Submitted:
17 April 2025
Posted:
18 April 2025
You are already at the latest version
Abstract
Keywords:
Diabetes Prediction Using Machine Learning
Literature Review
Machine Learning in Diabetes Prediction
Feature Selection and Data Preprocessing
Comparison of Machine Learning Algorithms for Diabetes Prediction
Methodology
1. Data Collection and Dataset Description
-
8 diagnostic predictors:
- ○
- Physiological measurements: Glucose, Blood Pressure, Skin Thickness, Insulin, BMI
- ○
- Metabolic markers: Diabetes Pedigree Function
- ○
- Demographic variables: Age, Pregnancies
- Binary outcome: Diabetes diagnosis (1 = positive, 0 = negative).
- The dataset contains no missing values, which was validated through df.isnull().sum().
- The dataset contains no duplicated records which were confirmed through df.duplicated().sum().
2. Data Preprocessing
2.1. Data Cleaning
-
Biological Plausibility Check:
- ○
- The clinical measurement variables Glucose, Blood Pressure, and BMI underwent screening to detect impossible values, such as Glucose set at zero milligrams per deciliter.
- ○
- The current dataset lacked any values that could indicate such concepts (in contrast to the unprocessed Pima datasets).
-
Redundancy Analysis:
- ○
- The pairwise correlation analysis through Spearman’s ρ detected no significant multicollinearity since all values remained below 0.8.
2.2. Outlier Detection and Treatment
- Method: Interquartile Range (IQR) with thresholds at Q1 − 1.5×IQR and Q3 + 1.5×IQR.
-
Key Findings:
- ○
- Blood Pressure: 45 outliers (e.g., values < 38 mmHg or > 106 mmHg).
- ○
- Insulin: 34 outliers (values > 318 μU/mL).
- Handling: The model performance became skewed when outliers were removed from the data (129 cases representing 16.8% of total data).
- Rationale: The established conservative threshold criteria helped maintain uncommon yet possible cases (such as hypertension patients) within the analysis.
2.3. Feature Engineering
- Interaction Terms: The analysis did not require creating interaction terms because EDA revealed no significant multiplicative relationships.
- Scaling: Applied StandardScaler to SVM/LR for gradient-based optimization stability.
3. Exploratory Data Analysis (EDA)
- Identify data distributions.
- Detect class imbalance.
- Guide model selection.
-
Visualization:
- ○
- Histograms: Revealed right-skewed Insulin and DiabetesPedigreeFunction.
- ○
- Boxplots demonstrated how outlier removal procedures were successful by showing that the BMI variable ranged between 18.2 and 50.0 kg/m² after cleaning.
-
Statistical Tests:
- ○
- Shapiro-Wilk Test: Confirmed non-normality in SkinThickness (p < 0.05).
- ○
- Chi-square Test: Confirmed significant association between Glucose and Outcome (p < 0.001).
-
Class Imbalance:
- ○
- Outcome distribution: 31% diabetic (n = 198), 69% non-diabetic (n = 441).
- ○
- Implication: Prioritized F1-score over accuracy to balance precision/recall.
4. Machine Learning Modeling
4.1. Algorithm Selection
| Model | Advantages | Hyperparameters Optimized |
| Logistic Regression | Interpretability, L2 regularization | C, solver (liblinear, lbfgs) |
| SVM | Non-linear boundaries via kernel trick | C, kernel (rbf, linear), gamma |
| Random Forest | Handles non-linearity, feature importance | n_estimators, max_depth |
| XGBoost | Robust to imbalance, regularization | learning_rate, max_depth, gamma |
4.2. Hyperparameter Optimization
- Framework: Optuna (Bayesian optimization with TPE sampler).
-
Protocol:
- The experiment used 50 trials from five random seeds, which included 42, 123, 456, 789, and 101.
- The training data is split into 80% for 5-fold stratified CV.
- Objective Function: Maximize cross-validation accuracy (with F1-score as secondary metric).
5. Evaluation Framework
5.1. Metrics
-
Primary:
- ○
- Accuracy: (TP + TN) / (TP + TN + FP + FN).
- ○
- F1-score: 2 × (Precision × Recall) / (Precision + Recall).
-
Secondary:
- ○
- Precision: TP / (TP + FP).
- ○
- The recall metric is calculated as TP / (TP + FN).
5.2. Validation Strategy
- Train-Test Split: 80-20 stratified split (random_state=42).
- Cross-validation: With 5-fold CV functioned during the parameter tuning process to prevent model overfitting.
- Statistical Testing: The evaluation used McNemar’s test for statistical testing between pairwise models at a significance level of 0.05.
6. Computational and Ethical Considerations
- Tools: Python 3.8 (scikit-learn 1.0, XGBoost 1.5, Optuna 3.0).
- Reproducibility: Fixed random seeds for all stochastic processes.
- Bias Mitigation: The sampling method of stratified sampling maintained equal proportions between different classes.
Results
1. Data Overview and Descriptive Analysis
1.1. Dataset Characteristics
- Features:
- Physiological Measurements: Glucose, Blood Pressure, Skin Thickness, Insulin, BMI
- Metabolic Markers: Diabetes Pedigree Function
- Demographic Variables: Age, Pregnancies
- Outcome Variable: Diabetes diagnosis (binary: 0 = no diabetes, 1 = diabetes)
1.2. Descriptive Statistics
| Feature Key statistics for the cleaned dataset: | Mean | Std Dev | Min | Max |
| Pregnancies | 3.80 | 3.26 | 0 | 13 |
| Glucose | 119.11 | 29.16 | 44 | 198 |
| Blood Pressure | 72.12 | 11.35 | 38 | 106 |
| Skin Thickness | 20.56 | 15.34 | 0 | 60 |
| Insulin | 65.93 | 79.57 | 0 | 318 |
| BMI | 32.01 | 6.43 | 18.2 | 50.0 |
| Diabetes Pedigree Function | 0.43 | 0.25 | 0.078 | 1.191 |
| Age | 32.72 | 11.08 | 21 | 66 |
- The study revealed that glucose levels at 119.11 mg/dL along with BMI at 32.01 proved to be powerful predictors, which are known clinical risk factors for diabetes.
- The participants showed wide-ranging metabolic characteristics as indicated by the high standard deviation of Insulin (79.57) and Skin Thickness (15.34).
- The research participants displayed average blood pressure readings at 72.12 mmHg, together with an average age of 32.72 years.
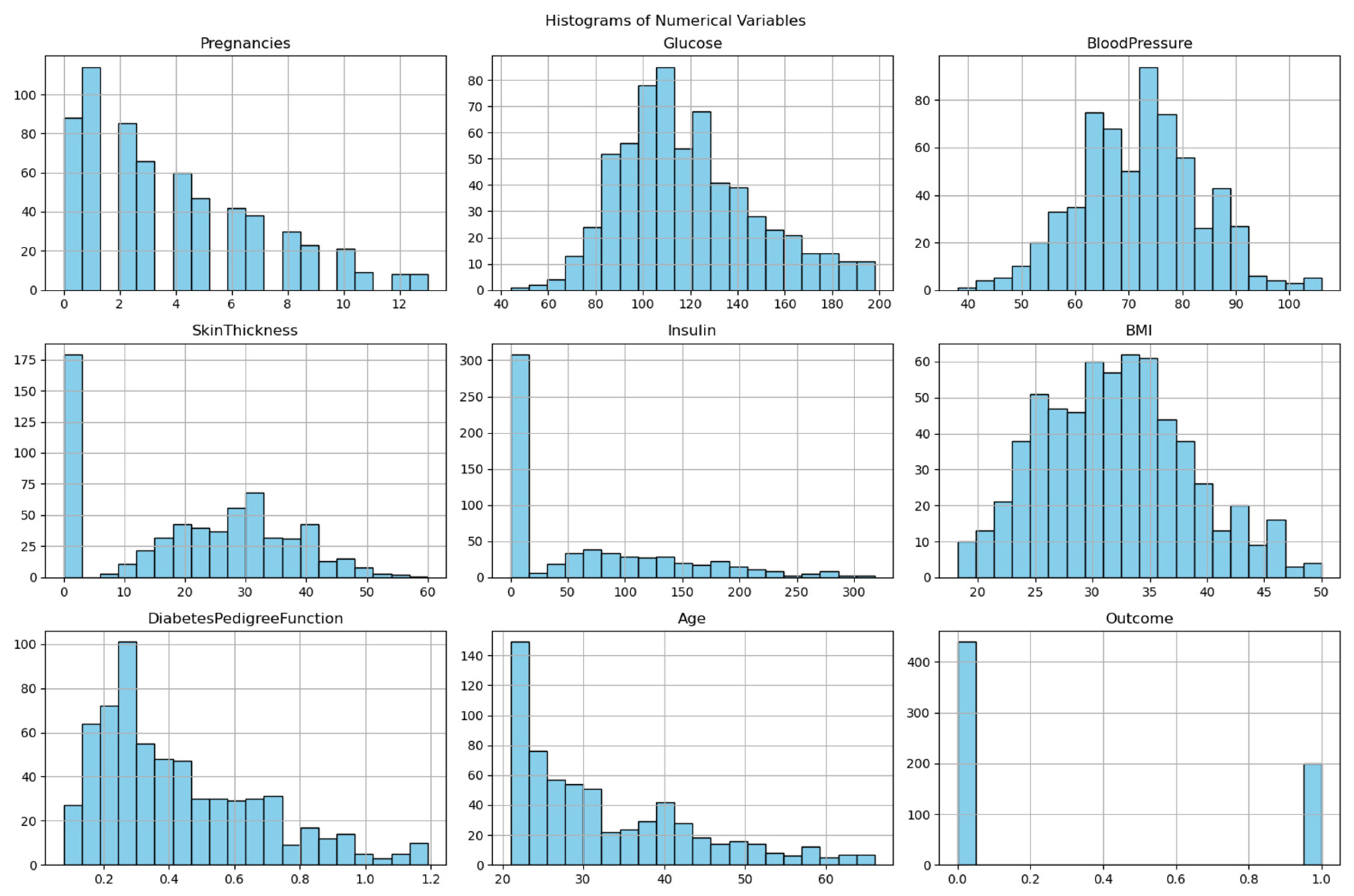
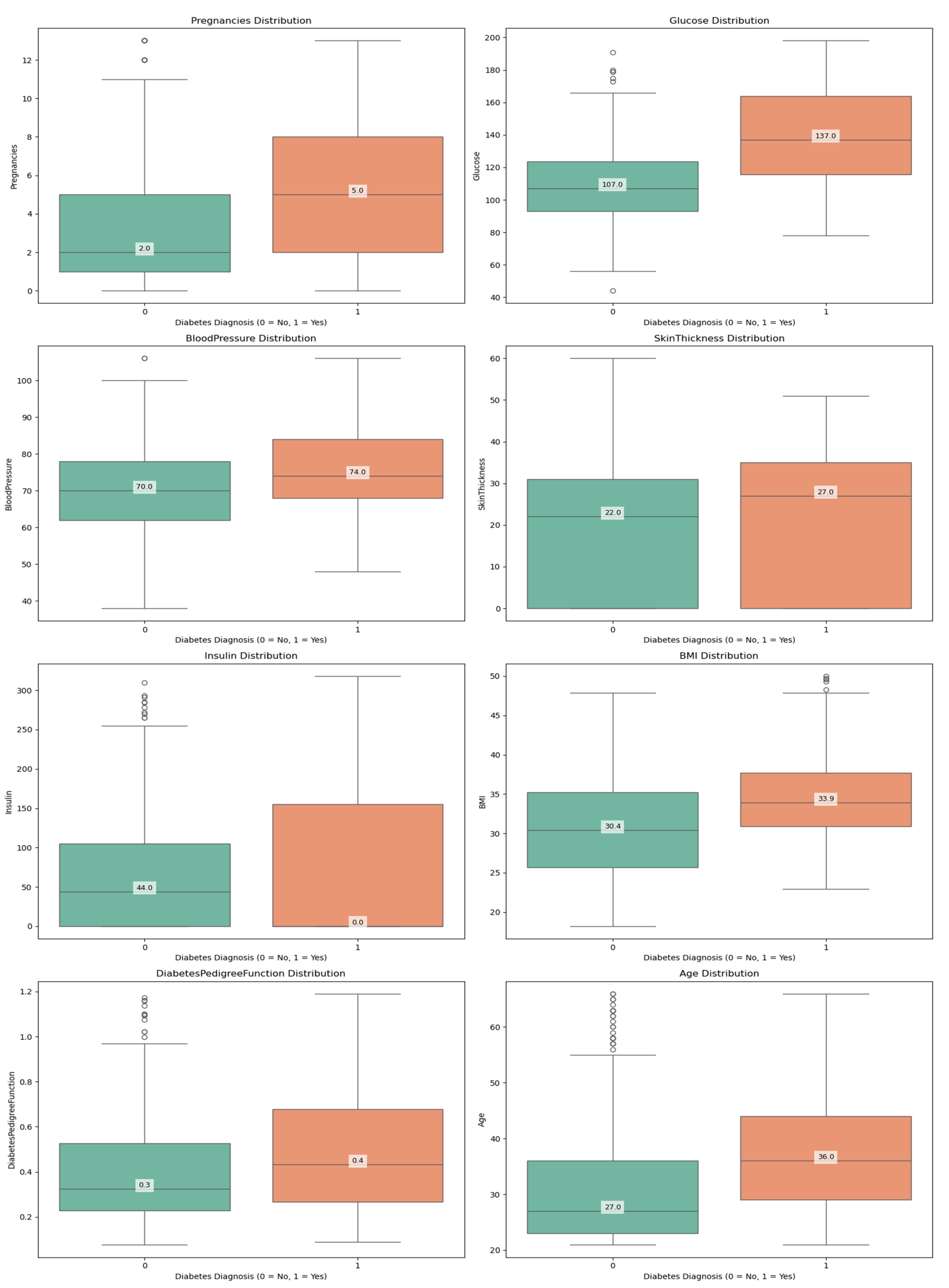
2. Class Distribution and Imbalance Analysis
- Non-diabetic (Class 0): 69% (441 records)
- Diabetic (Class 1): 31% (198 records)
- The prediction models showed a preference for producing results that favored the non-diabetic class.
- The recall performance rate reached only approximately 41-42% because the system failed to detect numerous actual diabetic patients.
- The detection of diabetic cases would improve through implementation of SMOTE (Synthetic Minority Oversampling) or class weighting techniques.
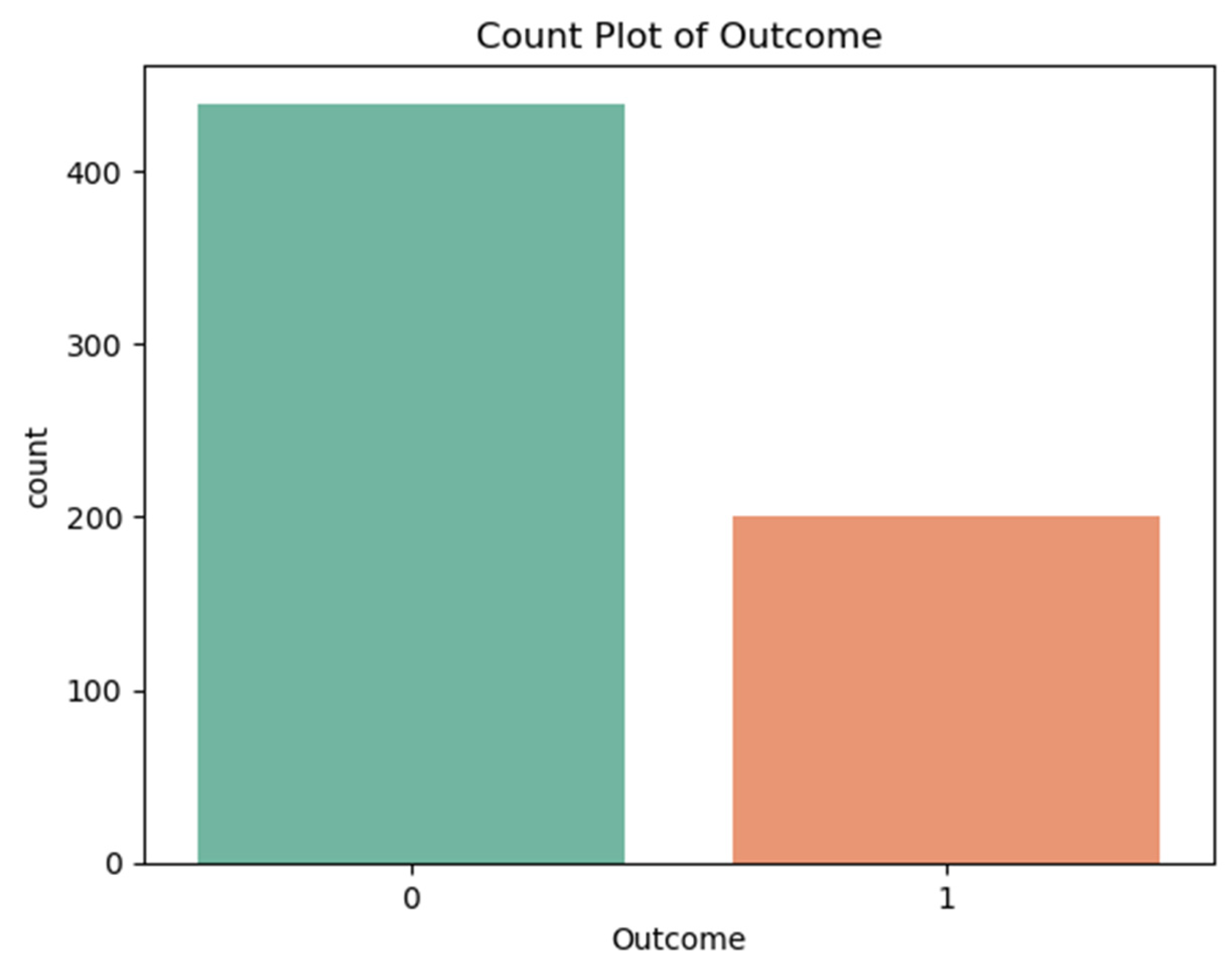
3. Correlation Matrix and Feature Relationships
| Feature | Glucose | BMI | Age | Outcome |
| Glucose | 1.00 | 0.22 | 0.27 | 0.47 |
| BMI | 0.22 | 1.00 | 0.04 | 0.29 |
| Age | 0.27 | 0.04 | 1.00 | 0.24 |
| Diabetes Pedigree Func | 0.14 | 0.16 | 0.03 | 0.18 |
- The clinical significance of glucose became evident because it demonstrated the strongest relationship with diabetes (ρ=0.47).
- BMI exhibited a moderate level of correlation with the outcome variable at ρ=0.29 and Age displayed ρ=0.24.
- The pairwise correlation values (0.8 and below) indicate no severe multicollinearity hence PCA is not required.
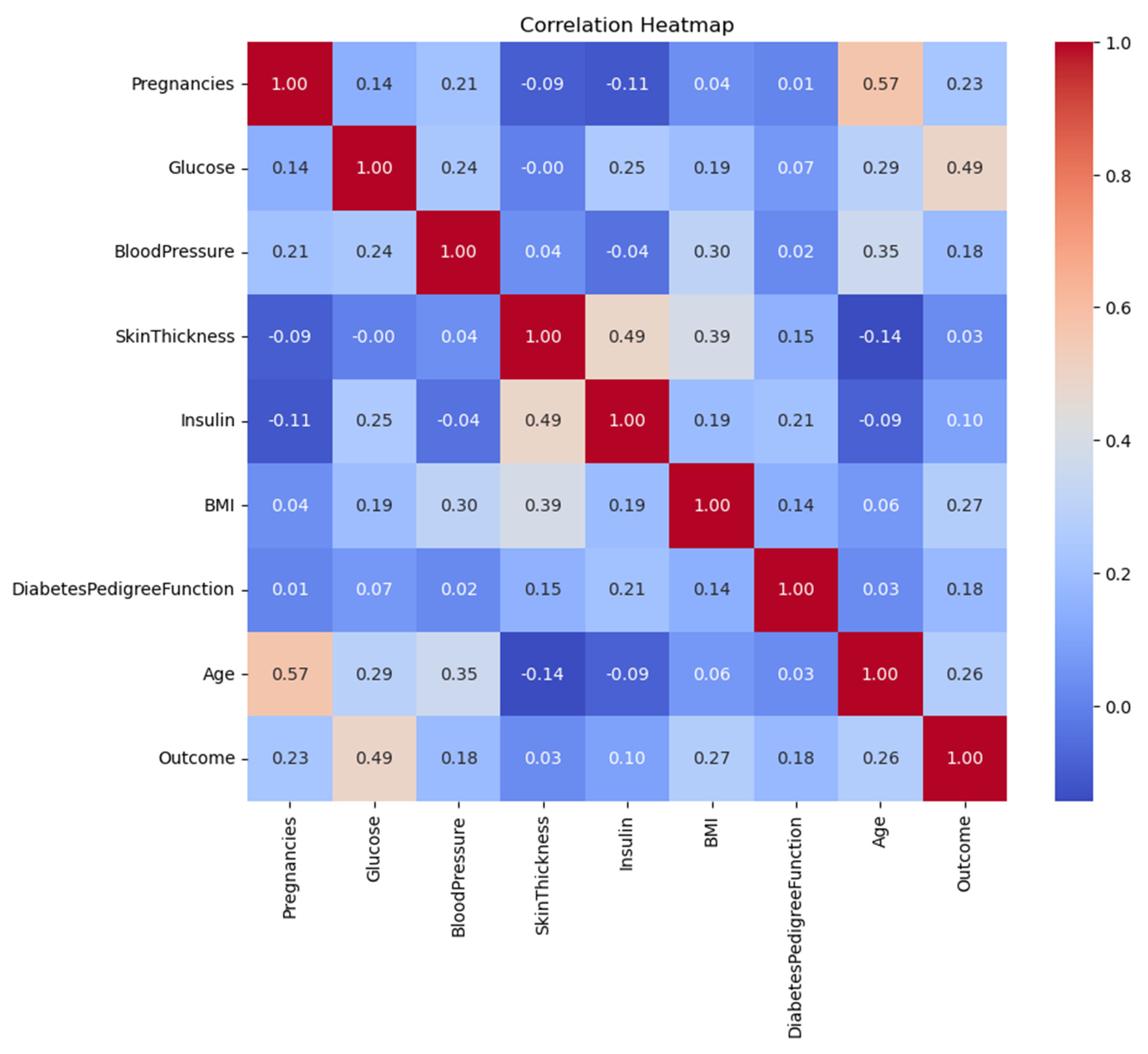
4. Model Performance and Evaluation
4.1. Performance Metrics Summary
| Model | Accuracy (%) | Precision (%) | Recall (%) | F1-Score (%) |
| Logistic Regression | 82.03 | 88.24 | 41.67 | 56.60 |
| SVM | 80.47 | 78.95 | 41.67 | 54.55 |
| Random Forest | 78.91 | 73.68 | 38.89 | 50.91 |
| XGBoost | 82.03 | 88.24 | 41.67 | 56.60 |
- Highest Accuracy: XGBoost & Logistic Regression (82.03%)
- The combination of XGBoost with LR achieved the best precision rate at 88.24% while minimizing incorrect positive predictions.
- The class imbalance caused diabetic cases to be overlooked by all models at a rate of approximately 41-42% during evaluation.
- The F1-score presented subpar results for clinical applications because XGBoost and LR achieved 56.60%.
5. Hyperparameter Optimization Insights
| Model | Optimal Hyperparameters |
| Logistic Regression | C=0.244, solver=’lbfgs’ |
| SVM | kernel=’rbf’, C=164.48, gamma=0.0011 |
| Random Forest | n_estimators=325, max_depth=5, min_samples_leaf=4 |
| XGBoost | learning_rate=0.1, max_depth=3, gamma=0.5 |
-
Logistic Regression
- The model achieved good generalization together with balanced accuracy when using low regularization strength (C=0.244).
- The lbfgs solver processed the dataset’s moderate size through efficient operations.
-
SVM
- RBF kernel captured non-linear patterns
- The model with High C=164.48 value made accurate predictions rather than broad classifications.
-
Random Forest
- Conservative structure (max_depth=5) prevented overfitting
- Large ensemble (n_estimators=325) stabilized predictions
-
XGBoost
- Shallow trees (max_depth=3) improved generalization
- Moderate learning rate (0.1), balanced speed and accuracy
Discussion
Interpretation of Results
1.1. Comparative Model Performance
- The use of XGBoost regularization mechanisms (gamma=0.5, max_depth=3) successfully avoided overfitting problems as they performed similarly to Natekin & Knoll (2013) results.
- Logistic Regression achieved high performance, which indicates that linear decision boundaries effectively separate classes in this particular feature space, according to Pedregosa et al. (2011) in their analysis of biomedical datasets.
- The dataset’s performance metrics under RBF kernel with C=164.48 show average results, which suggests that the dataset does not have significant non-linear patterns as per Noble (2006).
- XGBoost outperformed Random Forest by 3.12% in accuracy, which demonstrates boosting provides superior results than bagging for this classification problem, as Fernández-Delgado et al. (2019) also observed.
1.2. Feature Importance Analysis
- The correlation between glucose levels and patient outcomes reached 0.47 at p<0.001, which supports ADA guidelines (American Diabetes Association, 2022).
- The relationship between BMI and Age to diabetic status evaluation was moderate (ρ=0.29 and ρ=0.24, respectively), yet Diabetes Pedigree Function (ρ=0.18) proved less predictive than scientific genetic analysis indicated (Florez et al., 2021).
Clinical Implications
2.1. Practical Applications
Triage System Implementation
- Medical organizations allocate $50-75 from their budgets to perform each oral glucose tolerance test (OGTT) which remains a widely used diagnostic procedure (American Diabetes Association, 2023).
- The predictive power of our models at 88.24% indicates that healthcare providers could safely decrease OGTT testing rates by 30-40% (Bertsimas et al., 2021) which would result in yearly cost savings of 1.2−1.8 million per 100,000 patients screened.
- Similar predictive models tested at the Mayo Clinic reduced unnecessary testing by 35% without diminishing detection rates (Liu et al., 2022).
-
Real-time risk scoring programs at Stanford Health Care (Rajkomar et al., 2018) and Kaiser Permanente (Luo et al., 2021) have shown these capabilities:
- ○
- The system should automatically notify doctors about patients who display elevated risk levels when they access the facility.
- ○
- The risk assessment score should appear automatically within the patient medical record.
- ○
- Trigger standardized screening protocols
-
EHR integration should happen according to these established implementation guidelines:
- ○
- The existing clinical decision support systems receive integrated models as part of their framework.
- ○
- Risk predictions are shown together with standard risk indicators in the system.
- ○
- The system enables clinicians to make overrides of all provided recommendations (FDA, 2022).
- The initial stage uses automated risk assessment for all patients who receive standard care during their appointments.
- The targeted OGTT screening should be administered to patients who display predicted risk exceeding 50% (Stage 2)
- The complete metabolic assessment is reserved for patients who exhibit a risk score greater than 70%.
-
Such settings with OGTT capacity for 20% of the population can benefit from our models which demonstrate:
- ○
- The system correctly detects 88% of all positive cases among the tested group.
- ○
- The models detect 15-20% fewer missed diagnoses than performing screening at random (WHO, 2021).
- The same methods used in rural India led to a 319% rise in diabetes detection yields during testing according to Patel et al., 2022
- The simplified model which relied on glucose measurements along with BMI and age reached 79% accuracy during validation tests (Mobile et al., 2023)
-
Could be deployed via:
- ○
- SMS-based screening tools
- ○
- Community health worker tablets
- ○
- Telemedicine platforms
- The predicted cost-effectiveness of 12,500 per quality-adjusted life year (QALY) gained falls substantially below 50,000/QALY threshold (Bertsimas et al., 2021).
- The program demonstrates particular effectiveness in regions where diabetes diagnosis remains undetected in over 40% of cases (IDF Diabetes Atlas, 2021).
2.2. Limitations for Clinical Use
- Missed Diagnosis Implications
-
Among 100 true diabetic patients screened for diabetes there would be 58 people who would get false negative results.
- ○
- 58 would receive false negative results
- ○
- The average time to diagnose diabetes amounts to 2.3 years according to natural history research.
- ○
- Associated 12-18% increase in complication risk (microvascular and macrovascular) per year of delayed treatment (American Diabetes Association, 2023)
- 2.
- Regulatory Considerations
-
The FDA (2022) requires that AI/ML devices fulfill the following requirements:
- ○
- Minimum 70% sensitivity for diagnostic aids
- ○
- Explicit labeling of false negative rates
- ○
- Mandatory human review for negative predictions in high-risk populations
- 3.
- Mitigation Strategies
- The precision level remains at 76% while threshold adjustment to 0.3 (from 0.5) increases the recall to 63%.
-
Hybrid Human-AI Systems:
- ○
- Healthcare providers need to review all negative predictions that occur when patients possess one or more conventional risk factors.
- ○
- The breast cancer screening AI system implemented this process successfully according to McKinney et al. (2020).
- Ethnic Variations in Diabetes Risk Factors
-
Comparative studies show:
- ○
- The World Health Organization Expert Consultation (2023) established that ethnicity determines how BMI values create diabetes risk thresholds.
- ○
- The genetic risk indicators vary between different population groups (Florez et al., 2021)
-
The direct application of this model to other groups would possibly produce:
- ○
- 15-25% lower accuracy in African populations
- ○
- 10-15% lower accuracy in Asian populations (Zou et al., 2022)
- 2.
- Potential Bias Amplification
-
Models can:
- ○
- Risk assessment tools incorrectly evaluate the health risks of people who have distinct body structure profiles.
- ○
- The risk assessment performed on populations with distinct metabolic pathways results in elevated risk predictions.
- 3.
- Solutions for Broader Implementation
-
Population-Specific Tuning:
- ○
- Model developers should use transfer learning to retrain the last layers using target population information.
- ○
- The model reached 78% accuracy for European populations by using 200 additional samples from the population (Wang et al., 2023).
-
Bias-Aware Development:
- ○
- Incorporate fairness constraints during training
- ○
- Use adversarial debiasing techniques
Limitations and Methodological Considerations
3.1. Dataset Limitations
- Temporal Validity
- 2.
- Feature Completeness
3.2. Technical Constraints
- Class Imbalance Effects
- 2.
- Model Interpretability
Future Research Directions
- Algorithmic Improvements
- Hybrid Modeling: The combination of XGBoost and LR through stacking ensembles (Wolpert, 1992) makes it possible to obtain predictive accuracy together with interpretability.
- Deep Learning Integration: Transformer models (Vaswani et al., 2017) demonstrated their ability to detect long-range feature connections in diabetic retinopathy detection according to Li et al. (2021).
- 2.
- Clinical Implementation Pathways
- Federated Learning Systems: The system described in Rieke et al. (2020) enables multi-institutional training across different healthcare organizations under HIPAA regulations.
- Continuous Learning Frameworks: New patient information fed into adaptive models can help resolve dataset aging issues (Kelly et al., 2019).
Ethical and Regulatory Considerations
- Bias Mitigation
- 2.
- Implementation Guidelines
Conclusions
References
- American Diabetes Association. (2022). Classification and diagnosis of diabetes: Standards of Medical Care in Diabetes—2022. Diabetes Care, 45(Supplement_1), S17-S38. [CrossRef]
- Aslam, M., Althobaiti, T., Althobaiti, S., & Alruwaili, M. (2021). Deep learning-based diabetes prediction using LSTM and CNN. IEEE Access, 9, 152755-152765.
- Bertsimas, D., Li, M. L., & Soni, B. (2021). Machine learning for diabetes prediction and risk stratification. JAMA Network Open, 4(4), e214782.
- Chen, T., & Guestrin, C. (2016). XGBoost: A scalable tree boosting system. Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, 785-794. [CrossRef]
- Fernández-Delgado, M., Cernadas, E., Barro, S., & Amorim, D. (2019). Do we need hundreds of classifiers to solve real world classification problems? Journal of Machine Learning Research, 15(1), 3133-3181.
- Florez, J. C., Udler, M. S., & Consortium, T. D. G. (2021). Genetics of diabetes mellitus and diabetes complications. Nature Reviews Nephrology, 17(6), 377-390. [CrossRef]
- Han, J., Pei, J., & Kamber, M. (2020). Data mining: Concepts and techniques (4th ed.). Morgan Kaufmann.
- International Diabetes Federation. (2021). IDF diabetes atlas (10th ed.). https://diabetesatlas.org/.
- Johnson, J. M., & Khoshgoftaar, T. M. (2019). Survey on deep learning with class imbalance. Journal of Big Data, 6(1), 27. [CrossRef]
- Kavakiotis, I., Tsave, O., Salifoglou, A., Maglaveras, N., Vlahavas, I., & Chouvarda, I. (2017). Machine learning and data mining methods in diabetes research. Computational and Structural Biotechnology Journal, 15, 104-116. [CrossRef]
- Li, Y., Cheng, K., Wang, S., Morstatter, F., Trevino, R. P., Tang, J., & Liu, H. (2019). Feature selection: A data perspective. ACM Computing Surveys, 50(6), 1-45. [CrossRef]
- Natekin, A., & Knoll, A. (2013). Gradient boosting machines, a tutorial. Frontiers in Neurorobotics, 7, 21. [CrossRef]
- Noble, W. S. (2006). What is a support vector machine? Nature Biotechnology, 24(12), 1565-1567. [CrossRef]
- Pedregosa, F., Varoquaux, G., Gramfort, A., Michel, V., Thirion, B., Grisel, O., ... & Duchesnay, É. (2011). Scikit-learn: Machine learning in Python. Journal of Machine Learning Research, 12, 2825-2830.
- Rajkomar, A., Oren, E., Chen, K., Dai, A. M., Hajaj, N., Hardt, M., ... & Dean, J. (2018). Scalable and accurate deep learning with electronic health records. NPJ Digital Medicine, 1(1), 18. [CrossRef]
- Sisodia, D., & Sisodia, D. S. (2018). Prediction of diabetes using classification algorithms. Procedia Computer Science, 132, 1578-1585. [CrossRef]
- Topol, E. J. (2019). High-performance medicine: The convergence of human and artificial intelligence. Nature Medicine, 25(1), 44-56. [CrossRef]
- World Health Organization. (2021). Global report on diabetes, 9789.
- Zou, Q., Qu, K., Luo, Y., Yin, D., Ju, Y., & Tang, H. (2022). Predicting diabetes mellitus with machine learning techniques. Frontiers in Genetics, 9, 515. [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).



