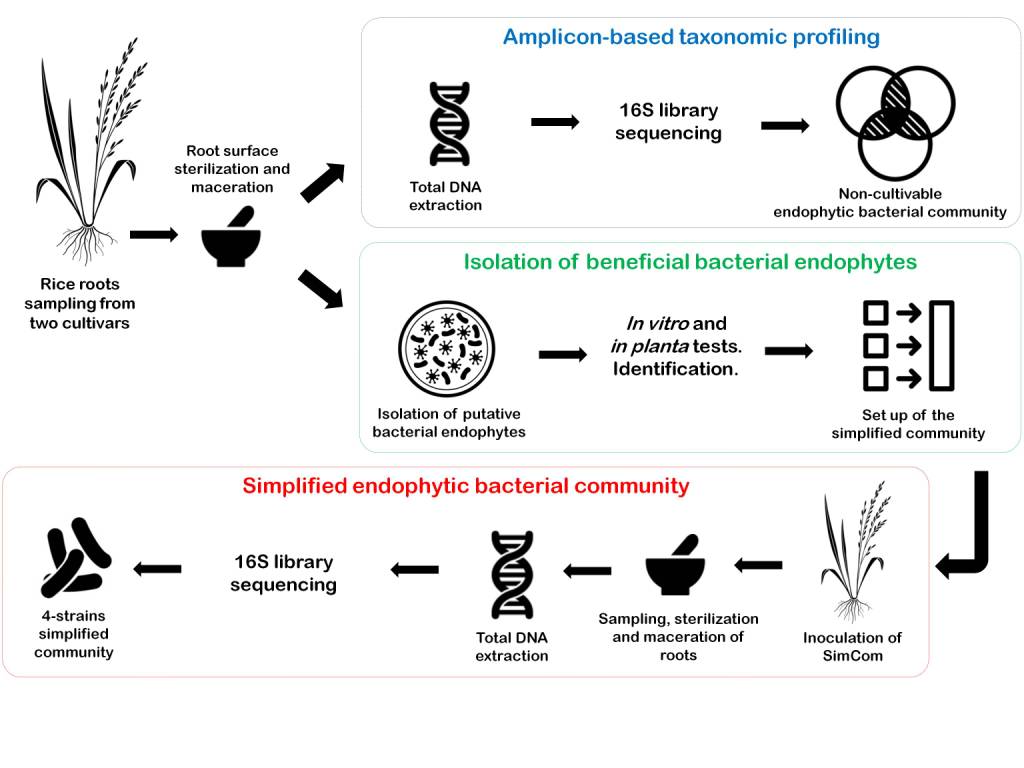
Rice is currently the most important food crop in the world and we are only just beginning to study the bacterial associated microbiome. It is of importance to perform screenings of the core rice microbiota and also to develop new plant-microbe models and simplified communities for increasing our understanding about the formation and function of its microbiome. In order to begin to address this aspect, we have performed the isolation of hundreds bacterial isolates obtained from endorhizosphere of two rice cultivars from Venezuela. The validation of plant-growth promoting bacterial activities in vitro has led us to select and characterize 15 isolates for in planta studies such as germination test, endophytism ability and plant growth promotion. Consequently, a set of 10 isolates was selected for the set-up of an endophytic consortium as a simplified model of the natural rice bacterial endomicrobiota. Upon inoculation, the colonization and abundance of each strain within the rice roots was tracked by a culture-independent technique in gnotobiotic conditions in a 30 days period. Four strains belonging to Pseudomonas, Agrobacterium and Delftia genera have shown a promising capacity for colonizing and coexistence in root tissues. On the other hand, a bacterial community taxonomic profiling of the rhizosphere and the endorhizosphere of both cultivars were obtained and are discussed. This study is part of a growing body of research on core crops microbiome and simplified microbiomes, which strengthens the formation process of the endophytic community leading to a better understanding of the rice microbiome.