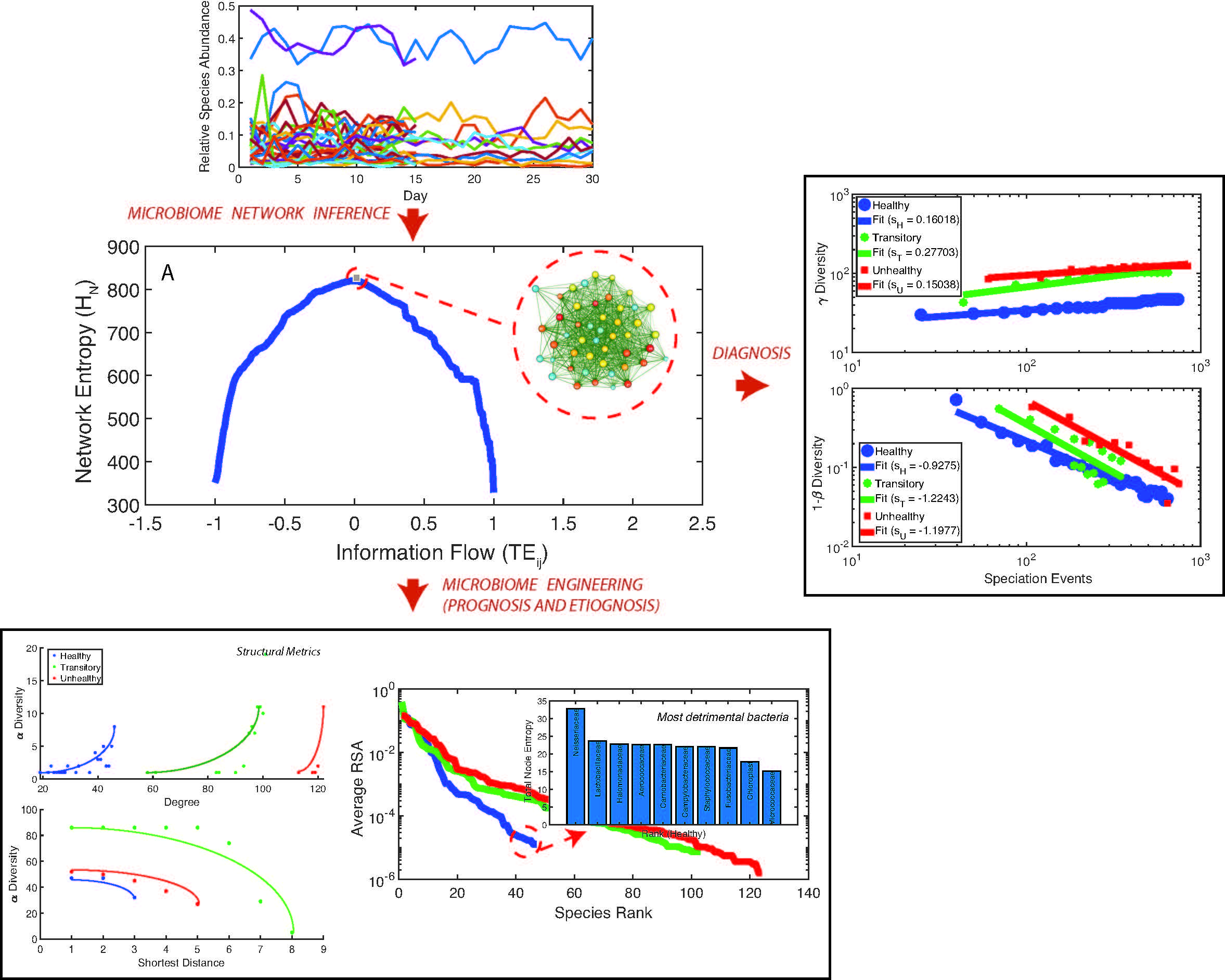
The human microbiome is an extremely complex ecosystem considering the amount of bacterial species, their interactions, and its variability over time. Here we untangle the complexity of the human microbiome for the Irritable Bowel Syndrome (IBS) that is the most prevalent functional gastrointestinal disorder in human populations. Based on a novel information theoretic network inference model we detect species interaction networks that are functionally and structurally different for healthy and unhealthy individuals. Healthy networks are characterized by a neutral symmetrical pattern of species interactions and scale-free topology versus random unhealthy networks. We detect an inverse scaling relationship between species total outgoing information flow, meaningful of node interactivity, and relative species abundance (RSA). The top ten interacting species are also the least relatively abundant for the healthy microbiome and the most detrimental. These findings support the idea about the diminishing role of network hubs and hubs should be defined considering the total outgoing information flow rather than the node degree. Macroecologically, the healthy microbiome is characterized by the highest total species diversity growth rate, the lowest species turnover, and the smallest variability of RSA for all species. This result challenges current views that posit a universal association between healthy states and the highest absolute species diversity in ecosystems. Additionally, we show how the transitory microbiome is unstable and microbiome criticality is not at the phase transition between healthy and unhealthy states. We stress out the importance of considering interacting pairs versus single node dynamics when characterizing the microbiome and of ranking these pairs in terms of their dynamics. Interactions (i.e. species collective behavior) shape transition from healthy to unhealthy states.\\ The macroecological characterization of the microbiome is useful for diagnostic purposes and disease etiognosis, while species-specific analyses can detect species that are more beneficial leading to personalized design of pre- and pro-biotic treatments and microbiome engineering.