Submitted:
23 April 2025
Posted:
24 April 2025
You are already at the latest version
Abstract
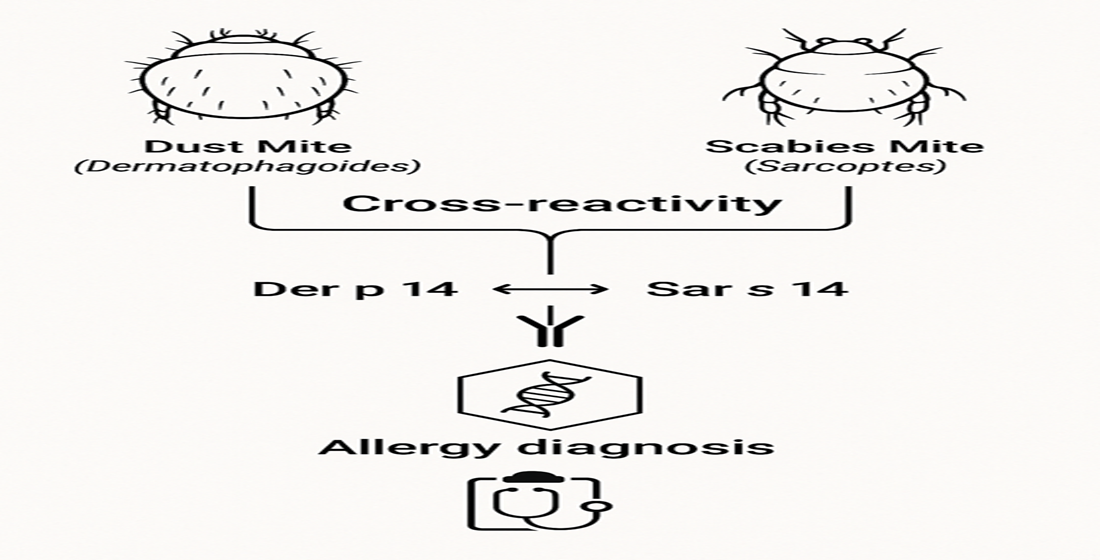
Keywords:
1. Introduction
2. Global Distribution and Health Implications of Different Mite Species
- House dust mites
- Storage mites
- Parasitic or ectoparasitic mites
| Category | Characteristics | Species | Family | Environments | Geographical distribution | Related pathologies | References |
|---|---|---|---|---|---|---|---|
| House Dust Mite | Present in domestic environments, where they feed on human dandruff and other organic debris | Dermatophagoides pteronyssinus | Pyroglyphidae | Temperate and humid climates | Europe (Italy, France, United Kingdom), Asia, and Oceania | Allergic rhinitis, asthma, and atopic dermatitis | Fernández-Caldas E, Iraola Calvo V. Mite allergens. Curr Allergy Asthma Rep. 2005 Sep;5(5):402-10. doi: 10.1007/s11882-005-0014-z. PMID: 16091214.) |
| Dermatophagoides farinae | Dry climates | North America and Central Asia | |||||
| Euroglyphus maynei | Humid climates | Western Europe, tropical regions of Asia | |||||
| Storage Mite | Present in agricultural environments and warehouses, where they feed on stored food products such as grain, flour, and seeds | Blomia tropicalis | Pyroglyphidae | Domestic environments | Latin America, Southeast Asia, and the Caribbean | Asthma and allergic rhinitis | |
| Lepidoglyphus destructor | Glycyphagidae | Granaries and farms | Northern Europe | Occupational allergies | |||
| Tyrophagus putrescentiae | Acaridae | Warehouses, humid houses | United States, United Kingdom, and Japan | Asthma, allergic rhinitis, and occupational allergies | |||
| Parasitic and Ectoparasitic Mites | Parasites of humans or animals | Sarcoptes scabiei | Sarcoptidae | Low-income areas, poor hygienic conditions | Sub-Saharan Africa, Southeast Asia, and Latin America | Scabies | |
| Demodex folliculorum | Demodecidae | Hair follicles | Ubiquitous | Dermatitis | |||
| Varroa destructor | Varroidae | Parasite of bees | Allergic reactions in beekeepers | ||||
| Ornithonyssus spp | Dermanyssidae | Parasites of birds | |||||
| Agricultural and Environmental Mites | Found in agricultural contexts, either as crop pests or as biological control agents | Tetranychus urticae | Tetranychidae | Temperate and warm climates | Mediterranean Basin and southeastern United States | Sensitization in agricultural workers | |
| Panonychus citri | Subtropical areas | Asia and Florida |
3. Discussion
3.1. Molecular Mechanisms of Cross-Reactivity
3.2. Main Proteins Involved in Cross-Reactivity
3.2.1. Tropomyosin
3.2.2. Paramyosin
3.2.3. Cysteine Proteases
3.2.4. Glutathione-S-Transferase (GST)
3.2.5. Lipid-Binding Protein Family (MD-2 Like)
3.2.6. SMIPPs (Scabies Mite Inactivated Protease Paralogs)
3.3. Clinical Implications
3.4. Diagnostic Challenges
3.5. Therapeutic Considerations
3.6. Bioinformatic Analysis
- An identity greater than 70% is considered a strong indicator of clinical cross-reactivity.
- Values between 50% and 70% can still lead to cross-reactivity, but they depend more on the three-dimensional structure of the allergen and the similarity of IgE-binding sites. Indeed, it is not always possible to establish a simple and direct relationship between sequence homology and cross-reactivity, as structural and conformational factors of allergenic proteins also influence their ability to bind IgE.
- For identity values below 50%, cross-reactivity is less likely but not excluded, especially for allergens with conserved IgE-binding sites [30].
4. Results
- Sequence info: The unique identifier code for the protein in the UniProt database.
- Name: Common name of the protein.
- Length: The length of the protein in terms of the number of amino acids.
- Opt: Optimized alignment score, reflecting the overall similarity between the sequences.
- Bits Score: The “bit” score, which helps assess the quality of the alignment; a higher score indicates a better alignment.
- E-Value: The E-value represents the expected number of alignments that would occur by chance with a score equal to or higher than the observed one, given the database and query size. A lower E-value indicates a statistically significant alignment.
- Identities (%): The percentage of identity between the sequences; it shows how many amino acid positions are exactly the same in the aligned sequences.
- Positives (%): The percentage of similarity, including conservatively substituted amino acids with similar chemical properties.
- Gaps (%): The percentage of gaps in the alignment, indicating where the sequences have been extended or truncated to optimize alignment.
| Sequence Info | Allergen Name | Lenght | Opt | Bits Score | E Value |
|---|---|---|---|---|---|
| AAF14270 | Eur m 14 | 1668 | 6992 | 1599.7 | 0 |
| AAM21322 | Der p 14 | 1662 | 6936 | 1586.9 | 0 |
| BAA04558 | Der f 14 | 349 | 1558 | 361.4 | 15e-100 |
| CAA31942 | Gal d vitellogenin | 1850 | 163 | 43.1 | 0.00055 |
4.1. Results of Sar s 14 Protein Analysis
-
Eur m 14 (Euroglyphus maynei):
- ○
- Identity: 59% (995/1662 residues), Overall similarity: 81%.
- ○
- Score: 1599.7 bits, E-value: 0.
- ○
- These data indicate a highly conserved sequence, with an extremely high level of homology and almost no significant differences. The strong positivity (81%) highlights the presence of conservative substitutions (amino acids with similar biochemical properties even if not identical).
-
Der p 14 (Dermatophagoides pteronyssinus):
- ○
- Identity: 59% (980/1657 residues), Overall similarity: 81%.
- ○
- Score: 1586.9 bits, E-value: 0.
- ○
- This result confirms a strong similarity with Sar s 14, suggesting a similar evolutionary role and a probable sharing of immunogenic regions relevant to the immune system.
-
Der f 14 (Dermatophagoides farinae):
- ○
- Identity: 65% (227/349 residues), Overall similarity: 83%.
- ○
- Score: 361.4 bits, E-value: 2e-100.
- ○
- In this case, the identity level is even higher (65%), with a very high positivity percentage. However, the lower bits score and E-value indicate a shorter sequence. This is because only a protein fragment was available in bioinformatics databases rather than the full amino acid sequence.
4.2. Comparison with Vitellogenins (Chicken Egg, Gallus domesticus)
-
Gal d vitellogenin:
- ○
- Score: 43.1 bits / 40.6 bits.
- ○
- Identity: 22%.
- ○
- Positivity: 46%.
- ○
- Gaps: ~15% (134/868 residues).
- ○
- The similarity is low, with only 22% sequence identity and multiple gaps. This suggests that while vitellogenins have a similar structural function, they share low homology with Sar s 14. The evolutionary distance between phyla explains these differences.
| Sequence Info | Allergen Name | Lenght | Opt | Bits Score | E Value |
|---|---|---|---|---|---|
| BAC53948 | Der f 1 | 321 | 793 | 170.6 | 8.1e-44 |
| P25780 | Eur m 1 | 321 | 785 | 168.9 | 2.5e-43 |
| CAQ68250 | Der p 1 | 302 | 763 | 164.5 | 5.1e-42 |
| AAQ24541 | Blo t 1 | 333 | 481 | 107.1 | 1e-24 |
| Sequence Info | Allergen Name | Lenght | Opt | Bits Score | E Value |
|---|---|---|---|---|---|
| O97370 | Eur m 3 | 261 | 657 | 156.4 | 9.1e-40 |
| P39675 | Der p 3 | 261 | 652 | 155.3 | 2e-39 |
| P49275 | Der f 3 | 259 | 642 | 153.0 | 9.7e-39 |
| AAM10779 | Blo t 3 | 266 | 583 | 139.5 | 1.1e-34 |
| AAP57077 | Der p 9 | 273 | 487 | 117.6 | 4.5e-28 |
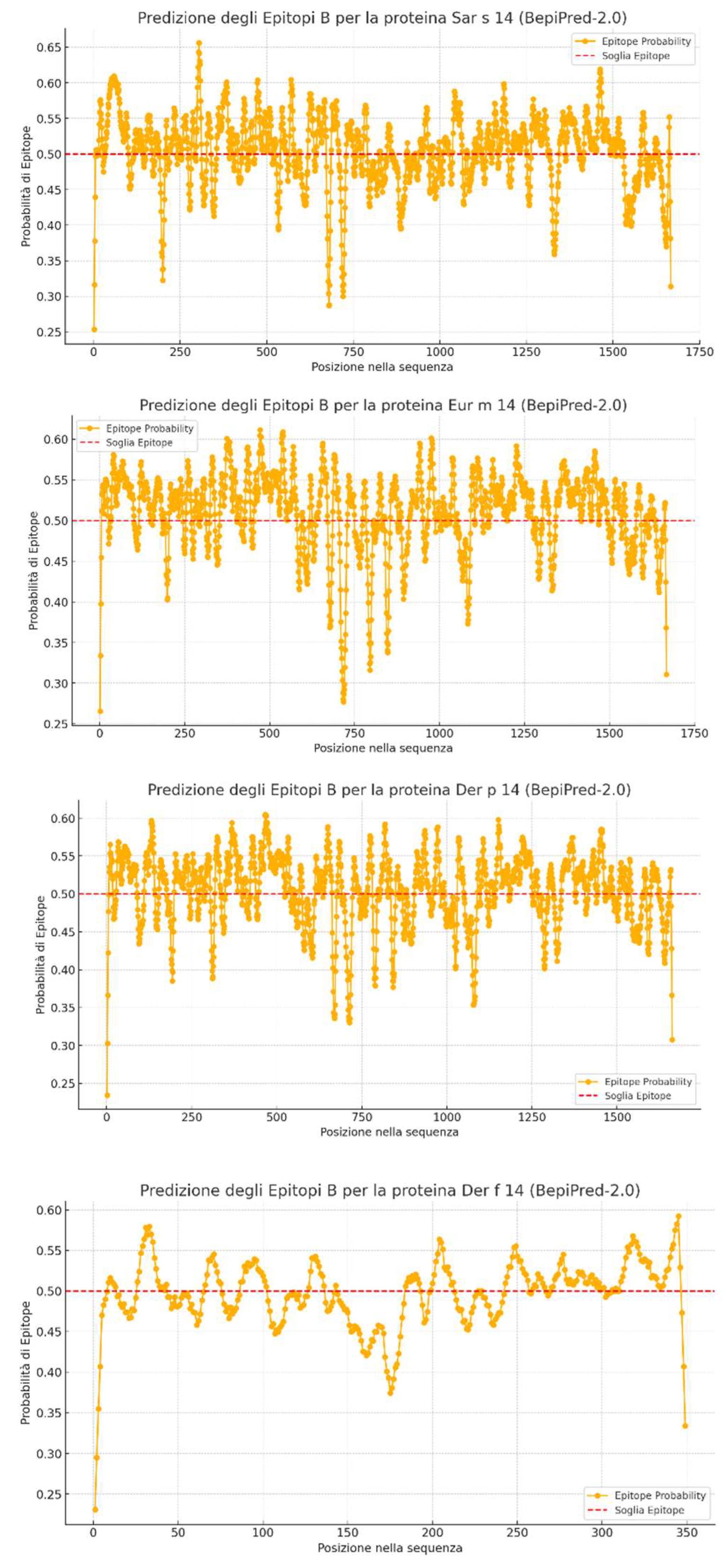
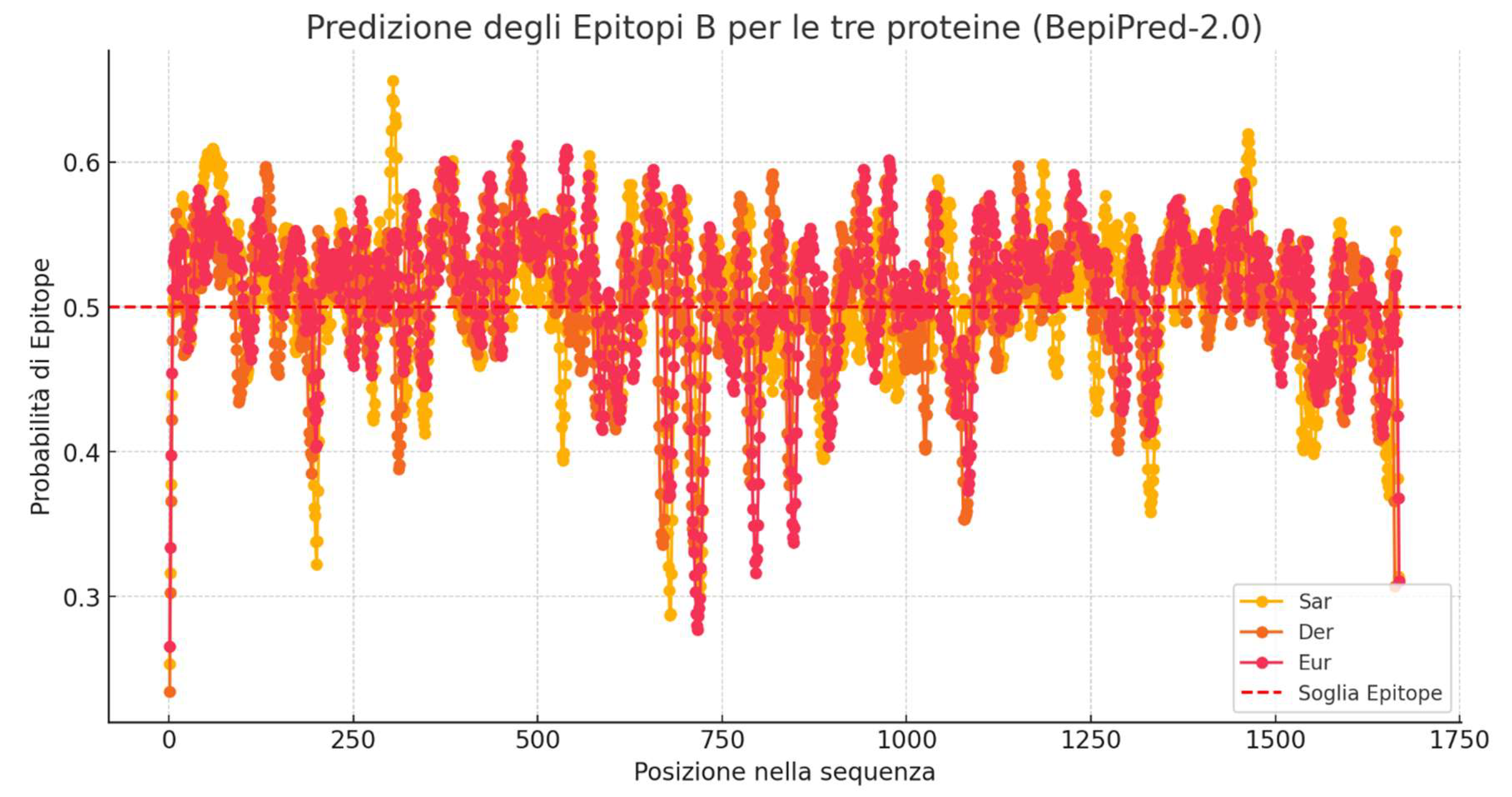
- The X-axis represents the position of amino acids within the protein sequence.
- The Y-axis shows the epitope prediction score.
- The dashed red line is the cutoff threshold set at 0.5: values above this threshold are considered potential epitopes.
- The highest average epitope probability was observed for Eur m 14 (0.5107), followed by Der p 14 (0.5053) and Sar s 14 (0.5047).
- This suggests that Eur m 14 has a higher tendency to contain antigenic epitopes than the other two proteins.
-
When considering residues with a probability above the 0.5 threshold, the distribution was:
- ○
- Eur m 14: 1132 antigenic residues (67.9%)
- ○
- Der p 14: 1007 antigenic residues (60.6%)
- ○
- Sar s 14: 979 antigenic residues (58.7%)
- ○
- This indicates that Eur m 14 has a wider distribution of epitopes and a greater likelihood of immune recognition.
-
The protein with the highest number of exposed residues was Sar s 14 (240 residues, 14.4%), followed by:
- ○
- Eur m 14 (237 residues, 14.2%)
- ○
- Der p 14 (231 residues, 13.9%)
- Although the differences are minimal, Sar s 14 shows higher surface exposure, potentially making it more accessible for antibody interaction.
-
Relative Surface Accessibility analysis revealed:
- ○
- Eur m 14: 0.1831
- ○
- Der p 14: 0.1783
- ○
- Sar s 14: 0.1757
- This means that Eur m 14 has a structural profile that makes it more accessible, potentially explaining its higher epitope density.
-
Helix Probability (α-helices formation tendency):
- ○
- Sar s 14: 0.2060
- ○
- Der p 14: 0.2046
- ○
- Eur m 14: 0.2013
-
Sheet Probability (β-sheet formation tendency):
- ○
- Eur m 14: 0.0934
- ○
- Sar s 14: 0.0920
- ○
- Der p 14: 0.0917
-
Coil Probability (flexible/disordered structure tendency):
- ○
- Eur m 14: 0.7052
- ○
- Der p 14: 0.7036
- ○
- Sar s 14: 0.7019
- ○
- Coil regions are often implicated in antigenic epitopes, which may further explain the higher antigenicity of Eur m 14.
- Based on:
- The highest average epitope probability
- The largest number of residues with epitope probability > 0.5
- The highest surface accessibility
- The strongest tendency to form flexible regions (coil structure)
- Positions with at least two proteins above the 0.5 threshold: Multiple regions along the sequence show high epitope probability in at least two of the three analyzed proteins. This suggests that certain sequence regions are likely antigenic in more than one protein, indicating conserved epitopes or structurally favorable antigenic sites.
- Positions where all three proteins exceed the 0.5 threshold: Several regions exhibit a probability above the critical threshold for all proteins. These areas are particularly significant as they represent highly conserved epitopes, which could serve as ideal targets for immunogenicity strategies or vaccine development.
5. Conclusions
References
- Arlian LG. Dust mites: update on their allergens and control. Curr Allergy Asthma Rep. 2001 Nov;1(6):581-6. [CrossRef]
- Sidenius, K.E., Hallas, T.E., Poulsen, L.K. and Mosbech, H. (2001), Allergen cross-reactivity between house-dust mites andother invertebrates. Allergy, 56: 723-733. [CrossRef]
- Huey-Jy Huang, Eszter Sarzsinszky, Susanne Vrtala, House dust mite allergy: The importance of house dust mite allergens for diagnosis and immunotherapy, Molecular Immunology, Volume 158, 2023, Pages 54-67, ISSN 0161-5890. [CrossRef]
- Hengge UR, Currie BJ, Jäger G, Lupi O, Schwartz RA. Scabies: a ubiquitous neglected skin disease. Lancet Infect Dis. 2006 Dec;6(12):769-79. [CrossRef]
- Govindarajan R, Rajamannar V, Kumar A, Samuel PP. Current status of mites and mite-borne diseases in India. J Vector Borne Dis. 2023 Jan-Mar;60(1):1-10. [CrossRef]
- Sunderkötter C, Wohlrab J, Hamm H. Scabies: Epidemiology, Diagnosis, and Treatment. Dtsch Arztebl Int. 2021 Oct 15;118(41):695-704. [CrossRef]
- Arora P, Rudnicka L, Sar-Pomian M, Wollina U, Jafferany M, Lotti T, Sadoughifar R, Sitkowska Z, Goldust M. Scabies: A comprehensive review and current perspectives. Dermatol Ther. 2020 Jul;33(4):e13746. [CrossRef]
- Arlian, L. G., Vyszenski-Moher, D. L., & Gilmore, A. M. (1988). Cross-antigenicity between sarcoptes scabiei and the house dust mite, dermatophagoides farinae (acari: sarcoptidae and pyroglyphidae). Journal of Medical Entomology, 25(4), 240-247. [CrossRef]
- Larry G Arlian, Diann L. Vyszenski-Moher, Salva G. Ahmed, Stephen A. Estes, Cross-Antigenicity Between the Scabies Mite, Sarcoptes scabiei, and the House Dust Mite, Dermatophagoides pteronyssinus, Journal of Investigative Dermatology, Volume 96, Issue 3, 1991, Pages 349-361, ISSN 0022-202X, . [CrossRef]
- Arlian LG, Feldmeier H, Morgan MS (2015) The Potential for a Blood Test for Scabies. PLoS Negl Trop Dis 9(10): e0004188. [CrossRef]
- Falk ES, Dale S, Bolle R, Haneberg B. Antigens Common to Scabies and House dust Mites. Allergy. 1981 May;36(4):233-8. [CrossRef]
- Sánchez-Borges M, et al. Scabies, crusted (Norwegian) scabies and the diagnosis of mite sensitisation. Allergol Immunopathol (Madr). 2017. [CrossRef]
- Smith AM, Benjamin D, Hozic N, et al.: The molecular basis of antigenic cross-reactivity between the group 2 mite allergens. J Allergy Clin Immunol 2001, 107:977–984.
- Fernández-Caldas E, Iraola Calvo V. Mite allergens. Curr Allergy Asthma Rep. 2005 Sep;5(5):402-10. [CrossRef]
- Schumann RJ, Morgan MS, Glass R, Arlian LG. Characterization of house dust mite and scabies mite allergens by use of canine serum antibodies. Am J Vet Res. 2001 Sep;62(9):1344-8. [CrossRef]
- Aalberse, R.C. (1998), Allergens from mites: implications of cross-reactivity between invertebrate antigens. Allergy, 53: 47-48. [CrossRef]
- Nisbet AJ, MacKellar A, Wright HW, Brennan GP, Chua KY, Cheong N, Thomas JE, Huntley JF. Molecular characterization, expression and localization of tropomyosin and paramyosin immunodominant allergens from sheep scab mites (Psoroptes ovis). Parasitology. 2006 Oct;133(Pt 4):515-23. [CrossRef]
- Falk ES, Bolle R. IgE antibodies to house dust mite in patients with scabies. Br J Dermatol. 1980 Sep;103(3):283-8. [CrossRef]
- K. Fischer, D.C. Holt, B.J. Currie, S.F. Walton, D.J. Kemp, Scabies mite inactivated protease paralogues, International Congress Series, Volume 1289, 2006, Pages 85-88, ISSN 0531-5131, ISBN 9780444522054, . [CrossRef]
- Rider SD, Jr., Morgan MS, Arlian LG (2017) Allergen homologs in the Euroglyphus maynei draft genome. PLoS ONE 12(8): e0183535. [CrossRef]
- Bajoghli AA, Bajoghli M, Adler S. Positive house dust mite skin test in a nonatopic patient with scabies. Ann Allergy Asthma Immunol. 2014 Dec;113(6):667. [CrossRef]
- Walton SF, Slender A, Pizutto S, Mounsey KE, Opresecu F, Thomas WR, Hales BJ, Currie BJ. Analysis of IgE binding patterns to house dust mite allergens in scabies-endemic communities: insights for both diseases. Clin Exp Allergy. 2015 Dec;45(12):1868-72; Erratum in: Clin Exp Allergy. 2016 Mar;46(3):508. doi: 10.1111/cea.12713. [CrossRef]
- Marjorie S. Morgan, Larry G. Arlian, Enzymatic Activity in Extracts of Allergy-Causing Astigmatid Mites, Journal of Medical Entomology, Volume 43, Issue 6, 1 November 2006, Pages 1200–1207, . [CrossRef]
- Wayne R. Thomas, Hierarchy and molecular properties of house dust mite allergens, Allergology International, Volume 64, Issue 4, 2015, Pages 304-311, ISSN 1323-8930, . [CrossRef]
- Panzer R, Krebs S. Mites, caterpillars and moths. J Dtsch Dermatol Ges. 2020 Aug;18(8):867-880. [CrossRef]
- Arlian LG, Morgan MS. Serum antibody to Sarcoptes scabiei and house dust mite prior to and during infestation with S. scabiei. Vet Parasitol. 2000 Jul 4;90(4):315-26. [CrossRef]
- Bigler, B. and Virchow, F. (2004), P-43. IgG antibodies against sarcoptic mite antigens in dogs cross-reacting with house dust and storage mite antigens. Veterinary Dermatology, 15: 54-54. [CrossRef]
- Taşkapan O, Harmanyeri Y. Atopy patch test reactions to house dust mites in patients with scabies. Acta Derm Venereol. 2005;85(2):123-5. [CrossRef]
- Aalberse RC. Clinically Significant Cross-Reactivities among Allergens. Int Arch Allergy Immunol. 1992;99(2-4):261-264. [CrossRef]
- Chapman MD, Pomés A, Breiteneder H, Ferreira F. Nomenclature and structural biology of allergens. J Allergy Clin Immunol. 2007 Feb;119(2):414-20. [CrossRef]
- Cantillo, Puerta, L.; Fernandez-Caldas, E.; et al. Tropomyosins in mosquito and house dust mite cross-react at the humoral and cellular level. Clin Exp Allergy 2018, 48, 1354-1363.
- ChatGPT as a bioinformatic partner. Gianluca Mondillo, Alessandra Perrotta, Simone Colosimo, Vittoria Frattolillo- medRxiv 2024.08.20.24312291; [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





