 or NaNO3 with the initial pH of 7.2. The concentrations of these adscititious salts were 1%, 3%, 5%, 7%, 9%, and 11%. During cultivation under 150 rpm at 30 °C, the bacterial growth and NH4+-N or nitrate nitrogen (NO3−-N) concentrations were measured at intervals.
2.5. SND Capability at 11% Na2SO4
To evaluate SND performance of the oxysalt-tolerant strain Y2, bacterial suspensions were added into the sterilized media containing different nitrogen sources and 11% Na2SO4. Sodium succinate was used as carbon source to achieve a C/N ratio of 15, the initial concentrations of nitrogen sources and pH values were 200 mg/L and 7.2. Strain Y2 was incubated at 150 rpm and 30 °C. Samples were collected at intervals to measure the bacterial growth and various nitrogen source concentrations.
2.6. Nitrogen Removal Enzymes and Functional Genes for the SND
Nitrogen removal enzymes for the SND mainly include ammonia monooxygenase (AMO), hydroxylamine oxidase (HAO), periplasm nitrate reductase (NAP), nitrite reductase (NIR). Ultrasonication was utilized to prepare acellular crude enzyme extracts [
28,
29]. The activities of AMO and HAO were determined in Tris-HCl buffer solutions containing the crude enzyme extracts, cytochrome
c, and specific substrates. The activities of NIR and NAP were determined in phosphate buffer solutions containing the crude enzyme extracts, NADH, and target substrates. Control experiments were conducted in the absence of either the crude enzyme extracts or cytochrome
c/NADH. Functional genes associated with nitrogen removal in the SND were detected. Genomic DNA of strain Y2 was extracted through the manufacturer’s protocol and was used for functional genes amplification. The primers and conditions for PCR amplification were based on previously established methodologies [
15].
2.7. Analytical Methods
The bacterial growth (OD
600) was estimated through determining the optical density of bacterial suspension at 600 nm. The concentrations of NH
4+-N, NO
2–-N, NO
3–-N, and TN were quantified through the standard methods [
30]. The concentrations of NH
2OH were determined referring to the reported spectrophotometric method [
31]. The concentrations of IN were determined by deducting the TN of centrifuged bacterial supernatants from the TN of uncentrifuged bacterial suspensions.
3. Results and Discussion
3.1. Identifying the Isolated Strain Y2
After a repeated-batch acclimation, eight robust salt-tolerant bacterial strains were isolated from saline aerobic sludge from a pharmaceutical WWTP. Among these isolated bacteria, strain Y2 with unique oxysalt-tolerant capabilities and high SND activities was used for further investigation. On the agar plate, strain Y2 forms beige colonies with a diameter of ca. 1 mm, and smooth, slightly raised surfaces (
Figure S1A). SEM image shows that strain Y2 appears short rods with the size of 0.4~0.6 × 2.0~4.0 µm (
Figure S1B).
Sequencing of the PCR-amplified 16S rRNA (1413 bp) of strain Y2 were conducted, and 16S rRNA were submitted to GenBank database (accession number: PQ241653). BLAST homology analysis indicates that strain Y2 shares over 99% similarity with
Marinobacter sp. A phylogenetic tree created using the NJ algorithm further classifies that strain Y2 is affiliated to
Marinobacter shengliensis (
Figure S1C). While
Marinobacter sp. are commonly found in wastewater treatment systems [
32,
33,
34], but there are few reports on their role in the SND of saline wastewater.
3.2. Optimization of the SND at High Salinity
To assess the impacts of different culture conditions on NH
4+-N removal efficiency of strain Y2 at 11% Na
2SO
4 salinity, the response surface methodology was carried out in combination with a four-factor, three-level Box-Behnken design. This approach was employed to explore the interactions among the independent variables and to identify the optimal condition for nitrogen removal. The correlation among the four independent variables and NH
4+-N removal efficiency was represented by the following equation:
where
Y is the NH
4+-N removal efficiency (%),
X1,
X2,
X3, and
X4 indicate the coded values of C/N ratio, initial pH value, temperature, and shaking speed, respectively.
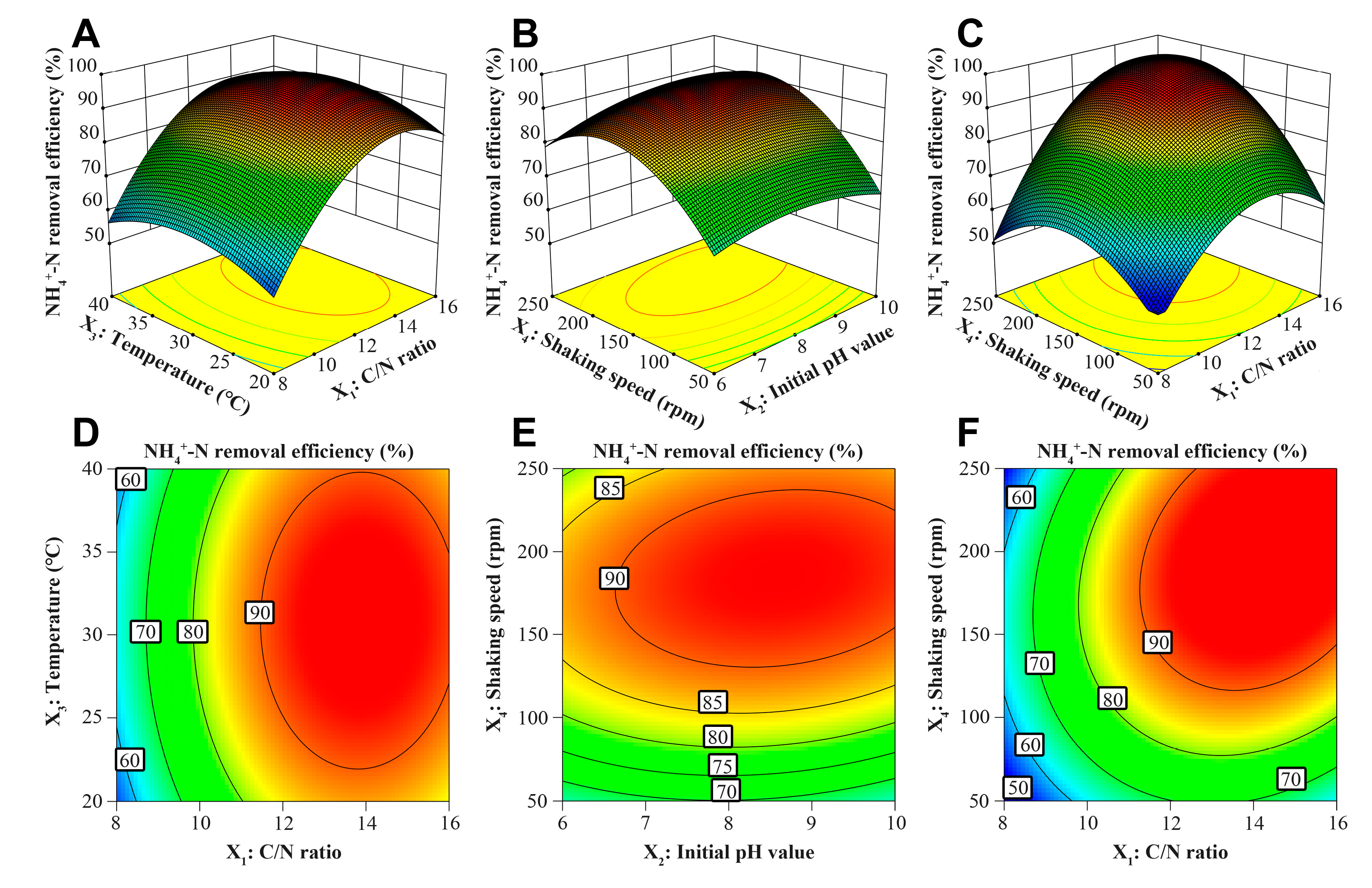
Variance analysis and significance testing were used to assess the rationality and validity of the response surface quadratic model (
Table S1). The model’s significance is corroborated by
F-value of 43.21 and low
p-value of < 0.0001. For the Lack of Fit term, the calculated
F-value and
p-value are 5.64 and 0.055, evidencing that the model exhibits a satisfactory degree of fit. The coefficient of determination (R
2 = 0.9774) illustrates that the fitted model accounts for 97.74% of the variability, thereby demonstrating the model’s high accuracy. In addition, according to the
F-value and
p-value of four independent variables, the impact of C/N ratio and shaking speed on the NH
4+-N removal performance is more substantial compared to that of initial pH value and temperature.
To further elucidate the relationship between NH
4+-N removal efficiencies and four independent variables, the response surface plots and corresponding contour plots were constructed to represent the influence of the SND conditions on nitrogen removal efficiency.
Figure 1A and 1D show that C/N ratio (8-16) has significantly more positive influence on NH
4+-N removal efficiency than temperature (20-40 °C), which is in accordance with the previously reported studies [
35,
36]. Under relatively high shaking speed (ca. 150-200 rpm), strain Y2 can well grow at 11% Na
2SO
4 salinity and efficiently remove NH
4+-N (> 90%) at relatively wide pH value range of 7-10 (
Figure 1B and 1E).
Figure 1C and 1F show that the interaction between C/N ratio and shaking speed is significant.
The optimal conditions for nitrogen removal by strain Y2 at 11% Na2SO4 salinity is C/N ratio of 13.97, initial pH value of 8.82, temperature of 30.96 °C, shaking speed of 171.89 rpm, corresponding to the predicted NH4+-N removal efficiency of 97.8%. To verify the precision of the prediction, nitrogen removal experiments were performed in triplicate under the optimal condition. The measured NH4+-N removal efficiency was 97.4±1.1%, demonstrating great concordance with the prediction.
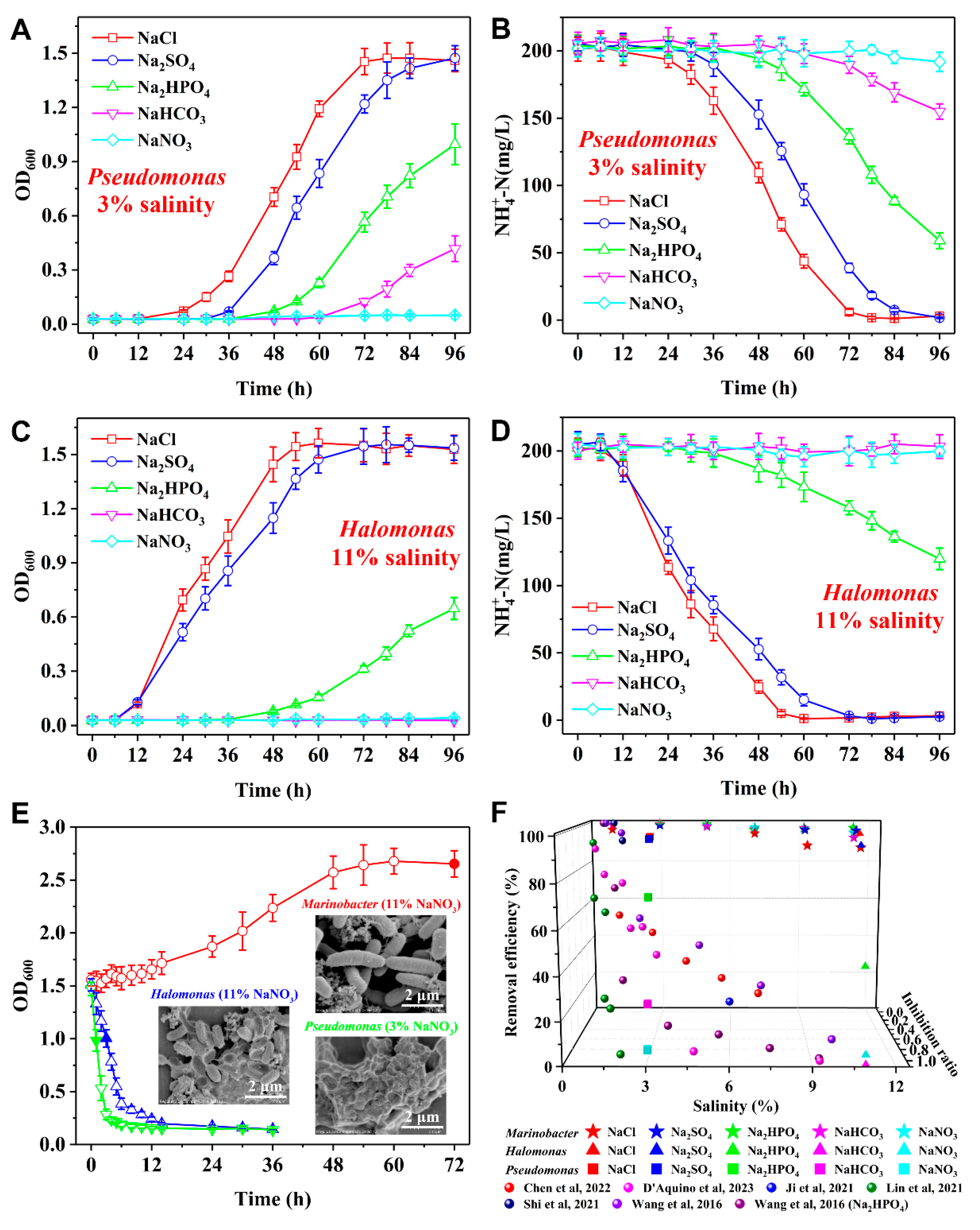
3.3. Oxysalt-Tolerant Capabilities
The proposed bacterium
Marinobacter sp. and other SND strains
Pseudomonas sp. and
Halomonas sp. were utilized to examine the oxysalt-tolerant capabilities. At 3% salinity,
Pseudomonas sp. can endure high osmotic pressure induced by NaCl, exhibiting vigorous growth with a short-term adaptation period (
Figure 2A). After 84 h, NH
4+-N is almost completely removed (
Figure 2B). When Na
2SO
4, Na
2HPO
4, NaHCO
3, and NaNO
3 replace NaCl, the duration of the adaptive phases is extended and the growth vitalities diminishes in sequence. Correspondingly, the rates and efficiencies of NH
4+-N removal decline owing to the toxicities of these oxysalts, with the respective inhibition ratios of 0.72, 0.82, 0.94, and 0.99. As another robust SND bacterium,
Halomonas sp. can more rapidly grow and efficiently remove NH
4+-N at 11% NaCl or Na
2SO
4 salinities (
Figure 2C and 2D), achieving high removal efficiencies of > 99% after 72 h. However, when Na
2HPO
4, NaHCO
3, and NaNO
3 replace NaCl and Na
2SO
4, both bacterial growth and NH
4+-N removal rates and efficiencies are significantly decreased. After 96 h, only 41% NH
4+-N can be removed by
Halomonas sp. at 11% Na
2HPO
4 salinity. In addition,
Halomonas sp. cannot grow at 11% NaHCO
3 or NaNO
3 salinities.
To further study the influence of oxysalts on these mature bacteria,
Pseudomonas sp.,
Halomonas sp., and
Marinobacter sp. were cultivated in the HN media containing 3%, 11%, and 11% NaCl, respectively. Subsequently, the grown bacteria were collected and were transferred to the HN media containing NaNO
3 at the same concentrations. As shown in
Figure 2E, the bacterial concentrations of
Pseudomonas sp. and
Halomonas sp. decreases immediately within 12 h. SEM images of the microbial samples acquired at the half decay period reveal that
Halomonas sp. is being wizened owing to water loss and
Pseudomonas sp. suffers from cell apoptosis and aggregation. In contrast,
Marinobacter sp. continues to grow, until nearly doubling the bacterial number while maintaining its initial shape, showing high oxysalt-tolerant capability. Notably, most of the reported halotolerant SND bacteria cannot efficiently remove nitrogen at > 3% Na
2SO
4 or Na
2HPO
4 salinities, and the SND bacteria that can work in saline wastewater containing NaHCO
3 and NaNO
3 are not reported (
Figure 2F) [
6,
23,
24,
25,
27,
37].
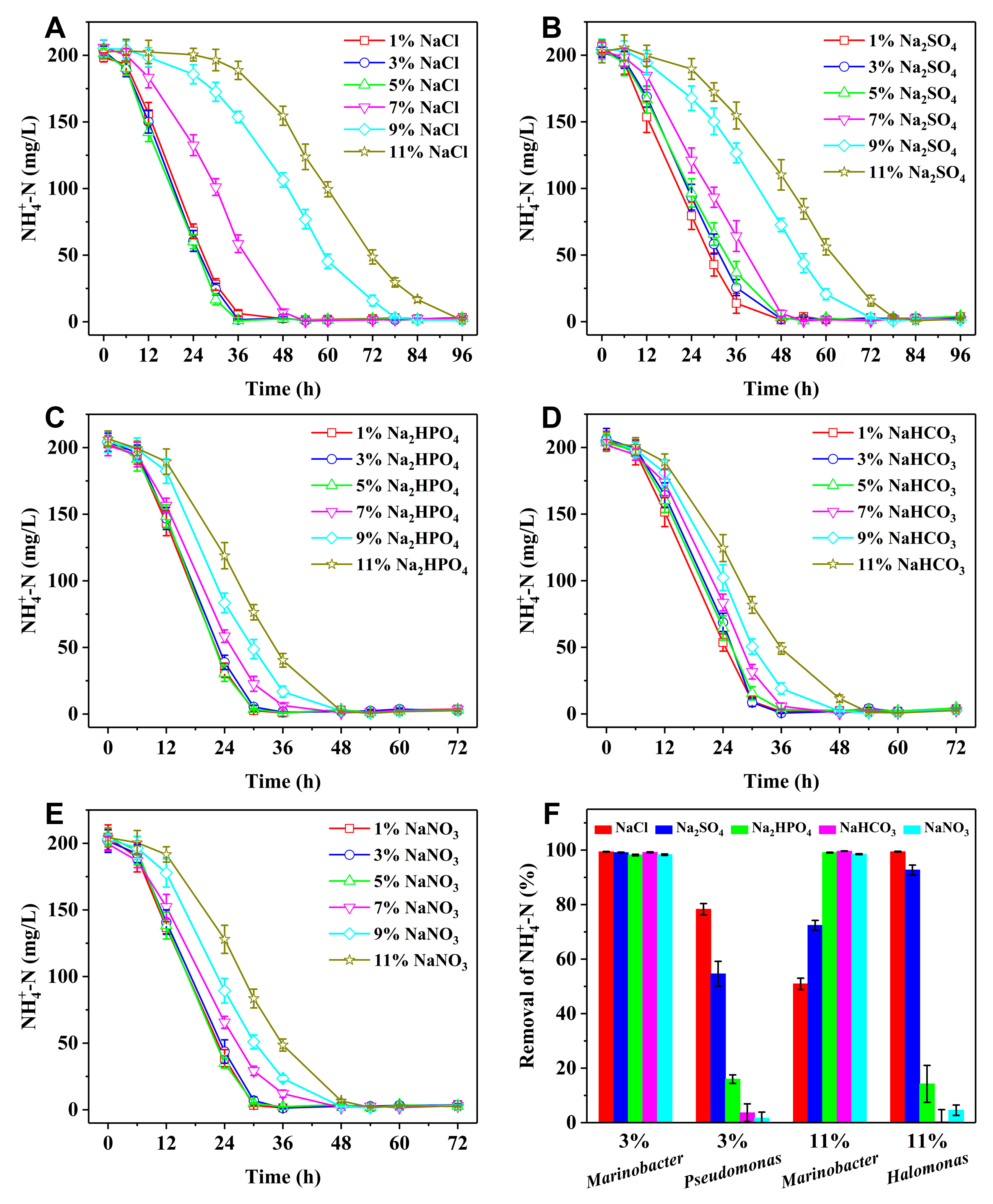
Figure 3 and
Figures S2 show the bacterial growth of
Marinobacter sp. in the HN media containing various salts, including NaCl, Na
2SO
4, Na
2HPO
4, NaHCO
3, and NaNO
3 at different salinities, as well as the corresponding NH
4+-N removal. It can be found that strain Y2 can vigorously grow and efficiently remove NH
4+-N at 1%-11% salinities of different salts. At 1%-5% salinities, the rates of bacterial growth and NH
4+-N removal are almost consistent. The rates of NH
4+-N removal are slightly higher in the presence of HPO
42−, HCO
3−, and NO
3− compared to Cl
− and SO
42−. After 36 h, all the NH
4+-N removal efficiencies exceed 90%, expect for SO
42−, which requires approximately 48 h. Beyond 5% salinity, both bacterial growth and NH
4+-N removal are inhibited by oxysalts, particularly HPO
42−, HCO
3−, and NO
3−, exhibiting significantly lower inhibition ratios. For example, after 48 h cultivation at 11% salinity, the NH
4+-N removal efficiencies of
Marinobacter sp. increase markedly when substituting from NaCl and Na
2SO
4 to Na
2HPO
4, NaHCO
3, and NaNO
3 (
Figure 3F). In contrast, the respective NH
4+-N removal efficiencies of
Pseudomonas sp. and
Halomonas sp. at 3% and 11% salinities are dramatically decreased owing to the toxicity of oxysalts.
Figures S3 and S4 show the bacterial growth of
Marinobacter sp. in the AD media containing NaCl, Na
2SO
4, Na
2HPO
4, and NaHCO
3 at different salinities, and the corresponding NO
3–-N removal efficiencies. Being consistent with the NH
4+-N removal results, the inhibition ratios of NO
3–-N removal induced by oxysalts especially HPO
42− and HCO
3− are obviously lower than that induced by NaCl. After 60 h cultivation at 11% Na
2HPO
4 or NaHCO
3 salinities, strain Y2 achieves the NO
3–-N removal efficiencies exceeding 90%. In contrast, achieving a NO
3–-N removal efficiency of > 90% requires 96 h at 11% NaCl salinity. These results demonstrate that the robust SND bacterium
Marinobacter sp. possesses excellent oxysalt-tolerant capabilities.
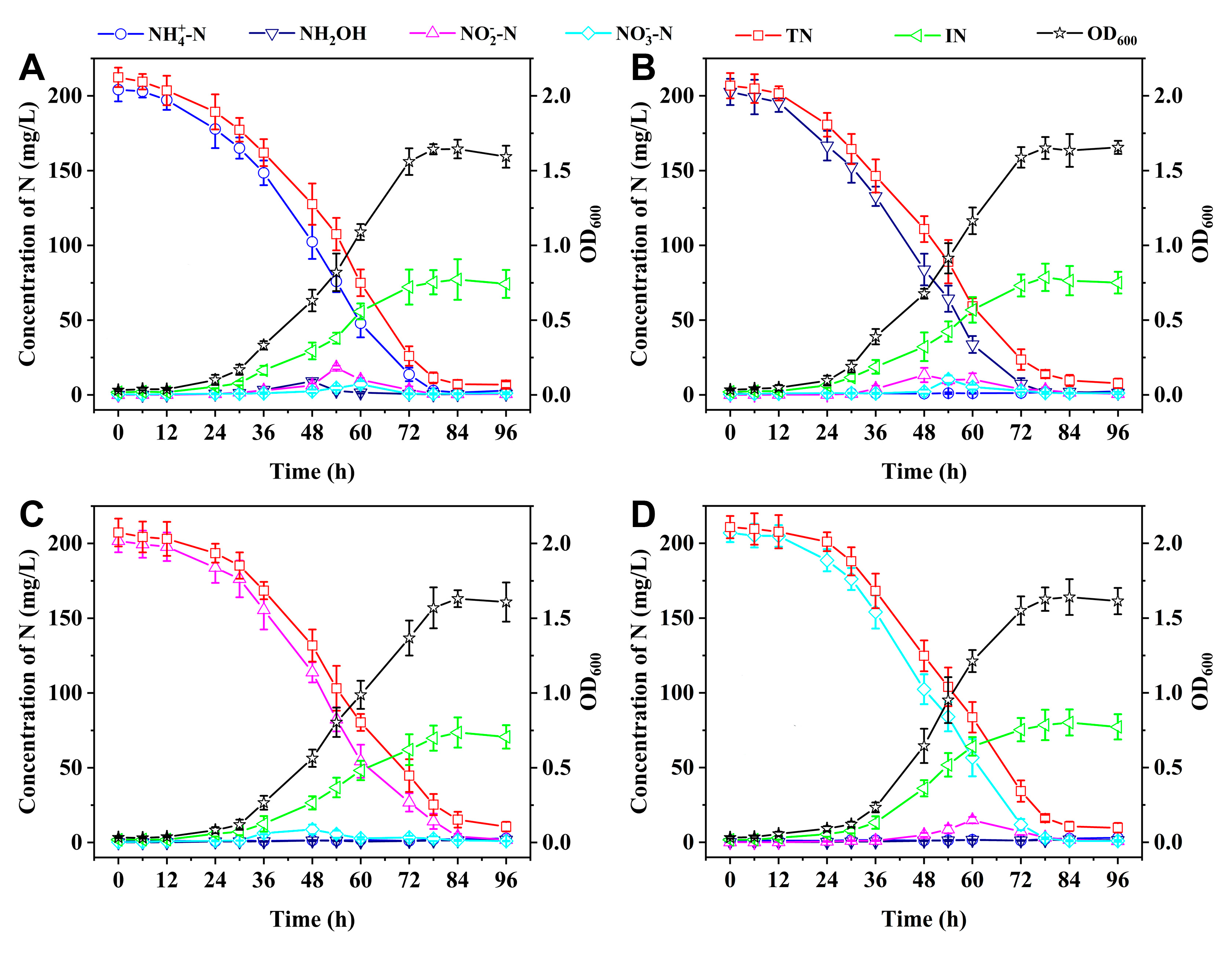
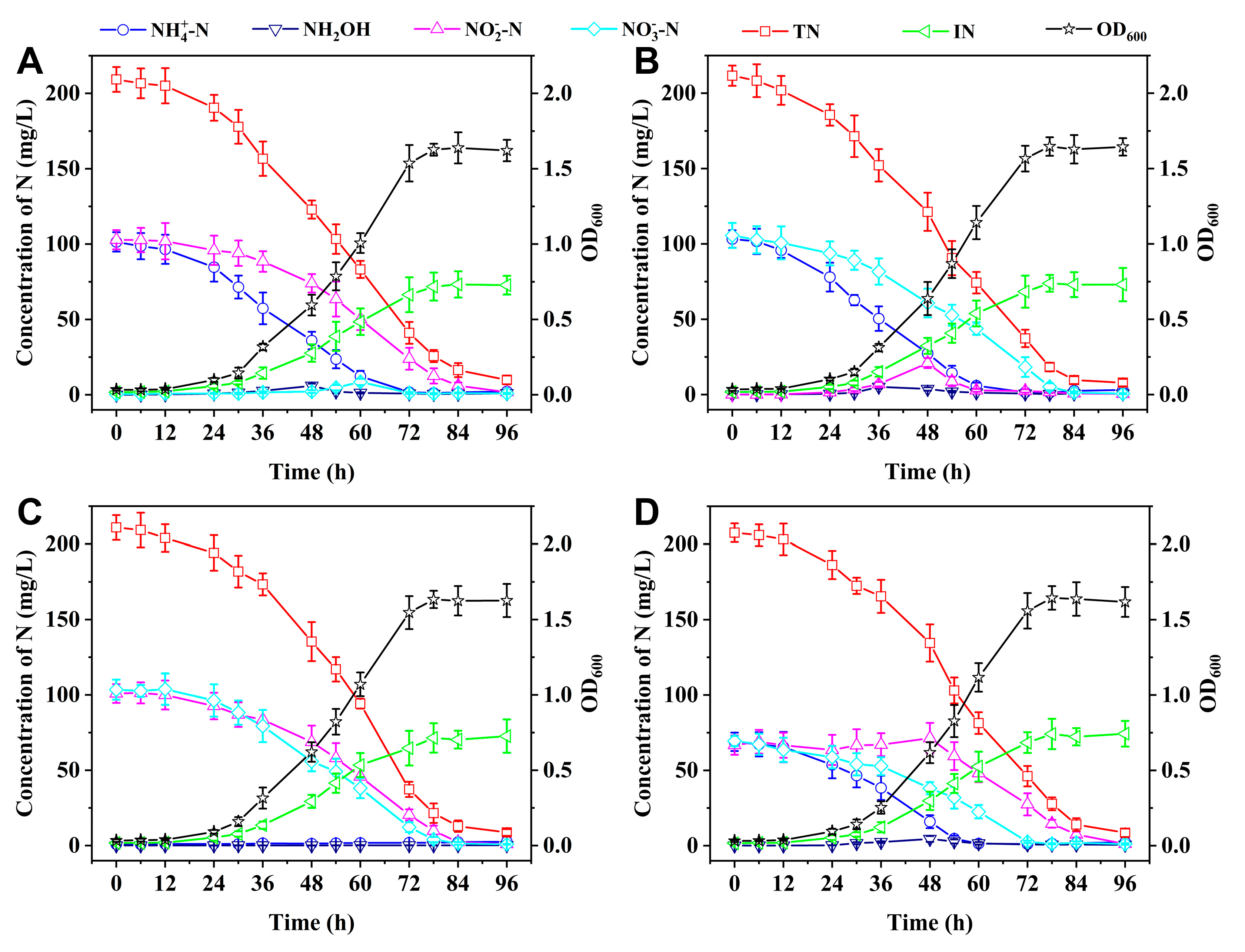
3.4. SND Performance Using Single Nitrogen Sources
The SND performance of the oxysalt-tolerant bacterium
Marinobacter sp. at 11% Na
2SO
4 salinity was evaluated using NH
4+-N, NH
2OH, NO
2–-N, NO
3–-N as single nitrogen sources with initial concentration of ca. 200 mg/L.
Figure 4 shows that strain Y2 can grow well in single nitrogen species and all the inorganic nitrogen species are barely detected in the SND systems after 96 h, with the TN removal rates exceeding 95%. In addition, NH
4+-N, NH
2OH, NO
3–-N are removed at slightly higher rates than NO
2–-N. For example, after 72 h, the respective removal efficiencies for NH
4+-N, NH
2OH, NO
3–-N are 93.3%, 96.4%, and 94.3%, while the NO
2–-N removal efficiency is 86.7%.
During the SND processes involving NH
4+-N, NH
2OH, NO
3–-N as single nitrogen species, a minor accumulation of NO
2–-N (< 20 mg/L) can be observed after 48-60 h followed by its rapid removal, which could be ascribed to slower removal rate of NO
2–-N than those of NH
4+-N, NH
2OH, and NO
3–-N. Similarly, more NO
2–-N was accumulated during the denitrification-anammox process at high salinity, which was ascribed to the enriched denitrifying community [
38,
39]. When
Halomonas sp. was utilized to remove NH
4+-N, NH
2OH, or NO
3–-N at 15% NaCl salinity, considerable NO
2–-N (ca. 50 mg/L) was progressively amassed until the complete elimination of these single nitrogen sources [
15]. In addition, a minor accretion of NO
3–-N is detected after 48 h during the NO
2–-N denitrification, indicating the conversion of NO
2–-N to NO
3–-N under aerobic condition. Nitrogen balance analysis reveals that 35%-38% of initial nitrogen sources are assimilated as the IN, while 55%-60% are removed through the SND processes.
3.5. SND Performance Using Mixed Nitrogen Sources
The SND capabilities of strain Y2 at 11% Na
2SO
4 salinity was further investigated using NH
4+-N, NO
2–-N, NO
3–-N as mixed nitrogen sources with initial TN concentration of ca. 200 mg/L. When NH
4+-N and NO
2–-N are used as mixed nitrogen source, they are simultaneously removed and NH
4+-N is removed at a higher rate than NO
2–-N (
Figure 5A). A minor quantity of NO
3–-N is accumulated after 60 h, which can be eliminated completely after 72 h. During simultaneous removal of NH
4+-N and NO
3–-N, they are nearly completely removed after 72 and 84 h, respectively, and NO
2–-N reaches the maximum accumulation of 21.0 mg/L after 48 h (
Figure 5B). When NO
2–-N and NO
3–-N are used as mixed nitrogen source, the removal rates of NO
2–-N and NO
3–-N are almost same, accomplishing complete removal after 84 h (
Figure 5C). When NH
4+-N, NO
2–-N, NO
3–-N are used as mixed nitrogen source, NH
4+-N and NO
3–-N are preferentially removed and NH
4+-N is removed at a higher rate than NO
3–-N. Until NH
4+-N is nearly entirely removed, NO
2–-N begins to undergo rapid denitrification under aerobic condition (
Figure 5D). The maximum assimilated IN falls within the range of 70-80 mg/L, indicating that the TN of 34%-36% contributes to the bacterial growth.
The results demonstrate that the isolated robust bacterium Marinobacter sp. is capable of efficiently conducting the SND processes in hypersaline wastewater containing oxysalts such as Na2SO4. During the SND processes, NH4+-N is oxidized to NH2OH, NO2–-N, and NO3–-N. Subsequently, the generated NO2–-N and NO3–-N can be transformed to N2, in which a small amount of NO2–-N is accumulated. The SND-related enzymes determine nitrogen removal efficiencies of the oxysalt-tolerant bacterium in such harsh oxysalt environments.
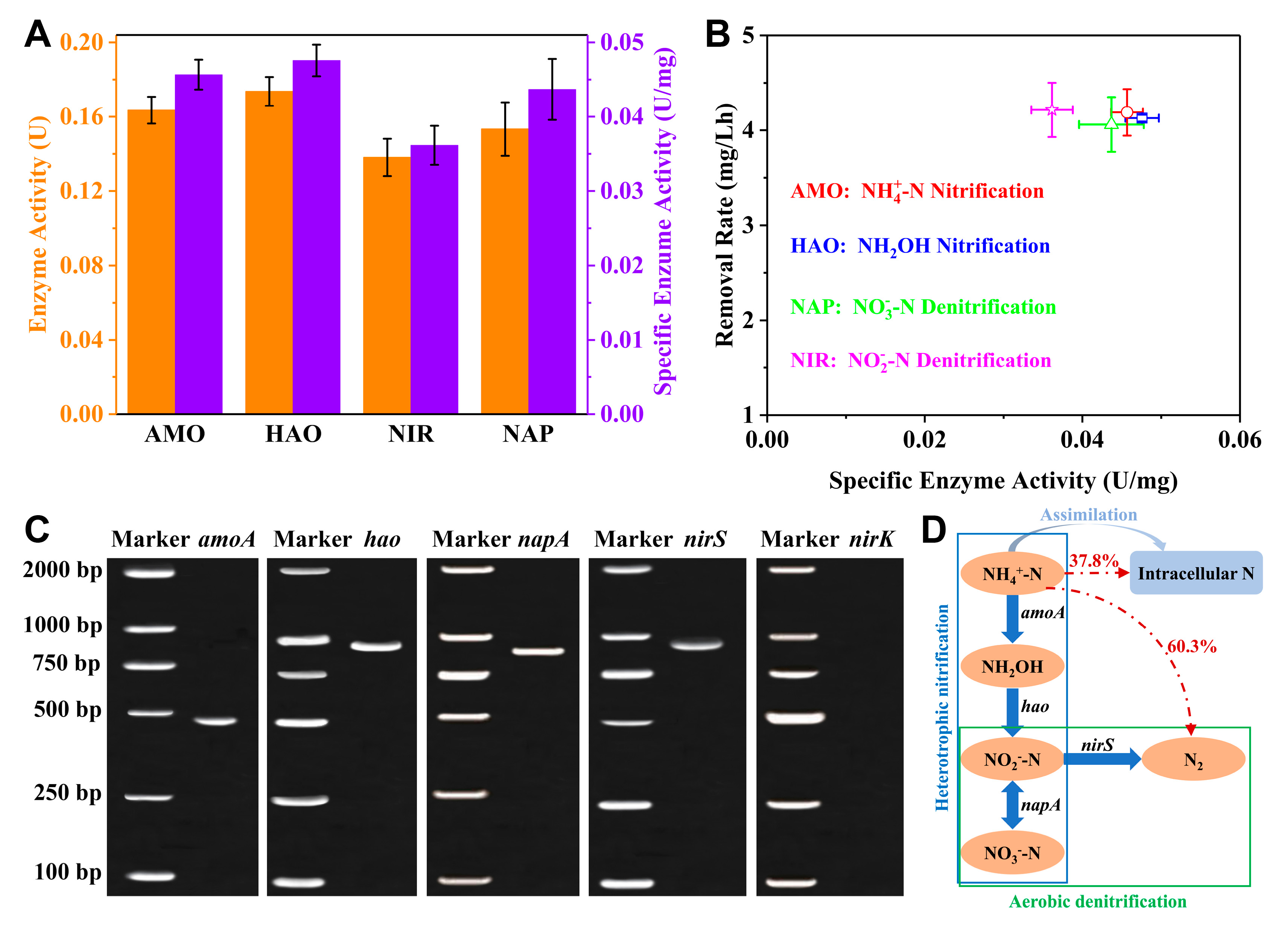
3.6. Nitrogen Removal Enzymes and Functional Genes for the SND
Enzymes associated with the SND including AMO, HAO, NAP, and NIR were extracted from strain Y2 cultivated at 11% Na
2SO
4 salinity and the enzyme activities and specific enzyme activities were measured. In the enzyme activity assays, cytochrome
c functioned as the electron acceptor, while NADH acted as the electron donor for target substrate removal, respectively. When only cytochrome
c/NADH and crude enzyme extracts coexist, the target substrates are removed significantly. During the nitrification process, AMO and HAO are the key enzymes responsible for NH
4+-N and NH
2OH oxidation. During the subsequent denitrification process, NO
3–-N and NO
2–-N are removed by the key enzymes of NAP and NIR, respectively [
28,
40,
41,
42].
The specific activities of AMO, HAO, NAP, NIR were determined according to the removal of target substrates. As shown in
Figure 6A, AMO, HAO, NAP have the specific activities of 0.046 ± 0.007, 0.048 ± 0.007, and 0.043 ± 0.007 U/mg protein, which are higher than NIR’s (0.036 ± 0.007 U/mg protein). The enzyme activities of AMO, HAO, NAP for the isolated bacterium
Marinobacter sp. at 11% Na
2SO
4 salinity are lower than those for the reported halotolerant bacterium
Halomonas sp. at 15% NaCl salinity [
15], which are consistent with the elimination rates of various nitrogen sources using different halotolerant bacteria (
Figure 6B). However, the enzyme activity of NIR for
Marinobacter sp. is higher than that for
Halomonas sp., resulting in significantly reduced NO
2–-N accumulation for
Marinobacter sp. compared to
Halomonas sp.
The SND-associated enzymes of strain Y2 at 11% Na
2SO
4 salinity are encoded by specific functional genes, e.g.,
amoA and
hao responsible for encoding the AMO and HAO in the nitrification process,
napA and
nirS/
nirK encoding the NAP and NIR in the denitrification process [43-45. As shown in
Figure 6C, the functional genes for SND, like
amoA,
hao,
napA,
nirS, are amplified successfully. The functional genes
amoA and
hao are widely detected for the SND study of heterotrophic nitrifying bacteria. The functional gene
napA involves in the transformation of NO
3–-N to NO
2–-N under aerobic condition [
36,
46,
47]. The successful amplification of these nitrogen removal functional genes further demonstrates the SND capabilities of strain Y2 in such harsh oxysalt environments. According to the SND-associated enzymes and functional genes, the proposed nitrogen metabolism pathway is NH
4+-N → NH
2OH → NO
2–-N ↔ NO
3–-N → N
2 (
Figure 6D).
4. Conclusions
An oxysalt-tolerant SND bacterium Marinobacter sp. has been isolated from saline aerobic sludge, which can achieve 98% of nitrogen removal at 11% Na2SO4 salinity after optimizing SND conditions via response surface methodology. At > 5% salinities, the robust bacterium exhibits significantly reduced oxysalt-induced inhibition on bacterial growth and nitrogen removal rates compared to other SND strains Pseudomonas sp. and Halomonas sp. The proposed strain can achieve nitrogen removal efficiencies exceeding 90% with both single and mixed nitrogen sources at 11% Na2SO4. The excellent SND performance is attributed to the SND-associated functional genes and their encoded enzymes. The isolated oxysalt-tolerant SND bacterium provides a viable approach for nitrogen removal from hypersaline wastewater containing oxysalts.
Supplementary Materials
The following supporting information can be downloaded at: Preprints.org, Figure S1: (A) Colony morphology and (B) SEM image of strain Y2. (C) NJ algorithm phylogenetic tree of strain Y2 based on 16S rRNA gene sequences; Figure S2: Bacterial growth of Marinobacter sp. in the HN media at different salinities. Salts include NaCl (A), Na2SO4 (B), Na2HPO4 (C), NaHCO3 (D), and NaNO3 (E); Figure S3: Bacterial growth of Marinobacter sp. in the AD media at different salinities. Salts include NaCl (A), Na2SO4 (B), Na2HPO4 (C), and NaHCO3 (D); Figure S4: NO3–-N removal of Marinobacter sp. at different salinities. Salts include NaCl (A), Na2SO4 (B), Na2HPO4 (C), and NaHCO3 (D); Table S1: Analysis of variance for response surface quadratic model (Y) a.
Author Contributions
Conceptualization, Jie Hu, Bing Xu, Jie Gao and Jiabao Yan; Data curation, Jie Hu, Bing Xu and Jiabao Yan; Formal analysis, Jie Hu, Jie Gao and Guozhi Fan; Funding acquisition, Jie Hu, Bing Xu, Jiabao Yan and Guozhi Fan; Investigation, Jie Hu, Bing Xu, Jie Gao and Guozhi Fan; Methodology, Jie Hu, Jie Gao, Jiabao Yan and Guozhi Fan; Project administration, Bing Xu and Jiabao Yan; Resources, Bing Xu and Jiabao Yan; Software, Jie Hu and Jie Gao; Supervision, Bing Xu, Jiabao Yan and Guozhi Fan; Validation, Jie Hu, Bing Xu and Guozhi Fan; Visualization, Jie Hu, Bing Xu and Jie Gao; Writing – original draft, Jie Hu; Writing – review & editing, Jie Hu and Bing Xu. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Nature Science Foundation of Hubei Province (2023AFB385), Key Laboratory of Hubei Province for Coal Conversion and New Carbon Materials (Wuhan University of Science and Technology) (WKDM202301), Research and Innovation Initiatives of WHPU (2023Y27), Research Funding of Wuhan Polytechnic University (2024R2006) and Research Program Project of Hubei Provincial Department of Education (F2023009).
Data Availability Statement
The original contributions presented in this study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author(s).
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Chen, B.; Qaisar, M.; Xiao, J.; Li, W.; Li, J.; Cai, J. Combined acute effect of salinity and substrate concentration on simultaneous sulfide and nitrite removal process. Sep. Purif. Technol. 2023, 305, 122544. [Google Scholar] [CrossRef]
- Shen, N.; Guo, H.; Yao, T.; Xu, L.; Gao, Y.; Yang, P. Treatment of PickleWastewater under Varying Salinity Conditions within the Sequencing Batch Biofilm Reactor System. Water 2024, 16, 1312. [Google Scholar] [CrossRef]
- Liu, Y.; Yin, P.; Zhou, J.; Ma, Y.; Lai, X.; Lin, J.; Peng, H.; Shu, H.; Huang, W. Removal of Nitrogen and Phosphorus by a Novel Salt-Tolerant Strain Pseudomonas sediminis D4. Water 2025, 17, 502. [Google Scholar] [CrossRef]
- Zhang, Q.; Qin, S.; Yuan, C.; Fang, D.; Chen, W. A novel biofilm reactor startup with HN-AD bacteria inoculation for high ammonia-salt wastewater treatment: Performance, microbial characteristics and salt-tolerant mechanism. J. Water Process Eng. 2024, 59, 105032. [Google Scholar] [CrossRef]
- Li, P.; Wang, Y.; Liu, Y.; Wang, S.; Peng, Y. The Effect of Salinity on N2O Emissions during Domestic Wastewater Partial Nitrification Treatment in a Sequencing Batch Reactor. Water 2023, 15, 3502. [Google Scholar] [CrossRef]
- Lin, L.; Pratt, S.; Crick, O.; Xia, J.; Duan, H.; Ye, L. Salinity effect on freshwater Anammox bacteria: Ionic stress and ion composition. Water Res. 2021, 188, 116432. [Google Scholar] [CrossRef] [PubMed]
- Ng, K.K.; Shi, X.; Ng, H.Y. Evaluation of system performance and microbial communities of a bioaugmented anaerobic membrane bioreactor treating pharmaceutical wastewater. Water Res. 2015, 81, 311–324. [Google Scholar] [CrossRef]
- Ng, K.K.; Shi, X.; Ong, S.L.; Lin, C.-F.; Ng, H.Y. An innovative of aerobic bio-entrapped salt marsh sediment membrane reactor for the treatment of high-saline pharmaceutical wastewater. Chem. Eng. J. 2016, 295, 317–325. [Google Scholar] [CrossRef]
- Srivastava, A.; Parida, V.K.; Majumder, A.; Gupta, B.; Gupta, A.K. Treatment of saline wastewater using physicochemical, biological, and hybrid processes: Insights into inhibition mechanisms, treatment efficiencies and performance enhancement. J. Environ. Chem. Eng. 2021, 9, 105775. [Google Scholar] [CrossRef]
- Wang, J.; Zhou, J.; Wang, Y.; Wen, Y.; He, L.; He, Q. Efficient nitrogen removal in a modified sequencing batch biofilm reactor treating hypersaline mustard tuber wastewater: The potential multiple pathways and key microorganisms. Water Res. 2020, 177, 115734. [Google Scholar] [CrossRef]
- Han, F.; Zhao, C.; Zhang, W.; Jiao, T.; Zhang, Z.; Zhou, W. Responses of halophilic microbial communities to changes in salt composition: Comparison between autotrophic nitrification and heterotrophic ammonia assimilation biosystems. Bioresour. Technol. 2023, 386, 129500. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; He, Y.; Wang, B.; Weng, N.; Zhang, L.; Wang, K.; Tian, F.; Lyu, M.; Wang, S. Heterotrophic Nitrification–Aerobic Denitrification by Bacillus sp. L2: Mechanism of Denitrification and Strain Immobilization. Water 2024, 16, 416. [Google Scholar] [CrossRef]
- Wang, M.; He, J.; Zhang, J. Evaluating the performance and microbial community of aerobic granular sludge under different salinities. J. Water Process Eng. 2024, 58, 104891. [Google Scholar] [CrossRef]
- Cui, Y.; Cui, Y.-W.; Huang, J.-L. A novel halophilic Exiguobacterium mexicanum strain removes nitrogen from saline wastewater via heterotrophic nitrification and aerobic denitrification. Bioresour. Technol. 2021, 333, 125189. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Yan, J.; Wu, L.; Bao, Y.; Yu, D.; Li, J. Simultaneous nitrification and denitrification of hypersaline wastewater by a robust bacterium Halomonas salifodinae from a repeated-batch acclimation. Bioresour. Technol. 2021, 341, 125818. [Google Scholar] [CrossRef]
- Hu, J.; Yan, J.; Wu, L.; Bao, Y.; Yu, D.; Li, J. Insight into halotolerance of a robust heterotrophic nitrifying and aerobic denitrifying bacterium Halomonas salifodinae. Bioresour. Technol. 2022, 351, 126925. [Google Scholar] [CrossRef]
- Blázquez, E.; Baeza, J.A.; Gabriel, D.; Guisasola, A. Treatment of real flue gas desulfurization wastewater in an autotrophic biocathode in view of elemental sulfur recovery: Microbial communities involved. Sci. Total Environ. 2019, 657, 945–952. [Google Scholar] [CrossRef]
- Han, Y.-L.; Zhang, X.-Z.; Liu, H.-B.; Rittmann, B.E.; Zhao, H.-P. Novel Sulfate Reduction Coupled to Simultaneous Nitrification and Autotrophic Denitrification Process for Removing Nitrogen and Organics from Saline Wastewater. Environ. Sci. Technol. 2023, 57, 10733–10744. [Google Scholar] [CrossRef]
- Mukhopadhyay, R.; Adhikari, T.; Sarkar, B.; Barman, A.; Paul, R.; Patra, A.K.; Sharma, P.C.; Kumar, P. Fe-exchanged nano-bentonite outperforms Fe3O4 nanoparticles in removing nitrate and bicarbonate from wastewater. J. Hazard. Mater. 2019, 376, 141–152. [Google Scholar] [CrossRef]
- Xu, Z.; Wang, J.; Qiu, J.; Cao, H.; Xie, Y. Unexpectedly Enhanced Organics Removal in Persulfate Oxidation with High Concentration of Sulfate: The Origin and the Selectivity. Environ. Sci. Technol. 2023, 57, 14442–14451. [Google Scholar] [CrossRef]
- Cheng, Q.; Xu, L.; Cheng, F.; Pan, G.; Zhou, Q. Bicarbonate-rich wastewater as a carbon fertilizer for culture of Dictyosphaerium sp. of a giant pyrenoid. J. Clean. Prod. 2018, 202, 439–443. [Google Scholar]
- Glass, C.; Silverstein, J. Denitrification of high-nitrate, high-salinity wastewater. Water Res. 1999, 33, 223–229. [Google Scholar] [CrossRef]
- Wang, R.; Zheng, P.; Ding, A-q. ; Zhang, M.; Ghulam, A.; Yang, C.; Zhao, H.-P. Effects of inorganic salts on denitrifying granular sludge: The acute toxicity and working mechanisms. Bioresour. Technol. 2016, 204, 65–70. [Google Scholar] [CrossRef]
- Ji, B.; Zhang, H.; Zhou, L.; Yang, J.; Zhang, K.; Yuan, X.; Ma, J.; Qian, Y. Effect of the rapid increase of salinity on anoxic-oxic biofilm reactor for treatment of high-salt and high-ammonia-nitrogen wastewater. Bioresour. Technol. 2021, 337, 125363. [Google Scholar] [CrossRef] [PubMed]
- Shi, X.; Li, J.; Wang, X.; Zhang, X.; Tang, L. Effect of the gradual increase of Na2SO4 on performance and microbial diversity of aerobic granular sludge. J. Environ. Manage. 2021, 292, 112696. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Yun, Y.; Liu, G.; Li, C.; Li, X.; Hilal, M.; Yang, W.; Wang, M. Direct contact membrane distillation with softening Pre-treatment for effective reclaiming flue gas desulfurization wastewater. Sep. Purif. Technol. 2021, 277, 119637. [Google Scholar] [CrossRef]
- Chen, A.-l.; Su, X.; Xing, Z.-l.; Xu, F.-q.; Chen, S.-j.; Xiang, J.-x.; Li, J.; Liu, H.; Zhao, T.-t. Effect mechanism of individual and combined salinity on the nitrogen removal yield of heterotrophic nitrification-aerobic denitrification bacteria. Environ. Res. 2022, 214, 113834. [Google Scholar] [CrossRef]
- Chen, H.; Zhao, X.; Cheng, Y.; Jiang, M.; Li, X.; Xue, G. Iron Robustly Stimulates Simultaneous Nitrification and Denitrification Under Aerobic Conditions. Environ. Sci. Technol. 2018, 52, 1404–1412. [Google Scholar] [CrossRef]
- Zhao, B.; He, Y.L.; Hughes, J.; Zhang, X.F. Heterotrophic nitrogen removal by a newly isolated Acinetobacter calcoaceticus HNR. Bioresour. Technol. 2010, 101, 5194–5200. [Google Scholar] [CrossRef]
- APHA. Standard Methods for the Examination of Water and Wastewater; APHA: Washington, DC, USA, 1998. [Google Scholar]
- Frear, D.S.; Burrell, R.C. Spectrophotometric Method for Determining Hydroxylamine Reductase Activity in Higher Plants. Anal. Chem. 1955, 27, 1664–1665. [Google Scholar] [CrossRef]
- Anh, H.T.H.; Shahsavari, E.; Bott, N.J.; Ball, A.S. The application of Marinobacter hydrocarbonoclasticu as a bioaugmentation agent for the enhanced treatment of non-sterile fish wastewater. J. Environ. Manage. 2021, 291, 112658. [Google Scholar] [CrossRef] [PubMed]
- Dou, L.; Zhang, M.; Pan, L.; Liu, L.; Su, Z. Sulfide removal characteristics, pathways and potential application of a novel chemolithotrophic sulfide-oxidizing strain, Marinobacter sp. SDSWS8. Environ. Res. 2022, 212, 113176. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Jiang, S.; Zhang, J.; Zheng, J.; Li, P.; Wang, S.; Bi, R.; Gao, L. Phycoremediation and valorization of hypersaline pickled mustard wastewater via Chaetoceros muelleri and indigenous bacteria. Bioresour. Technol. 2024, 393, 130172. [Google Scholar] [CrossRef]
- Hu, X.; Su, J.; Ali, A.; Wang, Z.; Wu, Z. Heterotrophic nitrification and biomineralization potential of Pseudomonas sp. HXF1 for the simultaneous removal of ammonia nitrogen and fluoride from groundwater. Bioresour. Technol. 2021, 323, 124608. [Google Scholar] [CrossRef]
- Zhang, Y.; Xu, J.; Dong, X.; Wang, J.; Liu, C.; Liu, J. Optimization of nitrogen removal conditions based on response surface methodology and nitrogen removal pathway of Paracoccus sp. QD-19. Sci. Total Environ. 2024, 908, 168348. [Google Scholar] [CrossRef] [PubMed]
- D’Aquino, A.; Kalinainen, N.; Auvinen, H.; Andreottola, G.; Puhakka, J.A.; Palmroth, M.R.T. Effects of inorganic ions on autotrophic denitrification by Thiobacillus denitrificans and on heterotrophic denitrification by an enrichment culture. Sci. Total Environ. 2023, 901, 165940. [Google Scholar] [CrossRef]
- Ji, J.; Peng, Y.; Wang, B.; Mai, W.; Li, X.; Zhang, Q.; Wang, S. Effects of salinity build-up on the performance and microbial community of partial-denitrification granular sludge with high nitrite accumulation. Chemosphere 2018, 209, 53–60. [Google Scholar] [CrossRef]
- Li, W.; Li, H.; Liu, Y.-d.; Zheng, P.; Shapleigh, J.P. Salinity-Aided Selection of Progressive Onset Denitrifiers as a Means of Providing Nitrite for Anammox. Environ. Sci. Technol. 2018, 52, 10665–10672. [Google Scholar] [CrossRef]
- Asamoto, C.K.; Rempfert, K.R.; Luu, V.H.; Younkin, A.D.; Kopf, S.H. Enzyme-Specific Coupling of Oxygen and Nitrogen Isotope Fractionation of the Nap and Nar Nitrate Reductases. Environ. Sci. Technol. 2021, 55, 5537–5546. [Google Scholar] [CrossRef]
- Liang, X.; Gan, L.; He, T.; Chen, M.; Zhang, M.; Wu, Q. The coexisted nitrate and nitrite as a driving force for the aerobic denitrification of Peribacillus sp. EM-C3. Environ. Technol. Innovation 2023, 32, 103299. [Google Scholar] [CrossRef]
- Sparacino-Watkins, C.; Stolz, J.F.; Basu, P. Nitrate and periplasmic nitrate reductases. Chem. Soc. Rev. 2014, 43, 676–706. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.; Wang, H.; Wu, P.; Lei, Y.; Deng, L.; Wang, W. Enhanced narH gene expression contributing to nitrite accumulation in simultaneous nitrification and denitrification under Na+ stress instead of K+ stress. Chem. Eng. J. 2024, 479, 147637. [Google Scholar] [CrossRef]
- Shi, S.; He, X.; He, L.; Fan, X.; Shu, B.; Zhou, J.; He, Q. Overlooked pathways of endogenous simultaneous nitrification and denitrification in anaerobic/aerobic/anoxic sequencing batch reactors with organic supplementation. Water Res. 2023, 230, 119493. [Google Scholar] [CrossRef]
- Wen, X.; Cui, L.; Lin, H.; Zhu, W.; Shao, Z.; Wang, Y. Comparison of nitrification performance in SBR and SBBR with response to NaCl salinity shock: Microbial structure and functional genes. Environ. Res. 2024, 252, 118917. [Google Scholar] [CrossRef]
- Ke, X.; Liu, C.; Tang, S.-Q.; Guo, T.-T.; Pan, L.; Xue, Y.-P.; Zheng, Y.-G. Characterization of Acinetobacter indicus ZJB20129 for heterotrophic nitrification and aerobic denitrification isolated from an urban sewage treatment plant. Bioresour. Technol. 2022, 347, 126423. [Google Scholar] [CrossRef]
- Yang, L.; Ren, Y.-X.; Liang, X.; Zhao, S.-Q.; Wang, J.-p.; Xia, Z.-H. Nitrogen removal characteristics of a heterotrophic nitrifier Acinetobacter junii YB and its potential application for the treatment of high-strength nitrogenous wastewater. Bioresour. Technol. 2015, 193, 227–233. [Google Scholar] [CrossRef]
Figure 1.
Response surface plots along with the contour plots for NH4+-N removal efficiency of strain Y2 at 11% Na2SO4 salinity. The interaction between (A, D) C/N ratio and temperature, (B, E) initial pH value and shaking speed, (C, F) C/N ratio and shaking speed for NH4+-N removal efficiency.
Figure 1.
Response surface plots along with the contour plots for NH4+-N removal efficiency of strain Y2 at 11% Na2SO4 salinity. The interaction between (A, D) C/N ratio and temperature, (B, E) initial pH value and shaking speed, (C, F) C/N ratio and shaking speed for NH4+-N removal efficiency.
Figure 2.
(A, C) Bacterial growth and (B, D) NH4+-N removal of Pseudomonas sp. (A, B) and Halomonas sp. (C, D) at 3% (A, B) and 11% (C, D) salinities. The salts include NaCl, Na2SO4, Na2HPO4, NaHCO3, and NaNO3. (E) Bacterial growth of Marinobacter sp., Pseudomonas sp., and Halomonas sp. after being transferred to the HN media containing NaNO3. Insets are SEM images of Marinobacter sp., Pseudomonas sp., and Halomonas sp. after cultivation in the HN media containing NaNO3 for 72 h, 1 h, and 3 h, respectively. (F) Nitrogen removal performance of the reported nitrogen removal bacteria at various salinities of oxysalts.
Figure 2.
(A, C) Bacterial growth and (B, D) NH4+-N removal of Pseudomonas sp. (A, B) and Halomonas sp. (C, D) at 3% (A, B) and 11% (C, D) salinities. The salts include NaCl, Na2SO4, Na2HPO4, NaHCO3, and NaNO3. (E) Bacterial growth of Marinobacter sp., Pseudomonas sp., and Halomonas sp. after being transferred to the HN media containing NaNO3. Insets are SEM images of Marinobacter sp., Pseudomonas sp., and Halomonas sp. after cultivation in the HN media containing NaNO3 for 72 h, 1 h, and 3 h, respectively. (F) Nitrogen removal performance of the reported nitrogen removal bacteria at various salinities of oxysalts.
Figure 3.
(A-E) NH4+-N removal of Marinobacter sp. at various salinities. Salts include NaCl (A), Na2SO4 (B), Na2HPO4 (C), NaHCO3 (D), and NaNO3 (E). (F) NH4+-N removal efficiencies of Marinobacter sp., Pseudomonas sp., and Halomonas sp. after 48 h cultivation at various salinities. Salts include NaCl, Na2SO4, Na2HPO4, NaHCO3, and NaNO3.
Figure 3.
(A-E) NH4+-N removal of Marinobacter sp. at various salinities. Salts include NaCl (A), Na2SO4 (B), Na2HPO4 (C), NaHCO3 (D), and NaNO3 (E). (F) NH4+-N removal efficiencies of Marinobacter sp., Pseudomonas sp., and Halomonas sp. after 48 h cultivation at various salinities. Salts include NaCl, Na2SO4, Na2HPO4, NaHCO3, and NaNO3.
Figure 4.
SND performance of strain Y2 at 11% Na2SO4 with (A) NH4+-N, (B) NH2OH, (C) NO2–-N, and (D) NO3–-N as single nitrogen sources.
Figure 4.
SND performance of strain Y2 at 11% Na2SO4 with (A) NH4+-N, (B) NH2OH, (C) NO2–-N, and (D) NO3–-N as single nitrogen sources.
Figure 5.
SND performance of strain Y2 at 11% Na2SO4 with (A) NH4+-N and NO2–-N, (B) NH4+-N and NO3–-N, (C) NO2–-N and NO3–-N, (D) NH4+-N, NO2–-N and NO3–-N as mixed nitrogen sources.
Figure 5.
SND performance of strain Y2 at 11% Na2SO4 with (A) NH4+-N and NO2–-N, (B) NH4+-N and NO3–-N, (C) NO2–-N and NO3–-N, (D) NH4+-N, NO2–-N and NO3–-N as mixed nitrogen sources.
Figure 6.
(A) AMO, HAO, NIR, NAP enzymatic activity and specific activity. (B) The correlation of specific enzymatic activity with the maximum rate of nitrogen removal. (C) PCR amplification of SND-associated functional genes. (D) The proposed nitrogen removal pathway of strain Y2.
Figure 6.
(A) AMO, HAO, NIR, NAP enzymatic activity and specific activity. (B) The correlation of specific enzymatic activity with the maximum rate of nitrogen removal. (C) PCR amplification of SND-associated functional genes. (D) The proposed nitrogen removal pathway of strain Y2.
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).