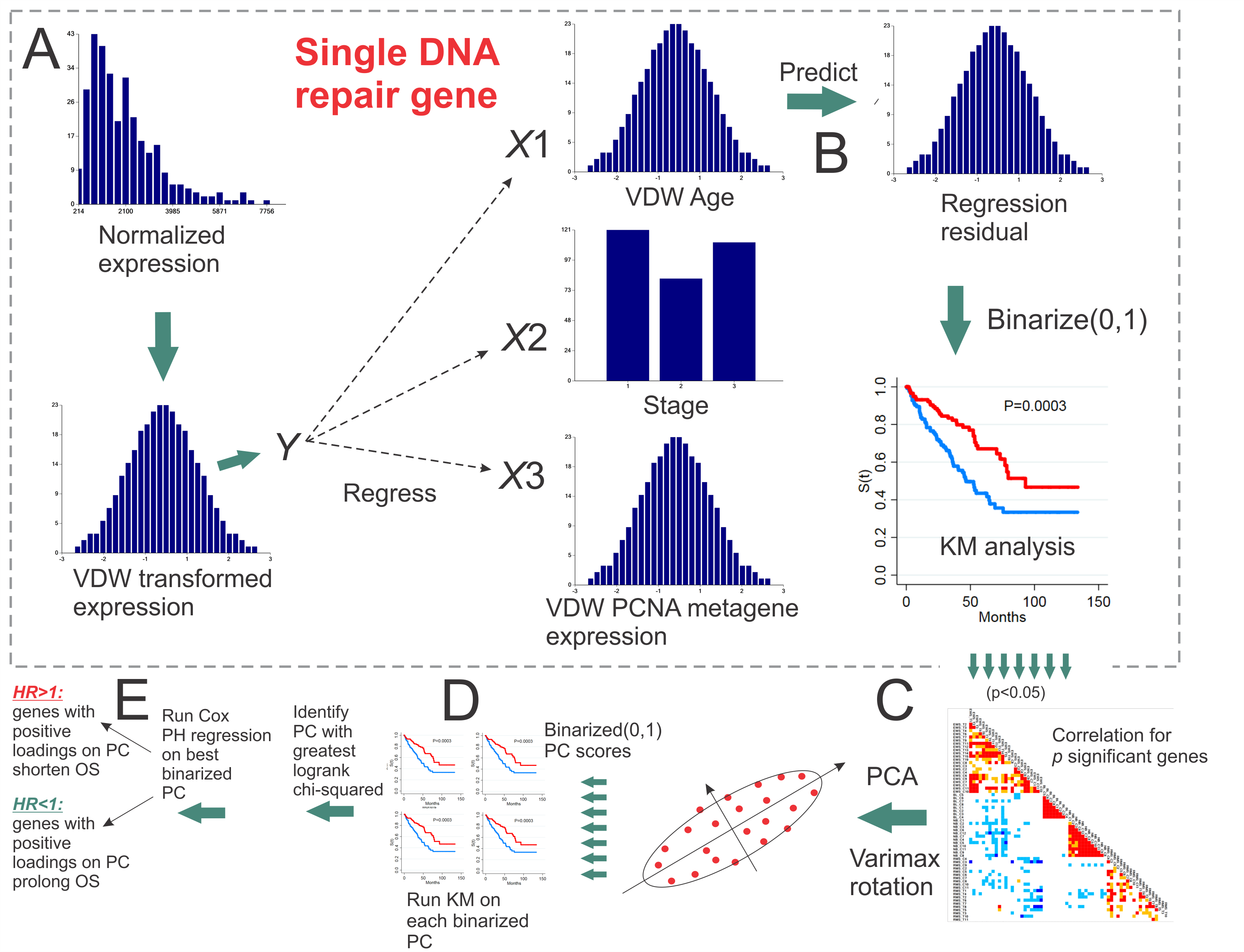
Removal of the proliferation component of gene expression by PCNA adjustment has been addressed in numerous survival prediction studies for breast cancer and all cancers in the TCGA. These studies indicate that widespread co-regulation of proliferation upwardly biases survival prediction when gene selection is performed on a genome-wide basis. In addition, removal of the correlative effects of proliferation does not reduce the random bias associated with survival prediction using random gene selection. Since most cancers become addicted to DNA repair as a result of forced cellular replication, increased oxidation, and repair deficiencies from oncogenic loss or genetic polymorphisms, we pursued an investigation to remove the proliferation component of expression in DNA repair genes to determine survival prediction. This translational hypothesis-driven focus on DNA repair genes is directly amenable to finding new sets of DNA repair genes that could potentially be studied for inhibition therapy. Overall survival (OS) prediction was evaluated in 18 cancers by using normalized RNA-Seq data for 126 DNA repair genes with expression available in TCGA. Transformations for normality and adjustments for age at diagnosis, stage, and PCNA metagene expression were performed for all DNA repair genes. We also analyzed genomic event rates (GER) for somatic mutations, deletions, and amplification in driver genes and DNA repair genes. After performing empirical p-value testing with use of randomly selected gene sets, it was observed that OS could be predicted significantly by sets of DNA repair genes for 61% (11/18) of the cancers. Interestingly, PARP1 was not a significant predictor of survival for any of the 11 cancers. Results from cluster analysis of GERs indicates that the most opportunistic cancers for inhibition therapy may be AML, colorectal, and renal papillary, because of potentially less confounding due to lower GERs for mutations, deletions, and amplifications in DNA repair genes. However, the most opportunistic cancer for inhibition therapy is likely to be AML, since it showed the lowest GERs for mutations, deletions, and amplifications in DNA repair genes. In conclusion, our hypothesis-driven focus to target DNA repair gene expression adjusted for the PCNA metagene as a means of predicting OS in various cancers resulted in statistically significant sets of genes.