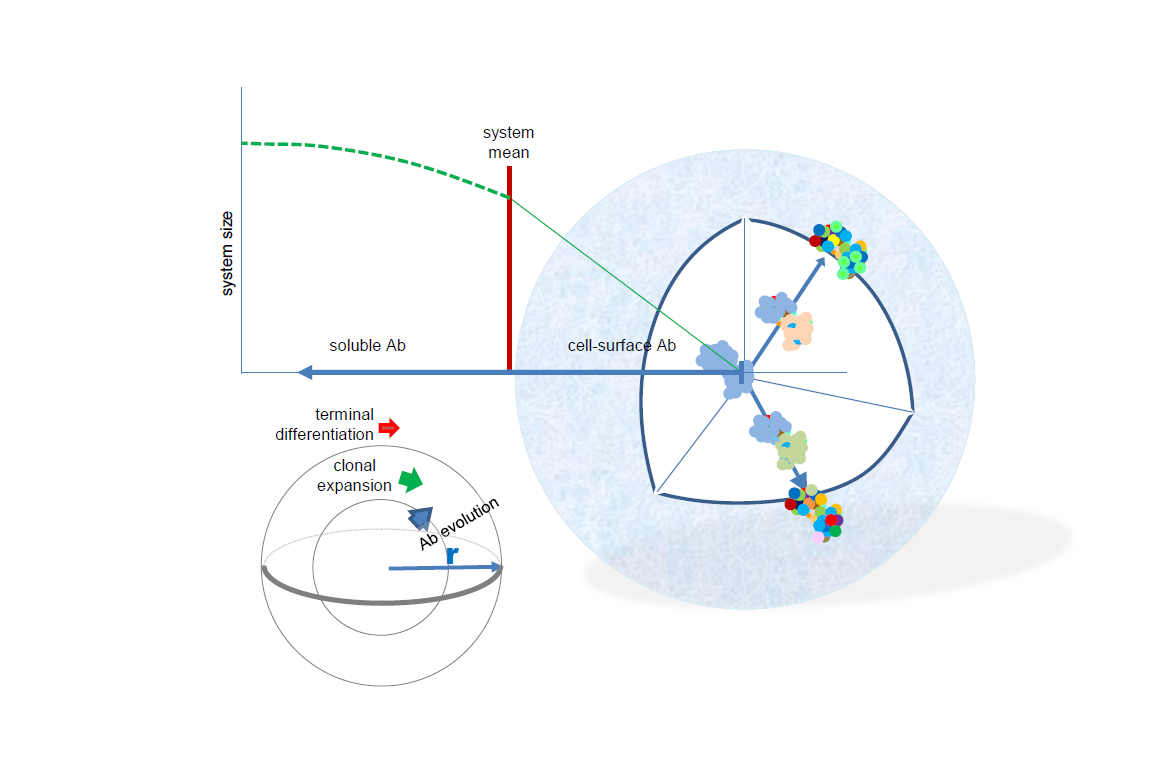
Based on the key molecule of humoral adaptive immunity, the antibody, evolution of the system comprises molecular genetic, cell biologic and immunologic mechanisms, and as a network the system is likely governed and can be characterized by physical rules as well. While deep sequencing can provide vast amounts of data related primarily to clonal relationships, functional interpretation of such data is hampered by the inherent limitations of converting sequence to structure to function. In this paper a novel model of structural interaction space, termed radial adjustment of system resolution, or RADARS, is proposed. The model is based on the radial growth of resolution of structural recognition, corresponding to increasing affinity of immune reactivity, and the virtual infinity of directions of growth, corresponding to the ability to respond to almost any molecular structure. Levels of interaction strength appear as shells of the sphere representing the system. B-cell development and immune responses can be readily interpreted in the model and quantitative properties of the antibody network can be inferred from the physical properties of a quasi-spherical system growing multi-radially. The system is described by double-Pareto distribution, sampling the lognormally distributed equilibrium constants at a rate of phi square. Finally, general strategies for merging antibody sequence space into structural space are outlined.