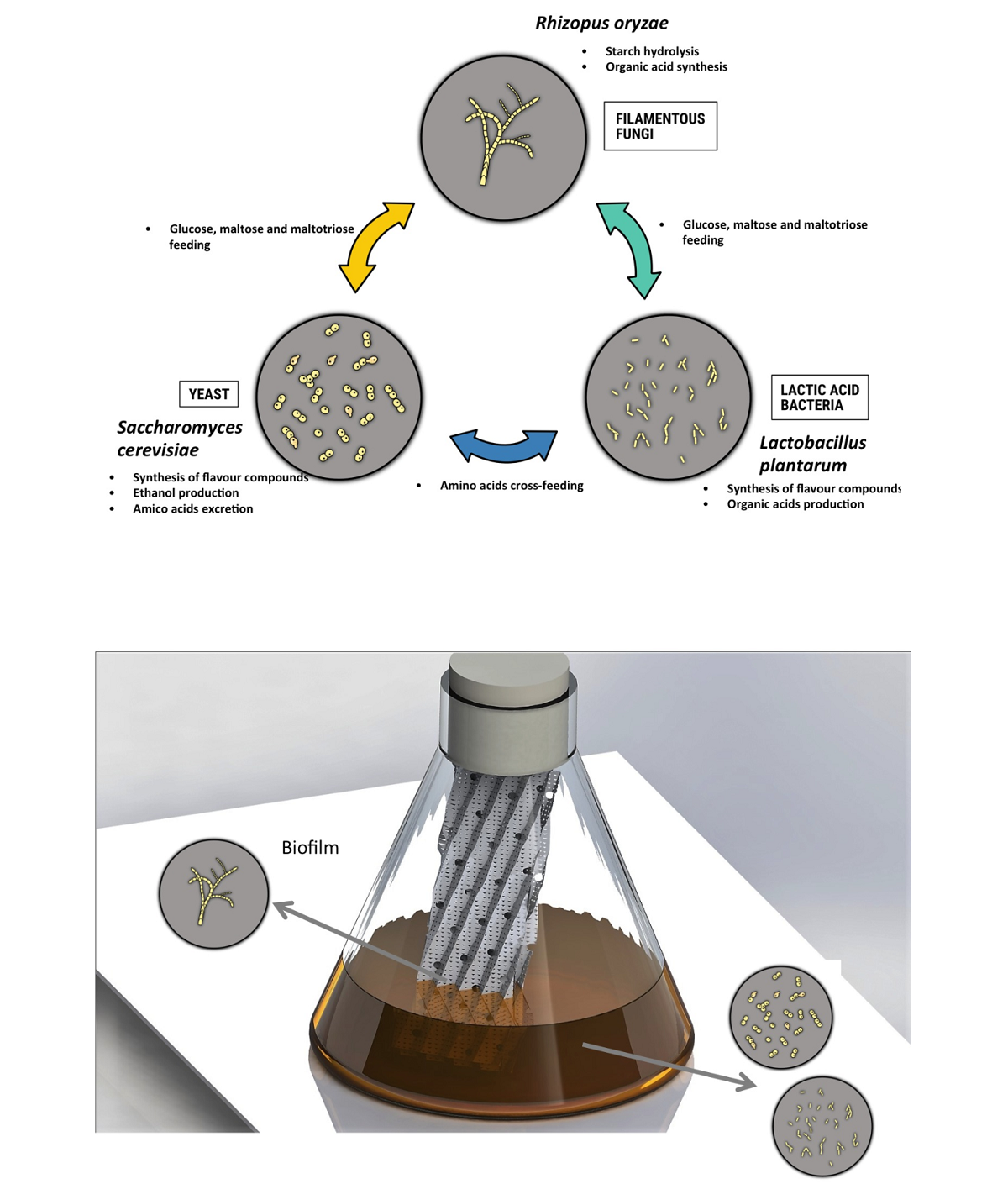
used traditional microbial starters revealed that effective fermentation requires three microbial strains with complementary metabolic activities: filamentous fungi (Rhizopus oryzae), yeast (Saccharomyces cerevisiae), and lactic acid bacteria (Lactobacillus plantarum). Relative to natural communities, modulation of the ratio of these three microorganisms led to significant differences not only in terms of ethanol and organic acid production, but also with the profile of volatile compounds. However, inoculation of an equal ratio of spores/cells of the three aforementioned microbial strains led to a flavor profile and ethanol yield similar to that obtained with natural communities. Compartmentalization of metabolic tasks through the use of a biofilm cultivation device allowed further improvement of the entire fermentation process, notably by increasing the amount of key components of the aroma profile of the fermented beverage (i.e., mainly phenylethyl alcohol, isobutyl alcohol, isoamyl alcohol, and 2-methyl-butanol) and reducing the amount of off-flavor compound. This study represents an initial step toward understanding interkingdom microbial interactions with a strong potential for application in the food biotechnology.