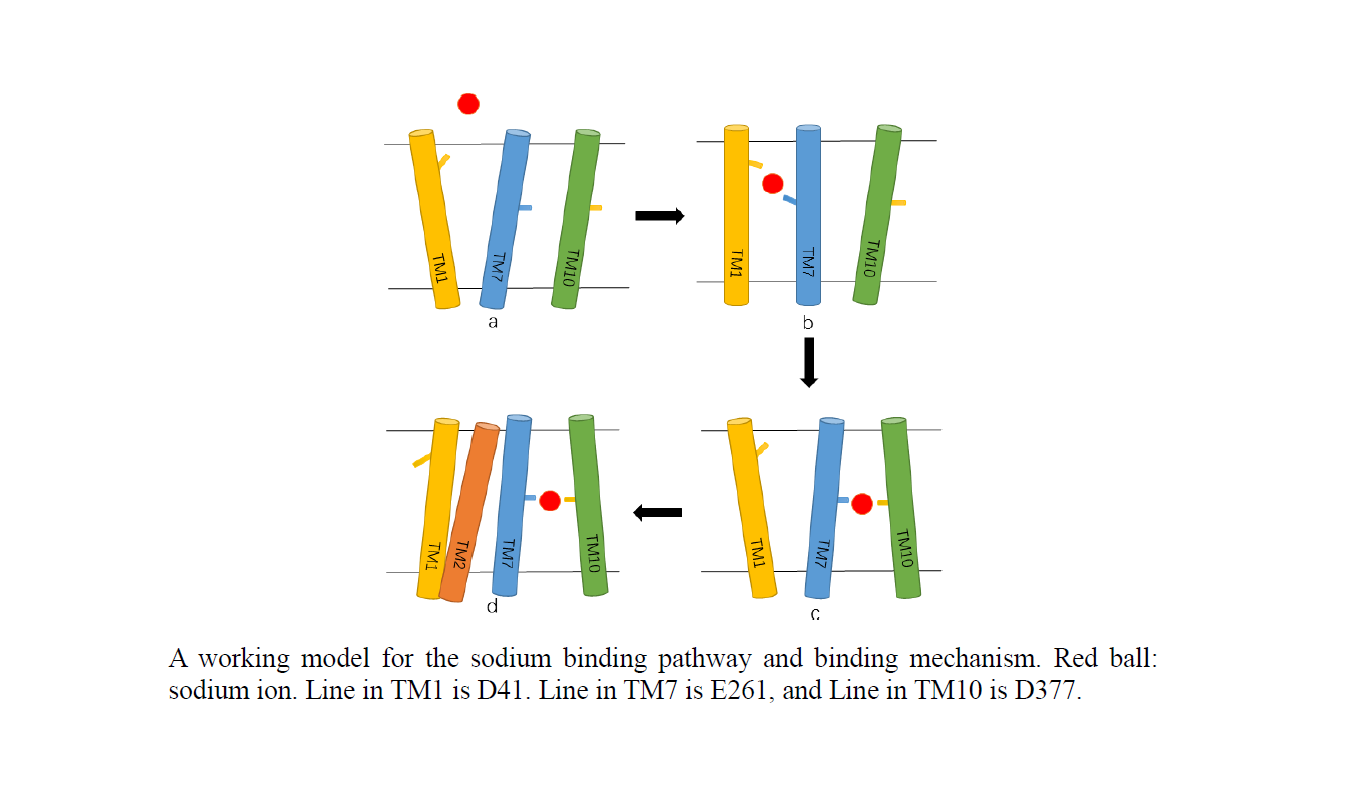
The multidrug resistance transporter NorM is an important drug resistance pump and plays a critical role in multidrug resistance in bacteria and mammals. In this study we carried out molecular dynamics simulation to study the mechanism of Na+ binding and dynamical structures of two long loops in the substrate-releasing process in substrate binding NorM. Our simulation study identified several key residues (D41, E261 D377) along the Na+ binding pathway and a multi-state ion-binding mechanism is proposed based on the simulation study. In this proposed model, the transport of Na+ is a multi-stage process with D41 being the first station for binding to Na+, followed by Na+ binding to the second station E262 and finally to the cation-binding site of E262 and D377. During the transport of Na+, the transmembrane components TM1, TM7 and TM2 are rearranged to facilitate the ion transport as well conformational changes of NorM to a closed state. Further, substrate-bound simulation revealed that Loop3-4 and Loop9-10 control the substrate-releasing process.